Details of the Target
General Information of Target
| Target ID | LDTP13256 | |||||
|---|---|---|---|---|---|---|
| Target Name | SUN domain-containing protein 2 (SUN2) | |||||
| Gene Name | SUN2 | |||||
| Gene ID | 25777 | |||||
| Synonyms |
FRIGG; KIAA0668; RAB5IP; UNC84B; SUN domain-containing protein 2; Protein unc-84 homolog B; Rab5-interacting protein; Rab5IP; Sad1/unc-84 protein-like 2 |
|||||
| 3D Structure | ||||||
| Sequence |
MTEPGASPEDPWVKASPVGAHAGEGRAGRARARRGAGRRGASLLSPKSPTLSVPRGCRED
SSHPACAKVEYAYSDNSLDPGLFVESTRKGSVVSRANSIGSTSASSVPNTDDEDSDYHQE AYKESYKDRRRRAHTQAEQKRRDAIKRGYDDLQTIVPTCQQQDFSIGSQKLSKAIVLQKT IDYIQFLHKEKKKQEEEVSTLRKDVTALKIMKVNYEQIVKAHQDNPHEGEDQVSDQVKFN VFQGIMDSLFQSFNASISVASFQELSACVFSWIEEHCKPQTLREIVIGVLHQLKNQLY |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Subcellular location |
Nucleus inner membrane
|
|||||
| Function |
As a component of the LINC (LInker of Nucleoskeleton and Cytoskeleton) complex, involved in the connection between the nuclear lamina and the cytoskeleton. The nucleocytoplasmic interactions established by the LINC complex play an important role in the transmission of mechanical forces across the nuclear envelope and in nuclear movement and positioning. Specifically, SYNE2 and SUN2 assemble in arrays of transmembrane actin-associated nuclear (TAN) lines which are bound to F-actin cables and couple the nucleus to retrograde actin flow during actin-dependent nuclear movement. Required for interkinetic nuclear migration (INM) and essential for nucleokinesis and centrosome-nucleus coupling during radial neuronal migration in the cerebral cortex and during glial migration. Required for nuclear migration in retinal photoreceptor progenitors implicating association with cytoplasmic dynein-dynactin and kinesin motor complexes, and probably B-type lamins; SUN1 and SUN2 seem to act redundantly. The SUN1/2:KASH5 LINC complex couples telomeres to microtubules during meiosis; SUN1 and SUN2 seem to act at least partial redundantly. Anchors chromosome movement in the prophase of meiosis and is involved in selective gene expression of coding and non-coding RNAs needed for gametogenesis. Required for telomere attachment to nuclear envelope and gametogenesis. May also function on endocytic vesicles as a receptor for RAB5-GDP and participate in the activation of RAB5.