Details of the Target
General Information of Target
| Target ID | LDTP04597 | |||||
|---|---|---|---|---|---|---|
| Target Name | Methylosome subunit pICln (CLNS1A) | |||||
| Gene Name | CLNS1A | |||||
| Gene ID | 1207 | |||||
| Synonyms |
CLCI; ICLN; Methylosome subunit pICln; Chloride channel, nucleotide sensitive 1A; Chloride conductance regulatory protein ICln; I(Cln); Chloride ion current inducer protein; ClCI; Reticulocyte pICln |
|||||
| 3D Structure | ||||||
| Sequence |
MSFLKSFPPPGPAEGLLRQQPDTEAVLNGKGLGTGTLYIAESRLSWLDGSGLGFSLEYPT
ISLHALSRDRSDCLGEHLYVMVNAKFEEESKEPVADEEEEDSDDDVEPITEFRFVPSDKS ALEAMFTAMCECQALHPDPEDEDSDDYDGEEYDVEAHEQGQGDIPTFYTYEEGLSHLTAE GQATLERLEGMLSQSVSSQYNMAGVRTEDSIRDYEDGMEVDTTPTVAGQFEDADVDH |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
PICln (TC 1.A.47) family
|
|||||
| Subcellular location |
Cytoplasm, cytosol
|
|||||
| Function |
Involved in both the assembly of spliceosomal snRNPs and the methylation of Sm proteins. Chaperone that regulates the assembly of spliceosomal U1, U2, U4 and U5 small nuclear ribonucleoproteins (snRNPs), the building blocks of the spliceosome, and thereby plays an important role in the splicing of cellular pre-mRNAs. Most spliceosomal snRNPs contain a common set of Sm proteins SNRPB, SNRPD1, SNRPD2, SNRPD3, SNRPE, SNRPF and