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| A2058 | Substitution: p.A419V | DBIA Probe Info | |||
| CAL62 | SNV: p.G609A | . | |||
| JURKAT | SNV: p.S553L | . | |||
| MCAS | SNV: p.R338C | DBIA Probe Info | |||
| SNGM | SNV: p.A427T | . | |||
| TOV21G | SNV: p.G555V | DBIA Probe Info | |||
| UACC257 | SNV: p.V629L | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K491(5.51); K533(10.00) | LDD0277 | [1] | |
|
AHL-Pu-1 Probe Info |
 |
C563(3.26) | LDD0168 | [2] | |
|
DBIA Probe Info |
 |
C601(20.31) | LDD0204 | [3] | |
|
5E-2FA Probe Info |
 |
H495(0.00); H325(0.00) | LDD2235 | [4] | |
|
m-APA Probe Info |
 |
N.A. | LDD2231 | [4] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C563(0.00); C601(0.00); C705(0.00) | LDD0038 | [5] | |
|
IA-alkyne Probe Info |
 |
C563(0.00); C601(0.00) | LDD0036 | [5] | |
|
Lodoacetamide azide Probe Info |
 |
C601(0.00); C563(0.00) | LDD0037 | [5] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [6] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [7] | |
|
Crotonaldehyde Probe Info |
 |
H269(0.00); H281(0.00) | LDD0219 | [7] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C017 Probe Info |
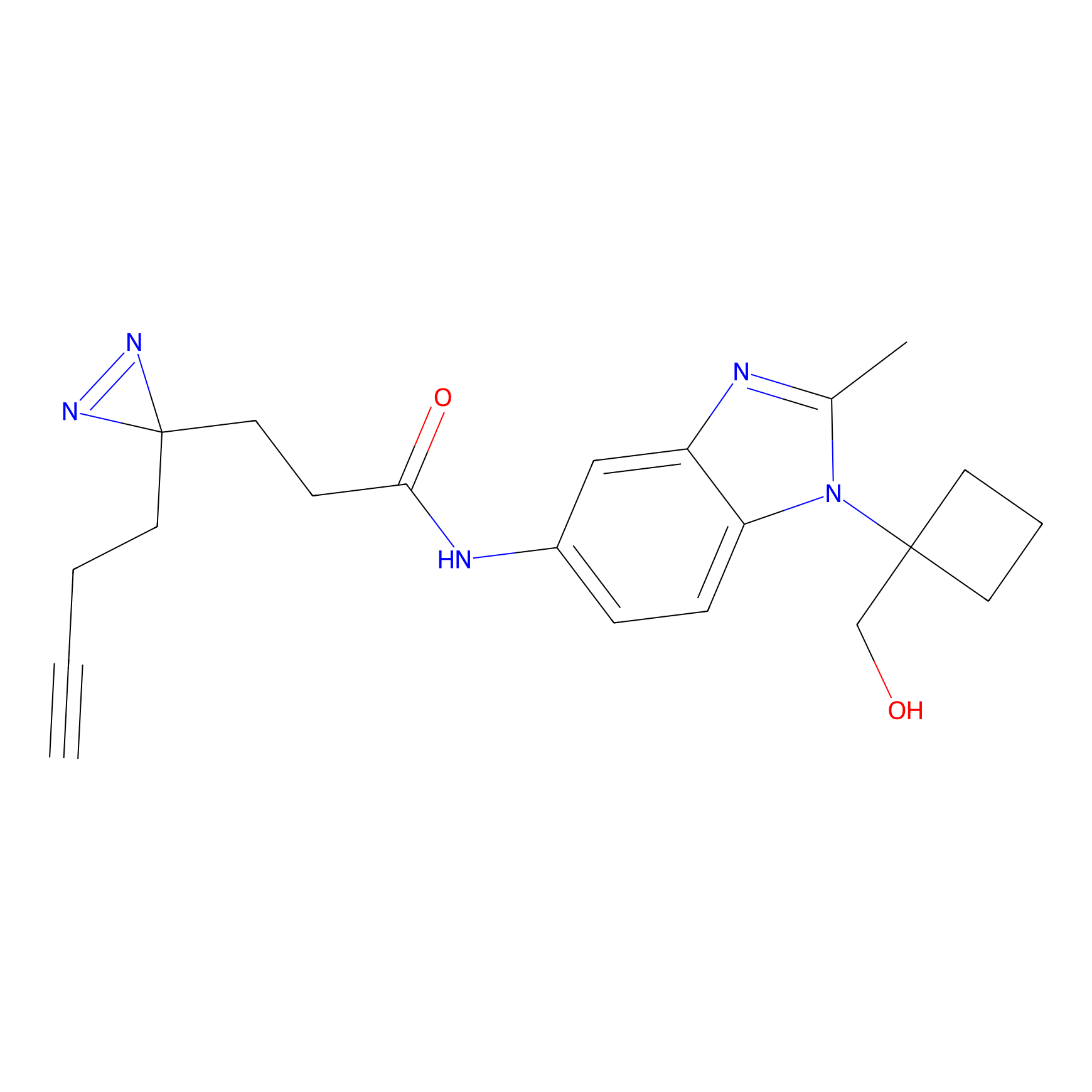 |
6.28 | LDD1725 | [8] | |
|
C040 Probe Info |
 |
7.52 | LDD1740 | [8] | |
|
C094 Probe Info |
 |
27.67 | LDD1785 | [8] | |
|
C106 Probe Info |
 |
17.88 | LDD1793 | [8] | |
|
C112 Probe Info |
 |
24.08 | LDD1799 | [8] | |
|
C158 Probe Info |
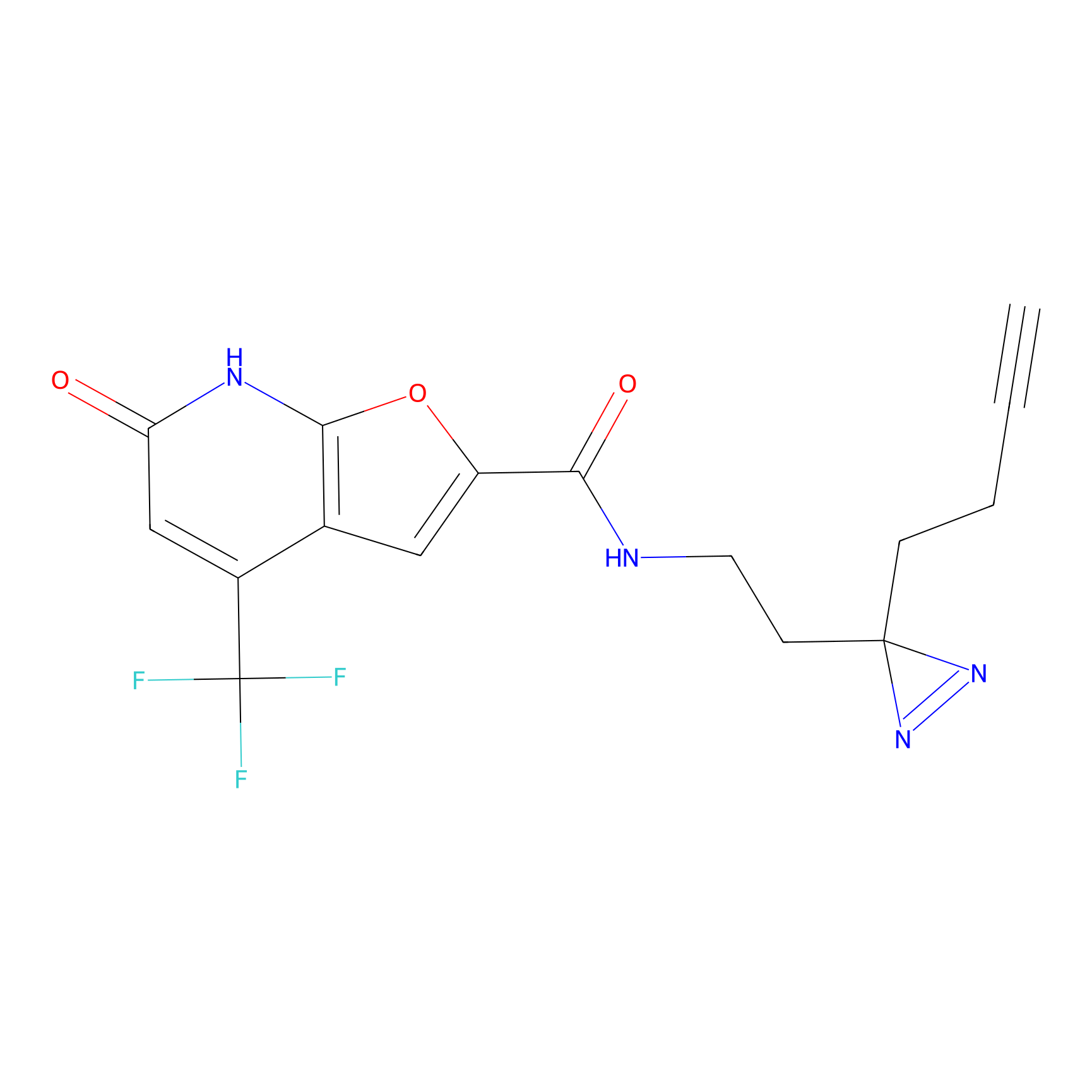 |
9.45 | LDD1838 | [8] | |
|
C201 Probe Info |
 |
24.93 | LDD1877 | [8] | |
|
C210 Probe Info |
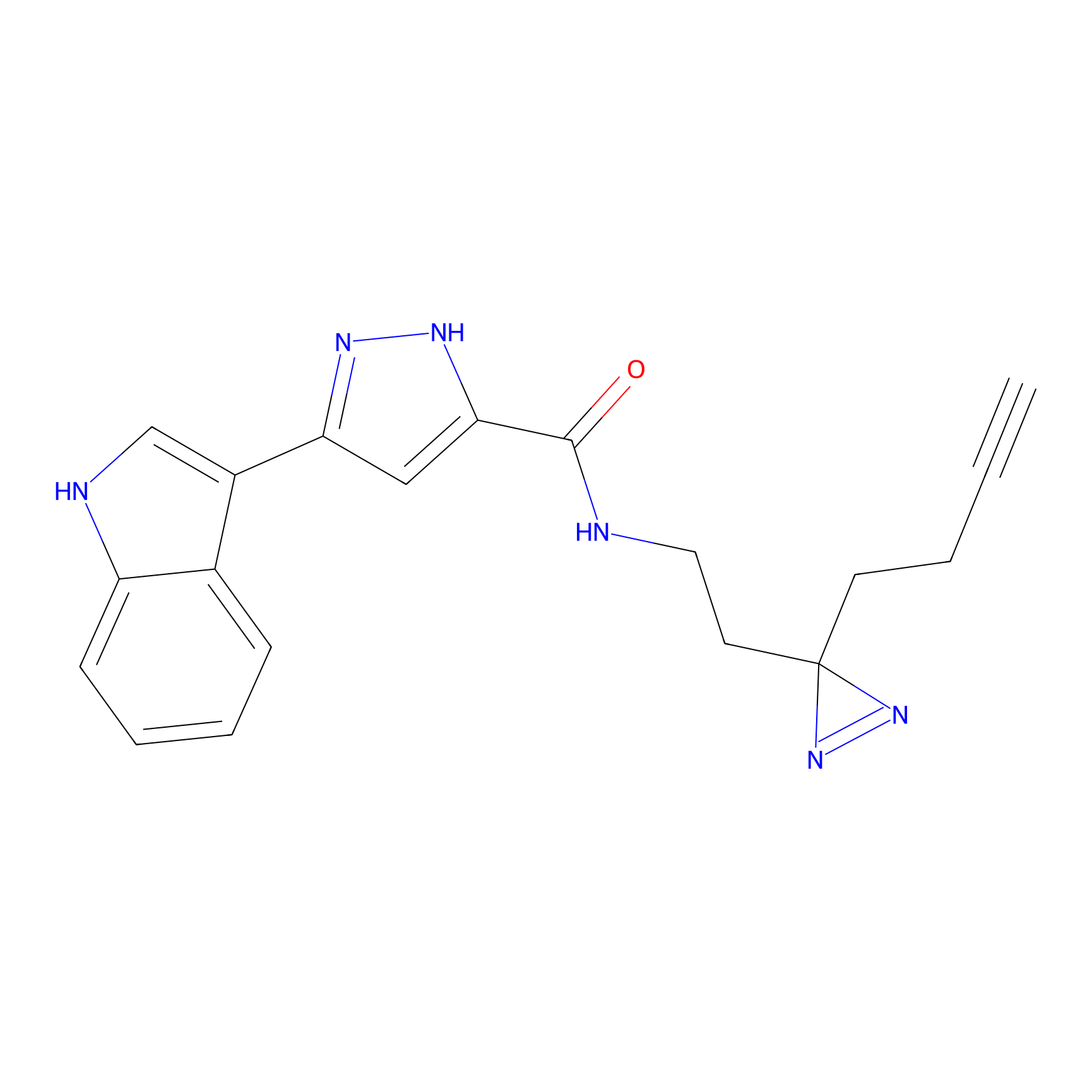 |
71.51 | LDD1884 | [8] | |
|
C211 Probe Info |
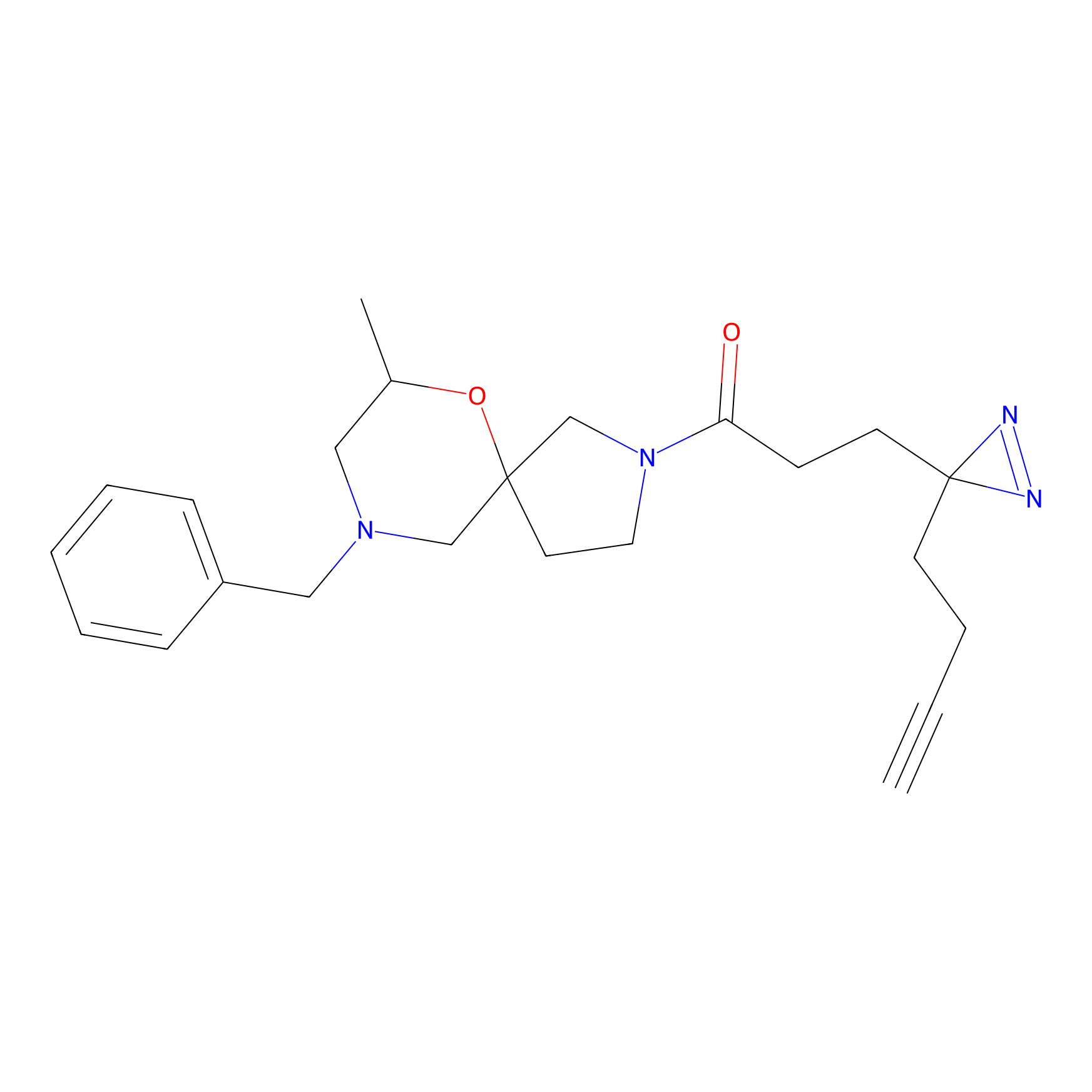 |
10.78 | LDD1885 | [8] | |
|
C212 Probe Info |
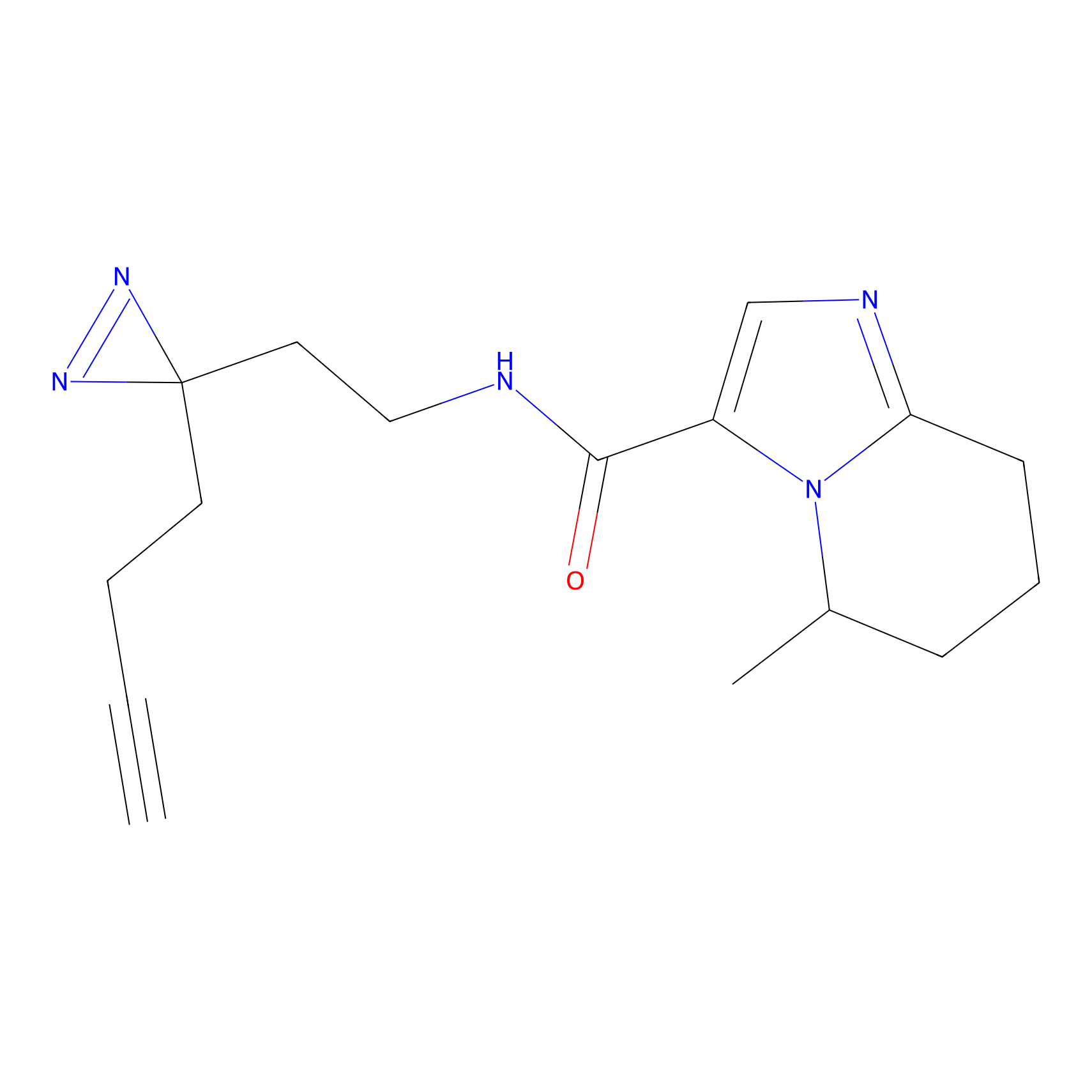 |
6.11 | LDD1886 | [8] | |
|
C213 Probe Info |
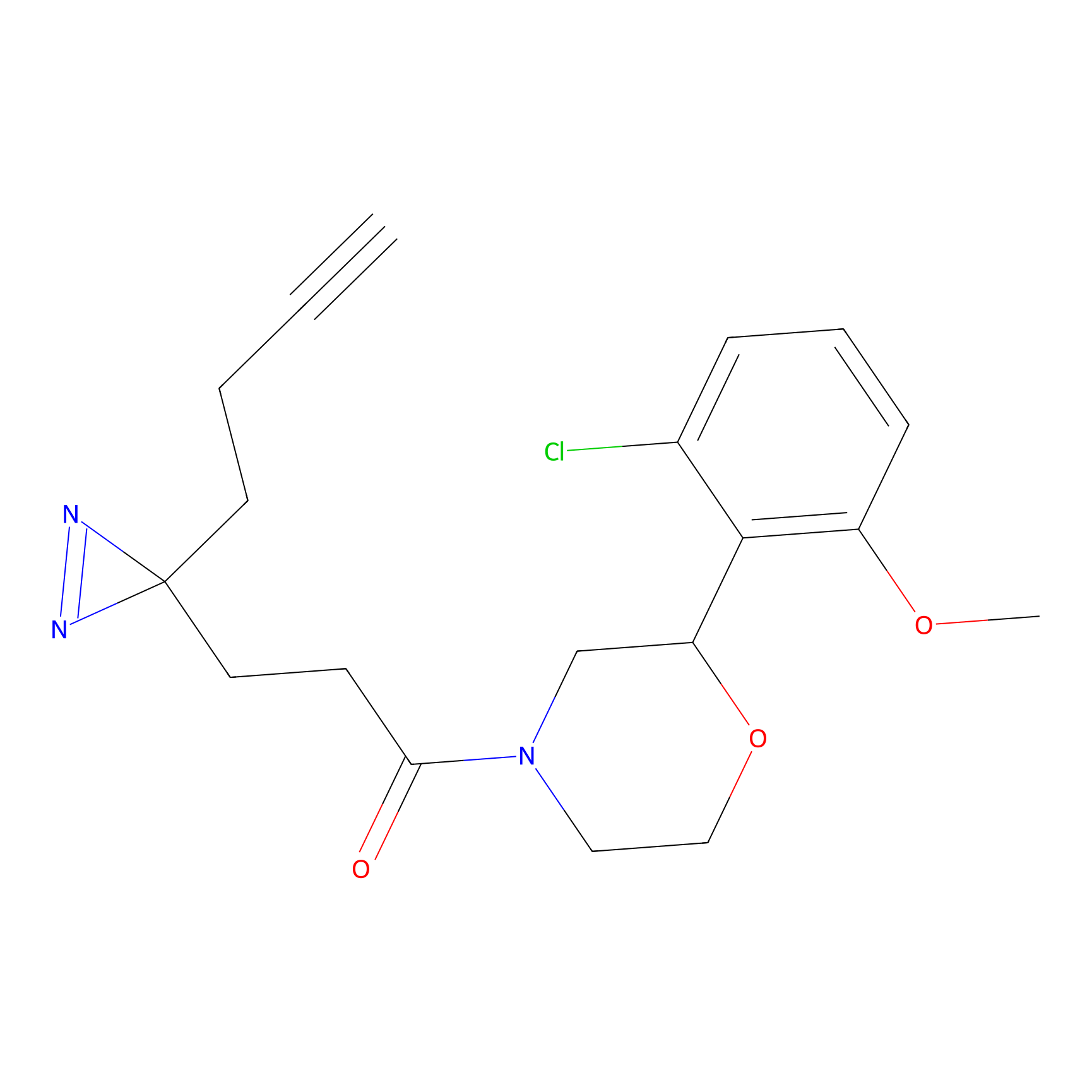 |
19.16 | LDD1887 | [8] | |
|
C214 Probe Info |
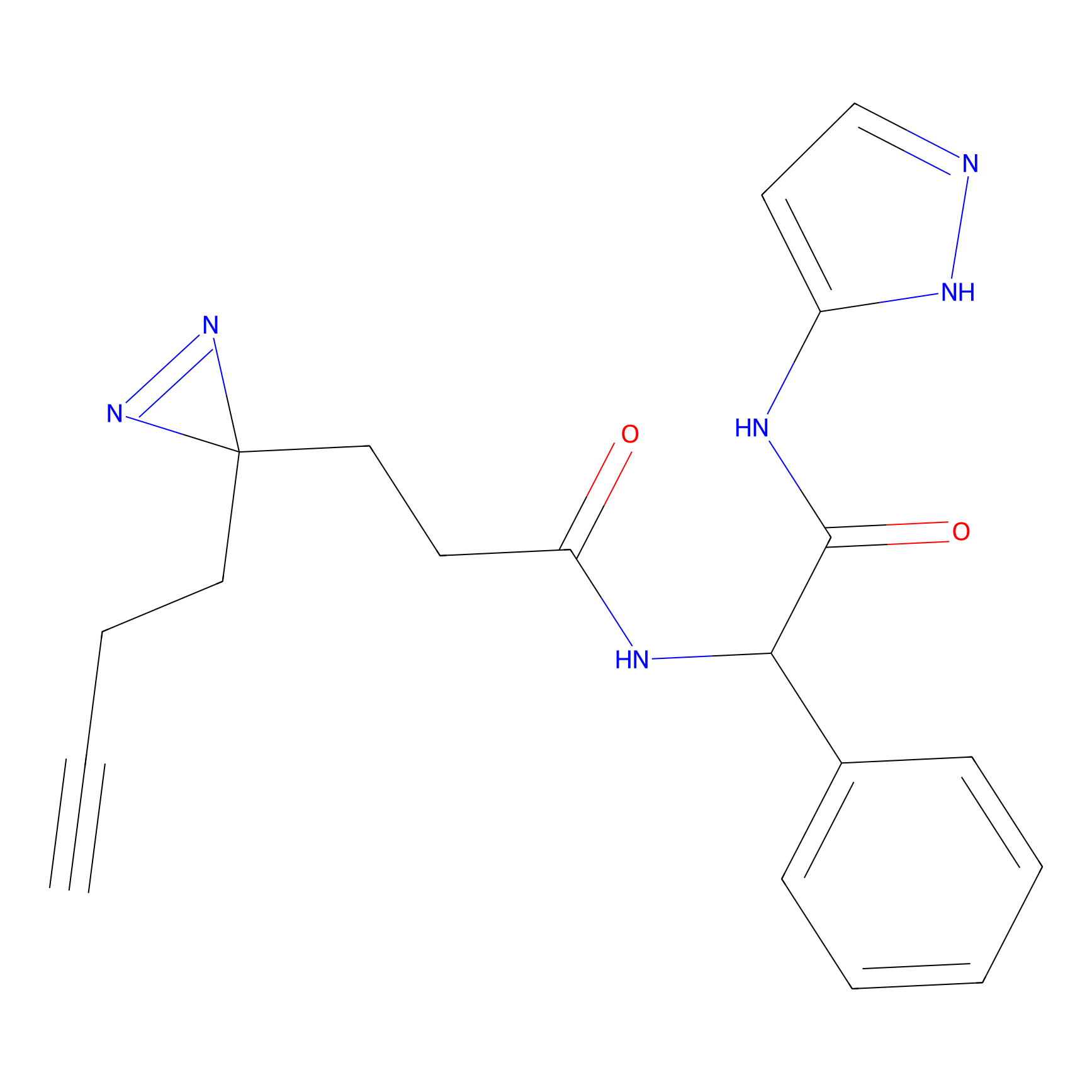 |
12.91 | LDD1888 | [8] | |
|
C228 Probe Info |
 |
15.35 | LDD1901 | [8] | |
|
C246 Probe Info |
 |
12.82 | LDD1919 | [8] | |
|
C285 Probe Info |
 |
19.56 | LDD1955 | [8] | |
|
C289 Probe Info |
 |
47.18 | LDD1959 | [8] | |
|
C348 Probe Info |
 |
10.34 | LDD2009 | [8] | |
|
C355 Probe Info |
 |
30.70 | LDD2016 | [8] | |
|
C356 Probe Info |
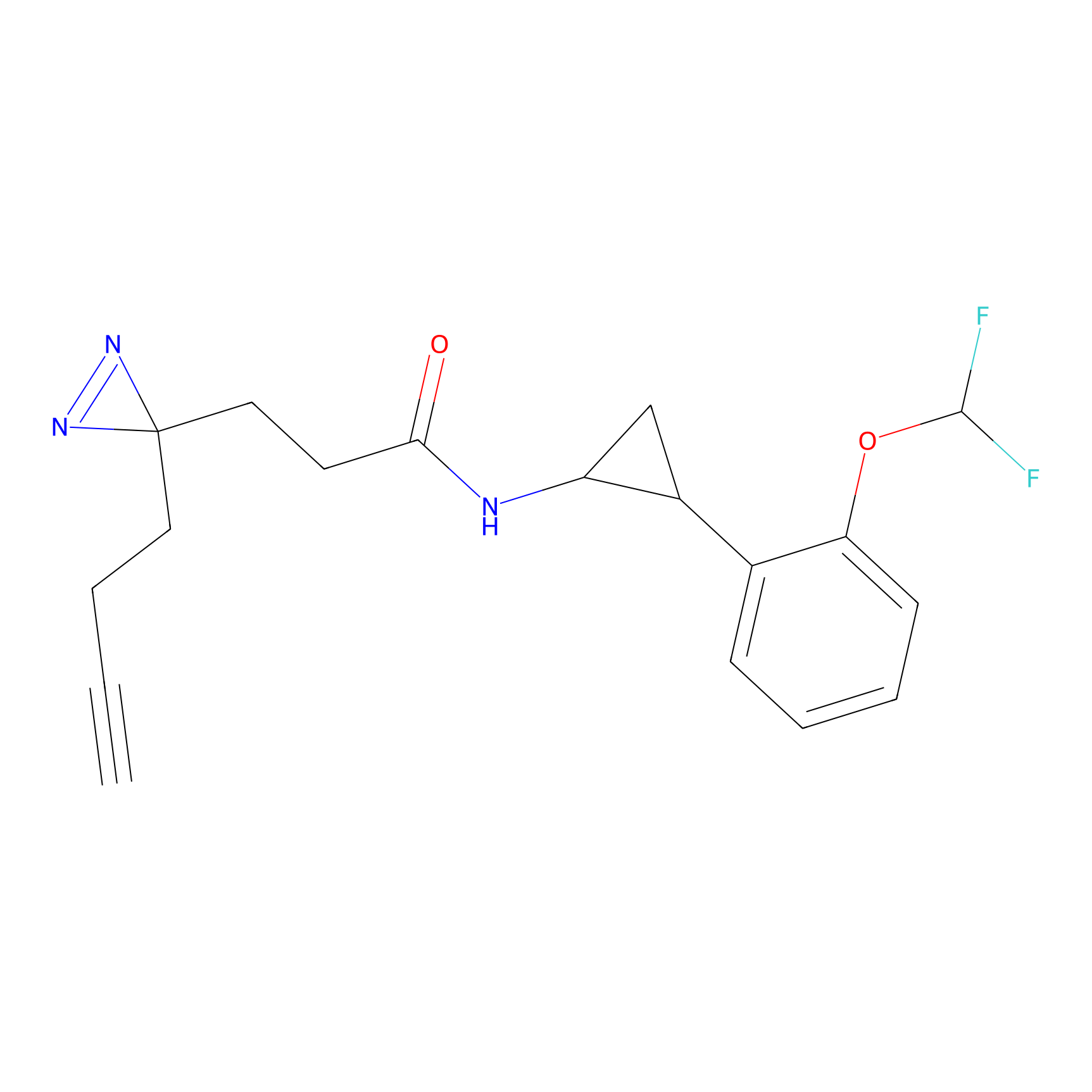 |
8.82 | LDD2017 | [8] | |
|
C397 Probe Info |
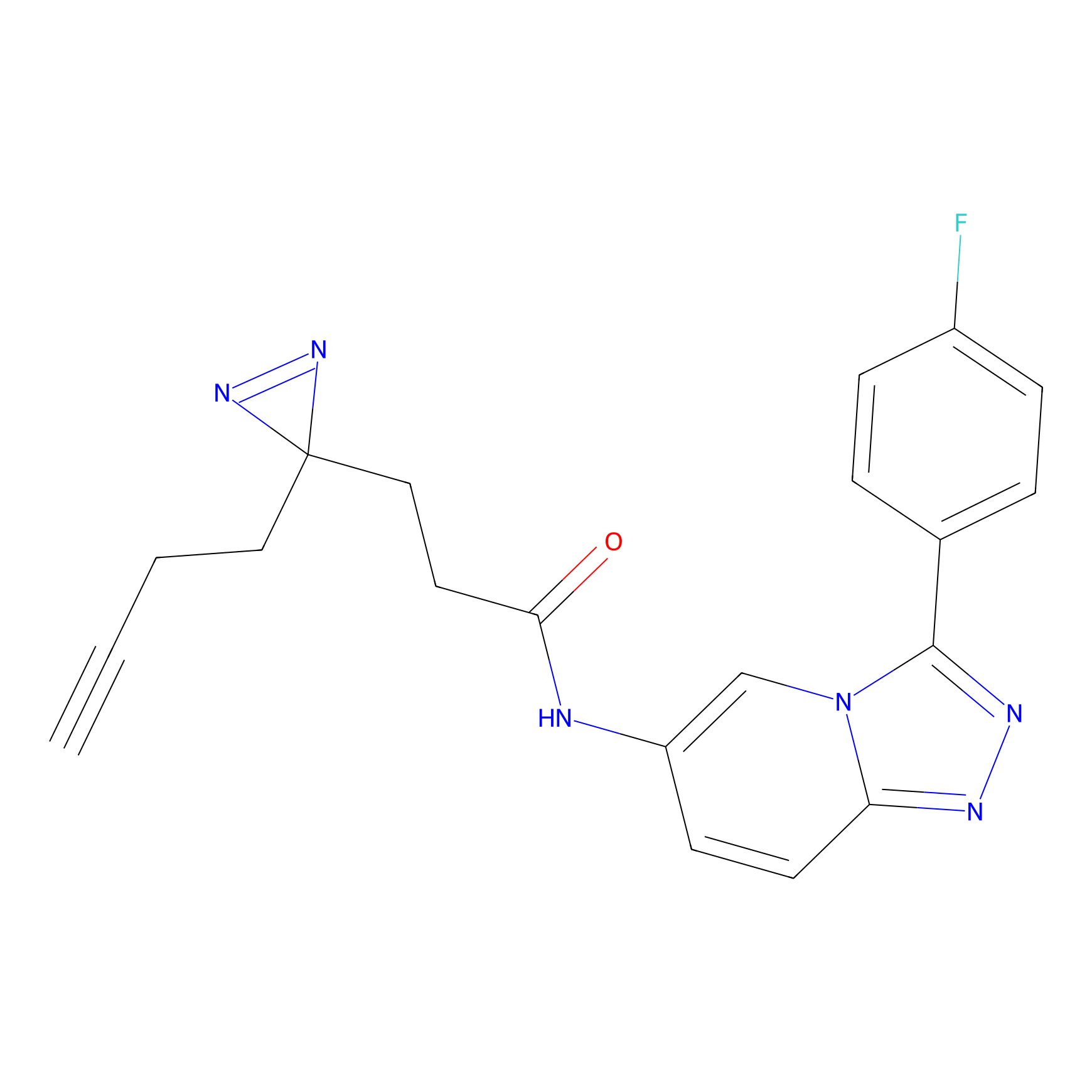 |
10.13 | LDD2056 | [8] | |
|
C400 Probe Info |
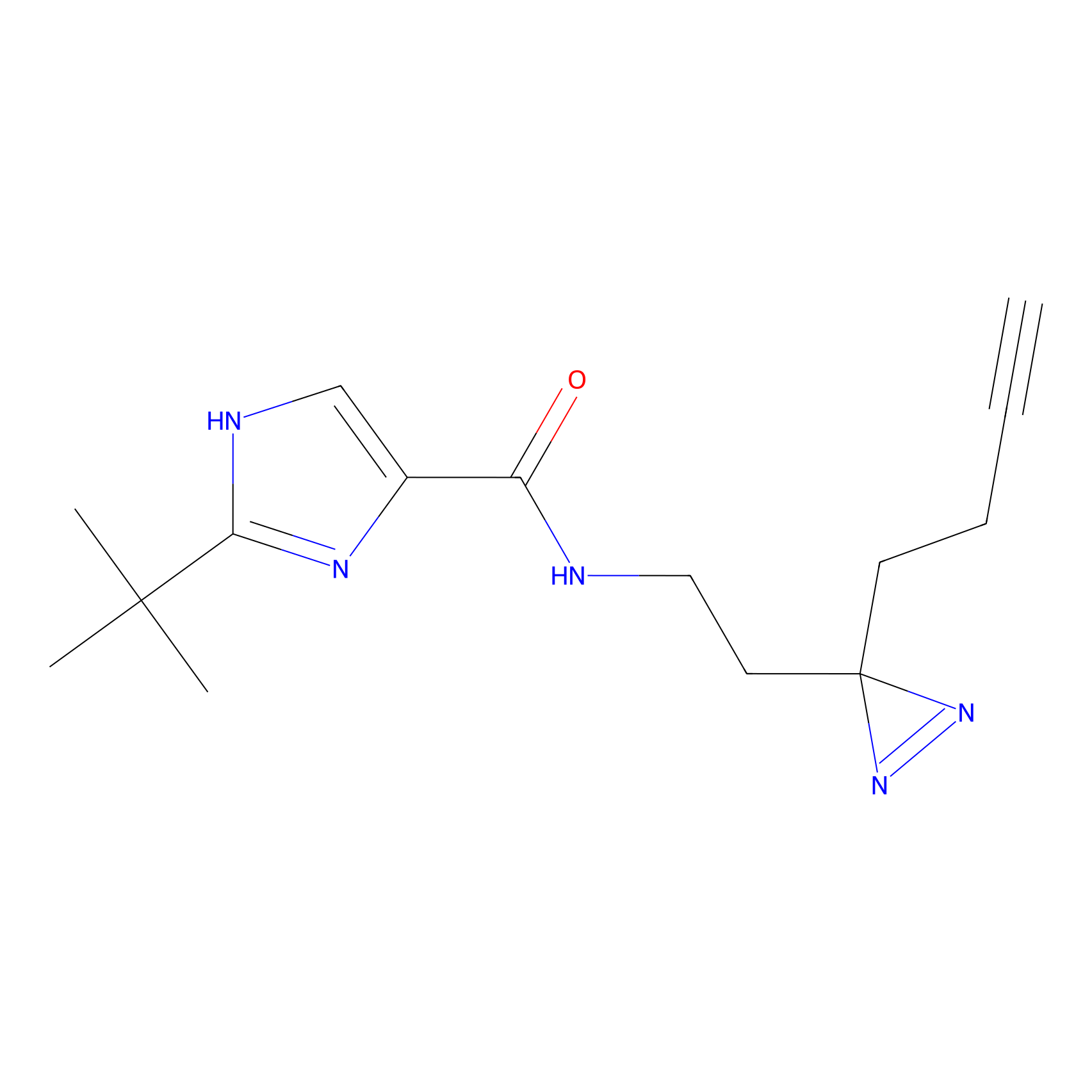 |
8.34 | LDD2059 | [8] | |
|
C403 Probe Info |
 |
18.90 | LDD2061 | [8] | |
|
C407 Probe Info |
 |
11.88 | LDD2064 | [8] | |
|
FFF probe11 Probe Info |
 |
17.52 | LDD0471 | [9] | |
|
FFF probe13 Probe Info |
 |
13.43 | LDD0475 | [9] | |
|
FFF probe2 Probe Info |
 |
20.00 | LDD0463 | [9] | |
|
FFF probe3 Probe Info |
 |
20.00 | LDD0464 | [9] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0136 | [10] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [11] | |
|
OEA-DA Probe Info |
 |
18.12 | LDD0046 | [12] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | HEK-293T | C563(3.26) | LDD0168 | [2] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C563(2.03) | LDD0169 | [2] |
| LDCM0215 | AC10 | HEK-293T | C705(0.99); C601(1.12) | LDD1508 | [13] |
| LDCM0226 | AC11 | HEK-293T | C705(0.90) | LDD1509 | [13] |
| LDCM0237 | AC12 | HEK-293T | C705(1.17) | LDD1510 | [13] |
| LDCM0259 | AC14 | HEK-293T | C705(0.88) | LDD1512 | [13] |
| LDCM0270 | AC15 | HEK-293T | C705(1.15) | LDD1513 | [13] |
| LDCM0277 | AC18 | HEK-293T | C705(1.02); C601(1.14) | LDD1516 | [13] |
| LDCM0278 | AC19 | HEK-293T | C705(1.08) | LDD1517 | [13] |
| LDCM0279 | AC2 | HEK-293T | C705(1.04); C601(1.20) | LDD1518 | [13] |
| LDCM0280 | AC20 | HEK-293T | C705(0.99) | LDD1519 | [13] |
| LDCM0281 | AC21 | HEK-293T | C705(1.05) | LDD1520 | [13] |