SNRPG that assemble in a heptameric protein ring on the Sm site of the small nuclear RNA to form the core snRNP (Sm core). In the cytosol, the Sm proteins SNRPD1, SNRPD2, SNRPE, SNRPF and SNRPG are trapped in an inactive 6S pICln-Sm complex by the chaperone CLNS1A that controls the assembly of the core snRNP. Dissociation by the SMN complex of CLNS1A from the trapped Sm proteins and their transfer to an SMN-Sm complex triggers the assembly of core snRNPs and their transport to the nucleus.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
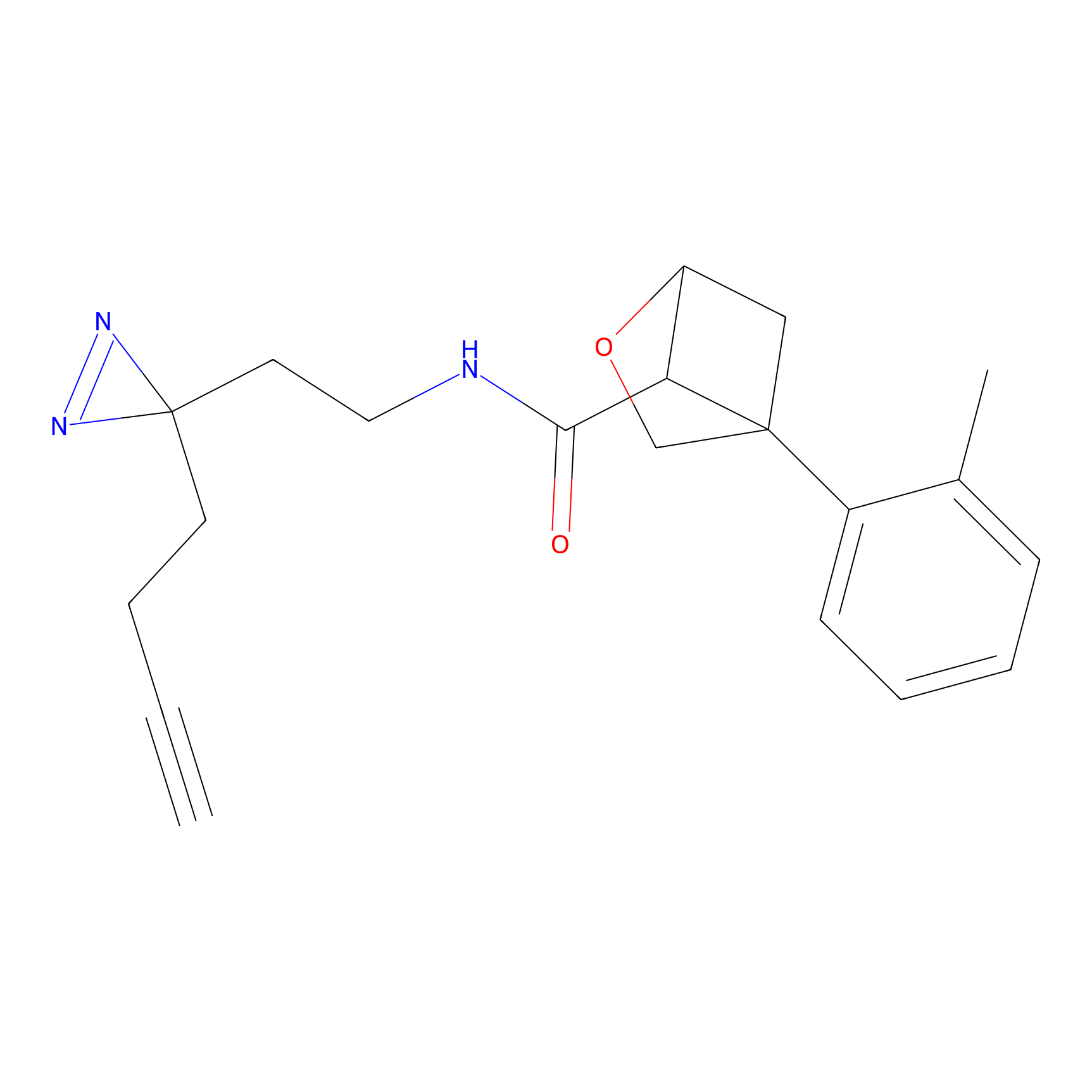
FBP2 Probe Info |
 |
3.71 | LDD0323 | [2] | |
|
TH211 Probe Info |