| LDCM0282 | AC22 | HEK-293T | C705(0.85) | LDD1521 | [13] |
| LDCM0283 | AC23 | HEK-293T | C705(0.98) | LDD1522 | [13] |
| LDCM0284 | AC24 | HEK-293T | C705(1.01); C601(1.38) | LDD1523 | [13] |
| LDCM0286 | AC26 | HEK-293T | C705(1.02); C601(1.18) | LDD1525 | [13] |
| LDCM0287 | AC27 | HEK-293T | C705(0.95) | LDD1526 | [13] |
| LDCM0288 | AC28 | HEK-293T | C705(0.90) | LDD1527 | [13] |
| LDCM0289 | AC29 | HEK-293T | C705(1.18) | LDD1528 | [13] |
| LDCM0290 | AC3 | HEK-293T | C705(1.07) | LDD1529 | [13] |
| LDCM0291 | AC30 | HEK-293T | C705(0.88) | LDD1530 | [13] |
| LDCM0292 | AC31 | HEK-293T | C705(0.89) | LDD1531 | [13] |
| LDCM0293 | AC32 | HEK-293T | C705(0.93); C601(1.43) | LDD1532 | [13] |
| LDCM0295 | AC34 | HEK-293T | C705(1.17); C601(1.07) | LDD1534 | [13] |
| LDCM0296 | AC35 | HEK-293T | C705(1.04) | LDD1535 | [13] |
| LDCM0297 | AC36 | HEK-293T | C705(0.89) | LDD1536 | [13] |
| LDCM0298 | AC37 | HEK-293T | C705(0.92) | LDD1537 | [13] |
| LDCM0299 | AC38 | HEK-293T | C705(1.03) | LDD1538 | [13] |
| LDCM0300 | AC39 | HEK-293T | C705(1.10) | LDD1539 | [13] |
| LDCM0301 | AC4 | HEK-293T | C705(0.92) | LDD1540 | [13] |
| LDCM0302 | AC40 | HEK-293T | C705(0.97); C601(1.10) | LDD1541 | [13] |
| LDCM0304 | AC42 | HEK-293T | C705(0.91); C601(1.29) | LDD1543 | [13] |
| LDCM0305 | AC43 | HEK-293T | C705(0.95) | LDD1544 | [13] |
| LDCM0306 | AC44 | HEK-293T | C705(0.94) | LDD1545 | [13] |
| LDCM0307 | AC45 | HEK-293T | C705(1.10) | LDD1546 | [13] |
| LDCM0308 | AC46 | HEK-293T | C705(1.14) | LDD1547 | [13] |
| LDCM0309 | AC47 | HEK-293T | C705(0.96) | LDD1548 | [13] |
| LDCM0310 | AC48 | HEK-293T | C705(0.96); C601(1.47) | LDD1549 | [13] |
| LDCM0312 | AC5 | HEK-293T | C705(0.81) | LDD1551 | [13] |
| LDCM0313 | AC50 | HEK-293T | C705(0.97); C601(1.10) | LDD1552 | [13] |
| LDCM0314 | AC51 | HEK-293T | C705(1.02) | LDD1553 | [13] |
| LDCM0315 | AC52 | HEK-293T | C705(1.20) | LDD1554 | [13] |
| LDCM0316 | AC53 | HEK-293T | C705(0.88) | LDD1555 | [13] |
| LDCM0317 | AC54 | HEK-293T | C705(0.85) | LDD1556 | [13] |
| LDCM0318 | AC55 | HEK-293T | C705(0.87) | LDD1557 | [13] |
| LDCM0319 | AC56 | HEK-293T | C705(0.94); C601(1.47) | LDD1558 | [13] |
| LDCM0321 | AC58 | HEK-293T | C705(1.07); C601(1.09) | LDD1560 | [13] |
| LDCM0322 | AC59 | HEK-293T | C705(1.11) | LDD1561 | [13] |
| LDCM0323 | AC6 | HEK-293T | C705(1.01) | LDD1562 | [13] |
| LDCM0324 | AC60 | HEK-293T | C705(0.91) | LDD1563 | [13] |
| LDCM0325 | AC61 | HEK-293T | C705(0.93) | LDD1564 | [13] |
| LDCM0326 | AC62 | HEK-293T | C705(0.87) | LDD1565 | [13] |
| LDCM0327 | AC63 | HEK-293T | C705(0.95) | LDD1566 | [13] |
| LDCM0328 | AC64 | HEK-293T | C705(0.89); C601(1.66) | LDD1567 | [13] |
| LDCM0334 | AC7 | HEK-293T | C705(0.99) | LDD1568 | [13] |
| LDCM0345 | AC8 | HEK-293T | C705(0.93); C601(1.24) | LDD1569 | [13] |
| LDCM0248 | AKOS034007472 | HEK-293T | C705(1.29) | LDD1511 | [13] |
| LDCM0275 | AKOS034007705 | HEK-293T | C705(0.84); C601(1.14) | LDD1514 | [13] |
| LDCM0102 | BDHI 8 | Jurkat | C601(20.31) | LDD0204 | [3] |
| LDCM0367 | CL1 | HEK-293T | C705(0.89) | LDD1571 | [13] |
| LDCM0368 | CL10 | HEK-293T | C705(0.98) | LDD1572 | [13] |
| LDCM0369 | CL100 | HEK-293T | C705(0.89); C601(1.18) | LDD1573 | [13] |
| LDCM0370 | CL101 | HEK-293T | C705(1.19) | LDD1574 | [13] |
| LDCM0372 | CL103 | HEK-293T | C705(0.95) | LDD1576 | [13] |
| LDCM0373 | CL104 | HEK-293T | C705(1.00); C601(1.13) | LDD1577 | [13] |
| LDCM0374 | CL105 | HEK-293T | C705(1.41) | LDD1578 | [13] |
| LDCM0376 | CL107 | HEK-293T | C705(0.92) | LDD1580 | [13] |
| LDCM0377 | CL108 | HEK-293T | C705(1.08); C601(1.41) | LDD1581 | [13] |
| LDCM0378 | CL109 | HEK-293T | C705(1.17) | LDD1582 | [13] |
| LDCM0379 | CL11 | HEK-293T | C705(1.09) | LDD1583 | [13] |
| LDCM0381 | CL111 | HEK-293T | C705(1.11) | LDD1585 | [13] |
| LDCM0382 | CL112 | HEK-293T | C705(1.17); C601(1.11) | LDD1586 | [13] |
| LDCM0383 | CL113 | HEK-293T | C705(1.09) | LDD1587 | [13] |
| LDCM0385 | CL115 | HEK-293T | C705(1.07) | LDD1589 | [13] |
| LDCM0386 | CL116 | HEK-293T | C705(1.05); C601(1.10) | LDD1590 | [13] |
| LDCM0387 | CL117 | HEK-293T | C705(0.84) | LDD1591 | [13] |