 |
Y38(6.32) | LDD0257 | [3] | |
|
BTD Probe Info |
 |
C73(0.97) | LDD2092 | [4] | |
|
AHL-Pu-1 Probe Info |
 |
C73(10.72) | LDD0168 | [5] | |
|
HHS-475 Probe Info |
 |
Y200(1.04) | LDD0264 | [6] | |
|
HHS-465 Probe Info |
 |
Y200(8.90) | LDD2237 | [7] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0223 | [8] | |
|
DBIA Probe Info |
 |
C73(1.29) | LDD0080 | [9] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [10] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0032 | [11] | |
|
IPIAA_L Probe Info |
 |
N.A. | LDD0031 | [12] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [10] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [13] | |
|
DA-P3 Probe Info |
 |
N.A. | LDD0178 | [14] | |
|
Compound 10 Probe Info |
 |
N.A. | LDD2216 | [15] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [15] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [16] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [17] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [18] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [19] | |
|
1c-yne Probe Info |
 |
K30(0.00); K5(0.00) | LDD0228 | [20] | |
|
W1 Probe Info |
 |
N.A. | LDD0236 | [21] | |
|
AOyne Probe Info |
 |
14.70 | LDD0443 | [22] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [13] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C285 Probe Info |
 |
24.59 | LDD1955 | [23] | |
|
C286 Probe Info |
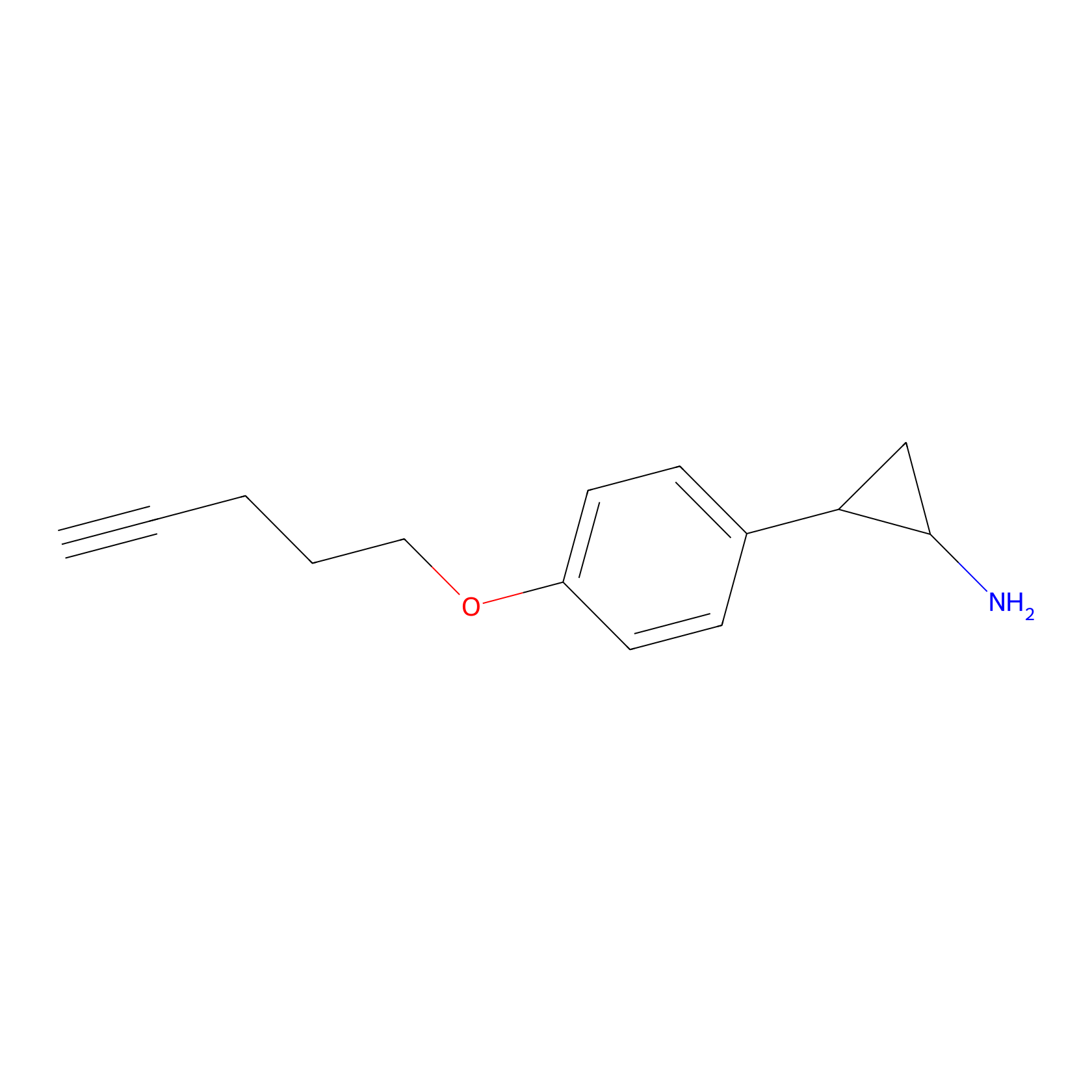
 |
5.13 | LDD1956 | [23] | |
|
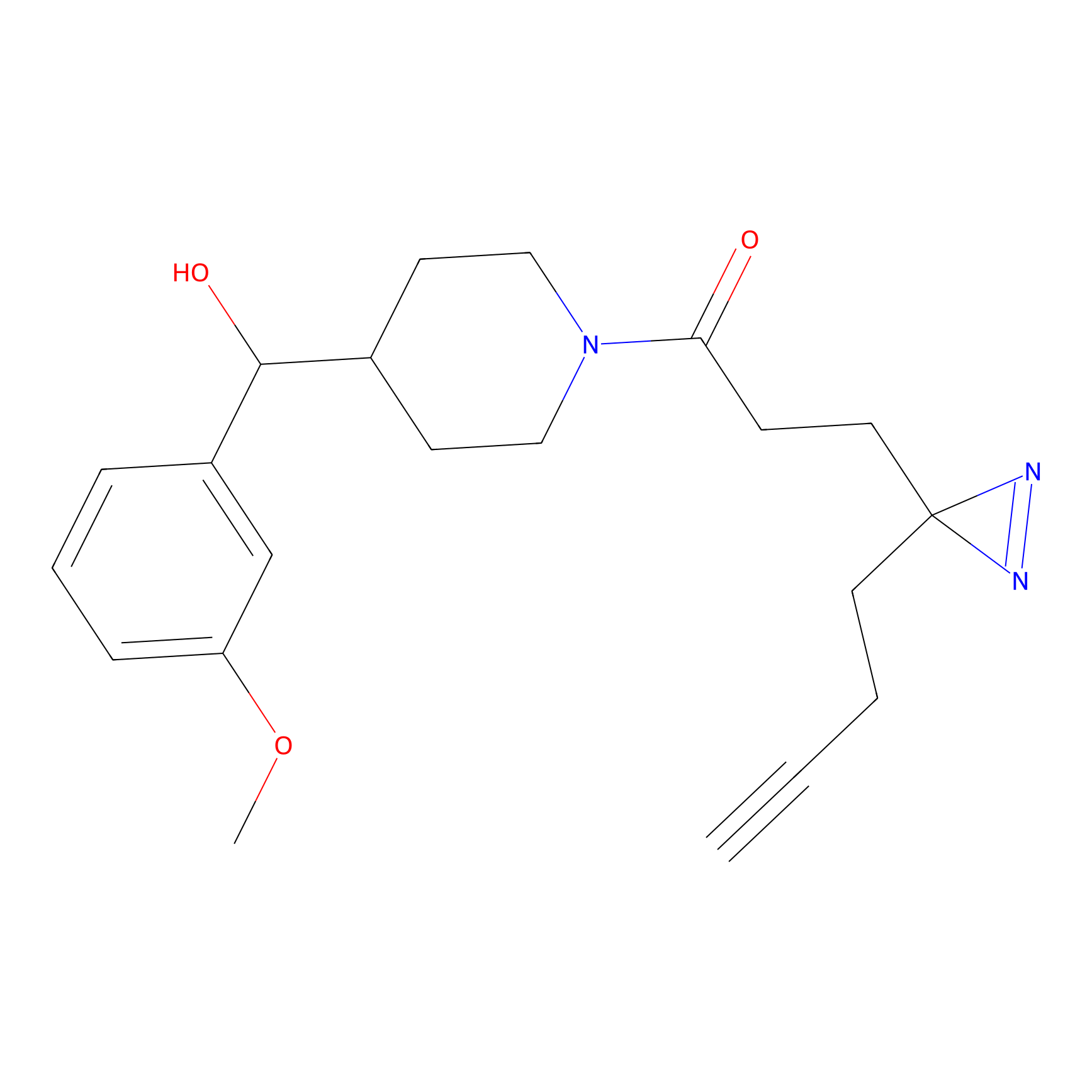
C287 Probe Info |
 |
15.14 | LDD1957 | [23] | |
|
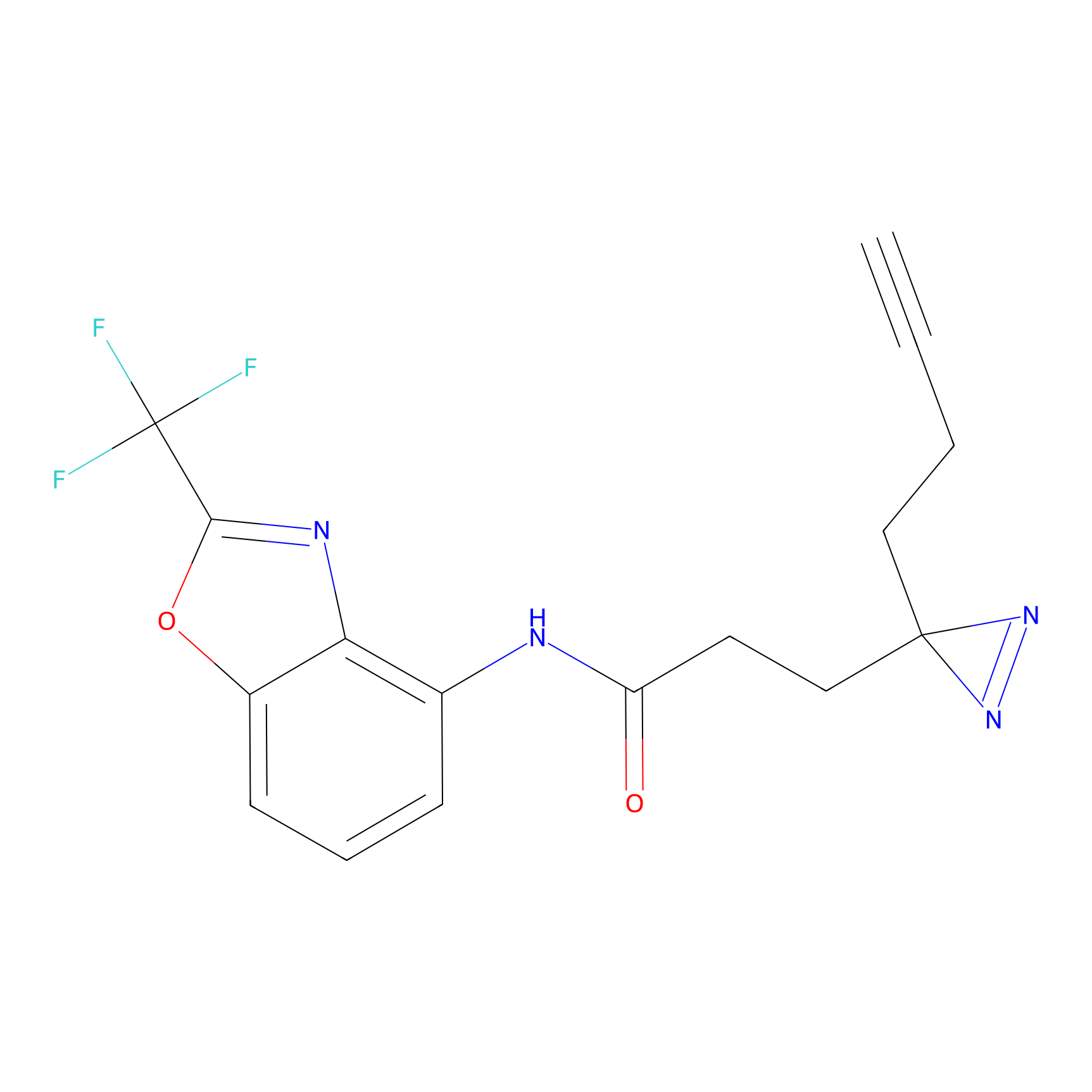
C288 Probe Info |
 |
7.26 | LDD1958 | [23] | |
|
C289 Probe Info |
 |
45.89 | LDD1959 | [23] | |
|
C296 Probe Info |
 |
12.55 | LDD1966 | [23] | |
|
C310 Probe Info |
 |
8.34 | LDD1977 | [23] | |
|
C322 Probe Info |
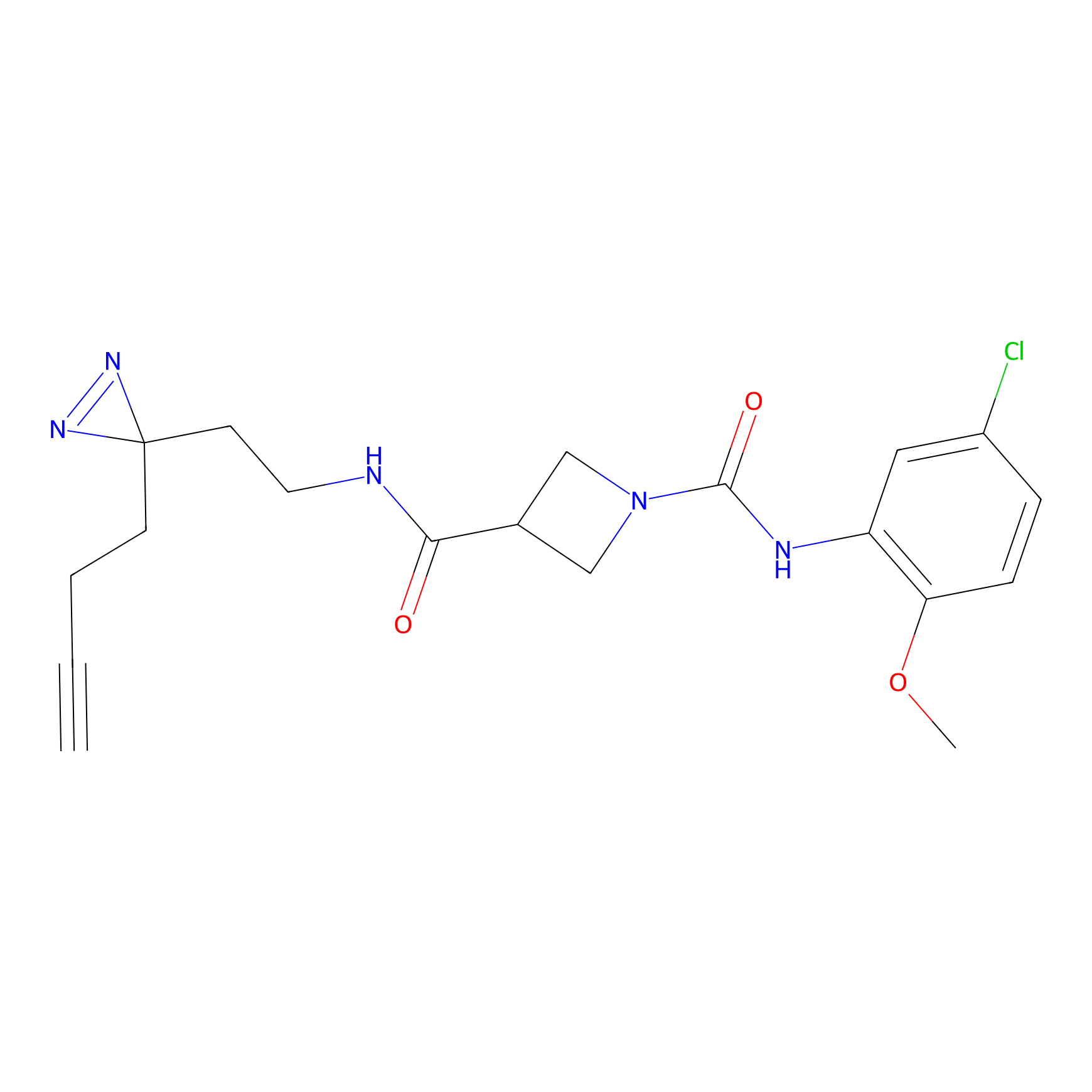 |
7.94 | LDD1988 | [23] | |
|
C338 Probe Info |
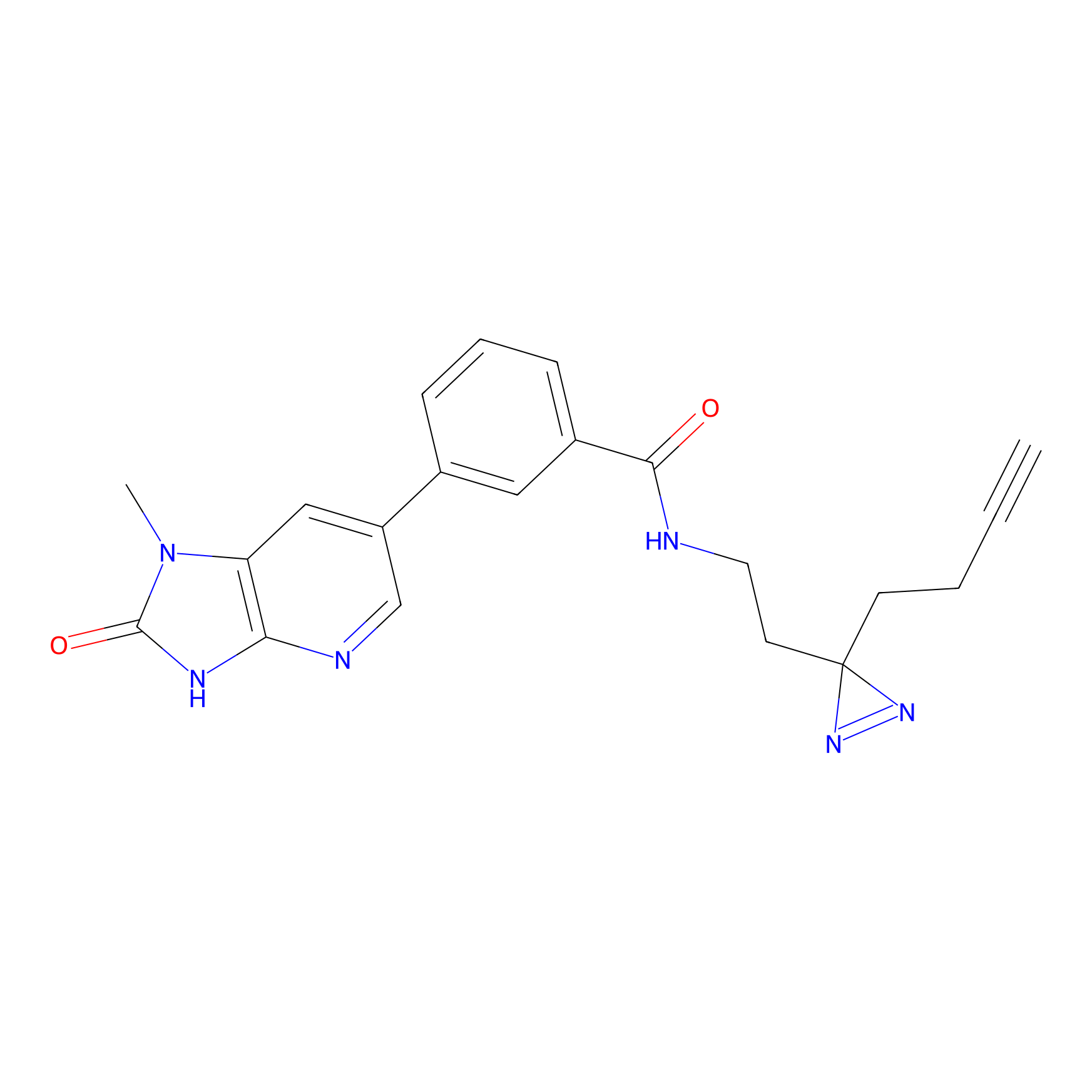 |
11.39 | LDD2001 | [23] | |