| LDCM0389 | CL119 | HEK-293T | C705(1.02) | LDD1593 | [13] |
| LDCM0390 | CL12 | HEK-293T | C705(0.95); C601(1.29) | LDD1594 | [13] |
| LDCM0391 | CL120 | HEK-293T | C705(1.09); C601(1.03) | LDD1595 | [13] |
| LDCM0392 | CL121 | HEK-293T | C705(1.07) | LDD1596 | [13] |
| LDCM0394 | CL123 | HEK-293T | C705(0.85) | LDD1598 | [13] |
| LDCM0395 | CL124 | HEK-293T | C705(1.06); C601(1.92) | LDD1599 | [13] |
| LDCM0396 | CL125 | HEK-293T | C705(1.21) | LDD1600 | [13] |
| LDCM0398 | CL127 | HEK-293T | C705(1.03) | LDD1602 | [13] |
| LDCM0399 | CL128 | HEK-293T | C705(1.11); C601(1.34) | LDD1603 | [13] |
| LDCM0400 | CL13 | HEK-293T | C705(1.00) | LDD1604 | [13] |
| LDCM0402 | CL15 | HEK-293T | C705(0.79) | LDD1606 | [13] |
| LDCM0403 | CL16 | HEK-293T | C705(0.94); C601(1.10) | LDD1607 | [13] |
| LDCM0405 | CL18 | HEK-293T | C705(1.01); C601(0.95) | LDD1609 | [13] |
| LDCM0406 | CL19 | HEK-293T | C705(1.03) | LDD1610 | [13] |
| LDCM0408 | CL20 | HEK-293T | C705(0.94) | LDD1612 | [13] |
| LDCM0409 | CL21 | HEK-293T | C705(0.99) | LDD1613 | [13] |
| LDCM0410 | CL22 | HEK-293T | C705(1.19) | LDD1614 | [13] |
| LDCM0411 | CL23 | HEK-293T | C705(1.20) | LDD1615 | [13] |
| LDCM0412 | CL24 | HEK-293T | C705(0.94); C601(1.10) | LDD1616 | [13] |
| LDCM0413 | CL25 | HEK-293T | C705(1.09) | LDD1617 | [13] |
| LDCM0415 | CL27 | HEK-293T | C705(1.00) | LDD1619 | [13] |
| LDCM0416 | CL28 | HEK-293T | C705(0.95); C601(0.95) | LDD1620 | [13] |
| LDCM0418 | CL3 | HEK-293T | C705(0.96) | LDD1622 | [13] |
| LDCM0419 | CL30 | HEK-293T | C705(1.02); C601(1.00) | LDD1623 | [13] |
| LDCM0420 | CL31 | HEK-293T | C705(1.09) | LDD1624 | [13] |
| LDCM0421 | CL32 | HEK-293T | C705(0.93) | LDD1625 | [13] |
| LDCM0422 | CL33 | HEK-293T | C705(0.74) | LDD1626 | [13] |
| LDCM0423 | CL34 | HEK-293T | C705(0.72) | LDD1627 | [13] |
| LDCM0424 | CL35 | HEK-293T | C705(0.77) | LDD1628 | [13] |
| LDCM0425 | CL36 | HEK-293T | C705(0.90); C601(1.18) | LDD1629 | [13] |
| LDCM0426 | CL37 | HEK-293T | C705(1.02) | LDD1630 | [13] |
| LDCM0428 | CL39 | HEK-293T | C705(1.05) | LDD1632 | [13] |
| LDCM0429 | CL4 | HEK-293T | C705(1.12); C601(1.00) | LDD1633 | [13] |
| LDCM0430 | CL40 | HEK-293T | C705(1.11); C601(1.07) | LDD1634 | [13] |
| LDCM0432 | CL42 | HEK-293T | C705(1.06); C601(0.94) | LDD1636 | [13] |
| LDCM0433 | CL43 | HEK-293T | C705(0.90) | LDD1637 | [13] |
| LDCM0434 | CL44 | HEK-293T | C705(0.73) | LDD1638 | [13] |
| LDCM0435 | CL45 | HEK-293T | C705(0.90) | LDD1639 | [13] |
| LDCM0436 | CL46 | HEK-293T | C705(1.01) | LDD1640 | [13] |
| LDCM0437 | CL47 | HEK-293T | C705(1.03) | LDD1641 | [13] |
| LDCM0438 | CL48 | HEK-293T | C705(0.90); C601(1.37) | LDD1642 | [13] |
| LDCM0439 | CL49 | HEK-293T | C705(0.85) | LDD1643 | [13] |
| LDCM0443 | CL52 | HEK-293T | C705(0.98); C601(1.37) | LDD1646 | [13] |
| LDCM0445 | CL54 | HEK-293T | C705(0.90); C601(1.01) | LDD1648 | [13] |
| LDCM0446 | CL55 | HEK-293T | C705(1.22) | LDD1649 | [13] |
| LDCM0447 | CL56 | HEK-293T | C705(0.99) | LDD1650 | [13] |
| LDCM0448 | CL57 | HEK-293T | C705(1.02) | LDD1651 | [13] |
| LDCM0449 | CL58 | HEK-293T | C705(0.89) | LDD1652 | [13] |
| LDCM0450 | CL59 | HEK-293T | C705(1.05) | LDD1653 | [13] |
| LDCM0451 | CL6 | HEK-293T | C705(1.05); C601(0.86) | LDD1654 | [13] |
| LDCM0452 | CL60 | HEK-293T | C705(0.96); C601(1.26) | LDD1655 | [13] |
| LDCM0453 | CL61 | HEK-293T | C705(0.96) | LDD1656 | [13] |
| LDCM0455 | CL63 | HEK-293T | C705(1.06) | LDD1658 | [13] |
| LDCM0456 | CL64 | HEK-293T | C705(0.97); C601(1.35) | LDD1659 | [13] |
| LDCM0458 | CL66 | HEK-293T | C705(0.90); C601(1.12) | LDD1661 | [13] |
| LDCM0459 | CL67 | HEK-293T | C705(1.17) | LDD1662 | [13] |
| LDCM0460 | CL68 | HEK-293T | C705(1.08) | LDD1663 | [13] |
| LDCM0461 | CL69 | HEK-293T | C705(0.76) | LDD1664 | [13] |
| LDCM0462 | CL7 | HEK-293T | C705(1.21) | LDD1665 | [13] |
| LDCM0463 | CL70 | HEK-293T | C705(1.03) | LDD1666 | [13] |
| LDCM0464 | CL71 | HEK-293T | C705(1.07) | LDD1667 | [13] |
| LDCM0465 | CL72 | HEK-293T | C705(0.95); C601(1.11) | LDD1668 | [13] |
| LDCM0466 | CL73 | HEK-293T | C705(1.02) | LDD1669 | [13] |
| LDCM0469 | CL76 | HEK-293T | C705(1.12); C601(0.97) | LDD1672 | [13] |
| LDCM0471 | CL78 | HEK-293T | C705(0.94); C601(1.03) | LDD1674 | [13] |
| LDCM0472 | CL79 | HEK-293T | C705(1.24) | LDD1675 | [13] |
| LDCM0473 | CL8 | HEK-293T | C705(0.86) | LDD1676 | [13] |
| LDCM0474 | CL80 | HEK-293T | C705(0.93) | LDD1677 | [13] |
| LDCM0475 | CL81 | HEK-293T | C705(0.86) | LDD1678 | [13] |
| LDCM0476 | CL82 | HEK-293T | C705(0.97) | LDD1679 | [13] |
| LDCM0477 | CL83 | HEK-293T | C705(1.10) | LDD1680 | [13] |
| LDCM0478 | CL84 | HEK-293T | C705(0.96); C601(1.16) | LDD1681 | [13] |
| LDCM0479 | CL85 | HEK-293T | C705(1.16) | LDD1682 | [13] |
| LDCM0481 | CL87 | HEK-293T | C705(1.03) | LDD1684 | [13] |
| LDCM0482 | CL88 | HEK-293T | C705(1.26); C601(1.44) | LDD1685 | [13] |
| LDCM0484 | CL9 | HEK-293T | C705(1.01) | LDD1687 | [13] |
| LDCM0485 | CL90 | HEK-293T | C705(0.80); C601(1.11) | LDD1688 | [13] |
| LDCM0486 | CL91 | HEK-293T | C705(1.17) | LDD1689 | [13] |
| LDCM0487 | CL92 | HEK-293T | C705(0.83) | LDD1690 | [13] |
| LDCM0488 | CL93 | HEK-293T | C705(0.82) | LDD1691 | [13] |
| LDCM0489 | CL94 | HEK-293T | C705(1.12) | LDD1692 | [13] |
| LDCM0490 | CL95 | HEK-293T | C705(0.89) | LDD1693 | [13] |
| LDCM0491 | CL96 | HEK-293T | C705(1.01); C601(1.43) | LDD1694 | [13] |
| LDCM0492 | CL97 | HEK-293T | C705(0.88) | LDD1695 | [13] |
| LDCM0494 | CL99 | HEK-293T | C705(0.94) | LDD1697 | [13] |
| LDCM0495 | E2913 | HEK-293T | C705(1.02) | LDD1698 | [13] |
| LDCM0468 | Fragment33 | HEK-293T | C705(0.99) | LDD1671 | [13] |
| LDCM0022 | KB02 | 22RV1 | C761(1.94) | LDD2243 | [14] |
| LDCM0023 | KB03 | 22RV1 | C761(1.63) | LDD2660 | [14] |
| LDCM0024 | KB05 | COLO792 | C726(1.98); C622(2.36) | LDD3310 | [14] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0224 | [7] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Ras-related protein Rab-5A (RAB5A) | Rab family | P20339 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Importin subunit alpha-1 (KPNA2) | Importin alpha family | P52292 | |||
| SUN domain-containing protein 2 (SUN2) | . | Q9UH99 | |||
Other
References