|
C413 Probe Info |
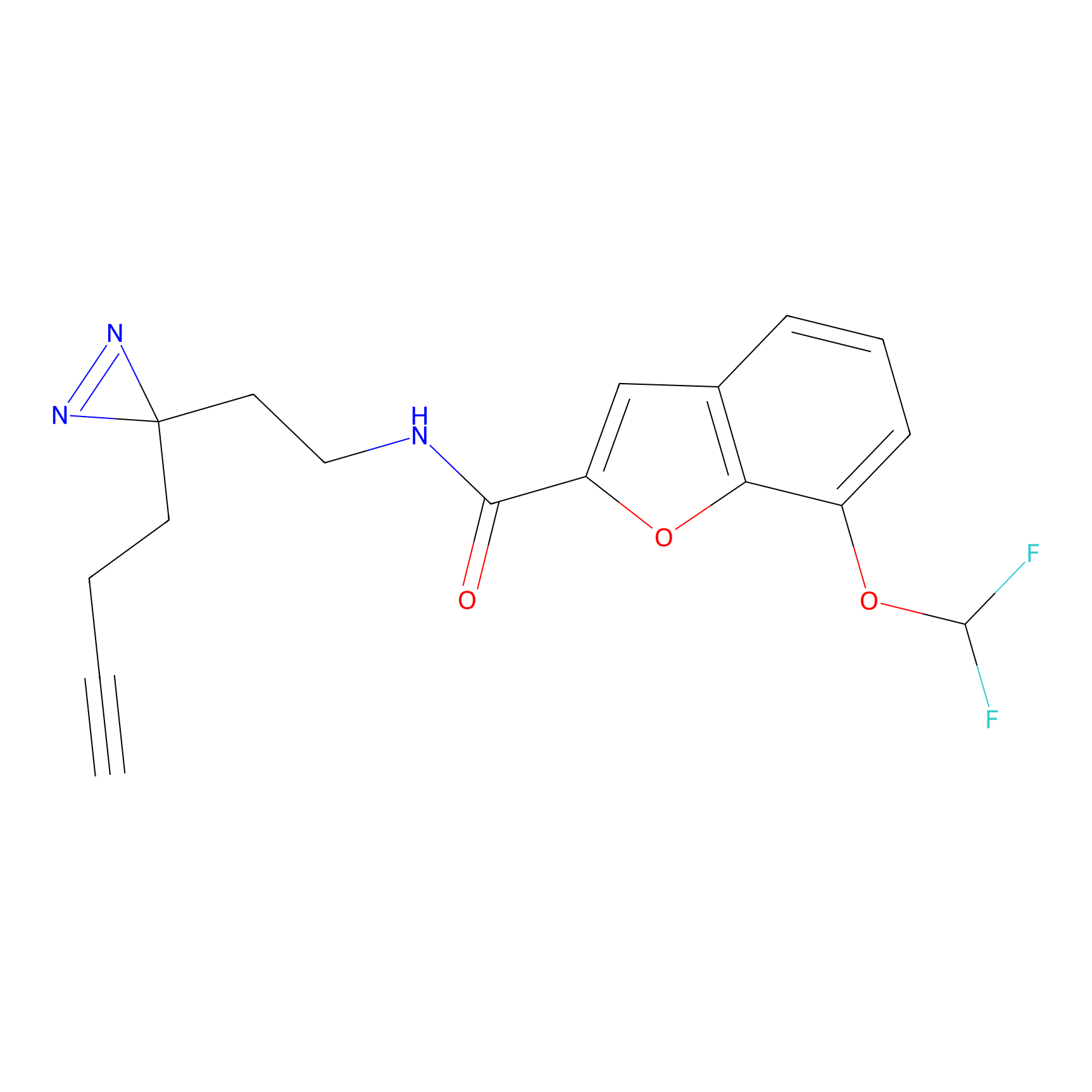 |
12.38 | LDD2069 | [23] | |
|
FFF probe11 Probe Info |
 |
5.61 | LDD0472 | [24] | |
|
FFF probe13 Probe Info |
 |
9.13 | LDD0476 | [24] | |
|
FFF probe2 Probe Info |
 |
15.72 | LDD0463 | [24] | |
|
FFF probe3 Probe Info |
 |
9.44 | LDD0464 | [24] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [25] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | HEK-293T | C73(10.72) | LDD0168 | [5] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C73(3.38) | LDD0169 | [5] |
| LDCM0214 | AC1 | HCT 116 | C73(1.06) | LDD0531 | [9] |
| LDCM0215 | AC10 | HCT 116 | C73(0.61) | LDD0532 | [9] |
| LDCM0216 | AC100 | HEK-293T | C73(1.03) | LDD0814 | [9] |
| LDCM0217 | AC101 | HEK-293T | C73(0.96) | LDD0815 | [9] |
| LDCM0218 | AC102 | HEK-293T | C73(1.03) | LDD0816 | [9] |
| LDCM0219 | AC103 | HEK-293T | C73(0.87) | LDD0817 | [9] |
| LDCM0220 | AC104 | HEK-293T | C73(0.89) | LDD0818 | [9] |
| LDCM0221 | AC105 | HEK-293T | C73(0.96) | LDD0819 | [9] |
| LDCM0222 | AC106 | HEK-293T | C73(0.84) | LDD0820 | [9] |
| LDCM0223 | AC107 | HEK-293T | C73(0.95) | LDD0821 | [9] |
| LDCM0224 | AC108 | HEK-293T | C73(0.96) | LDD0822 | [9] |
| LDCM0225 | AC109 | HEK-293T | C73(0.97) | LDD0823 | [9] |
| LDCM0226 | AC11 | HCT 116 | C73(0.55) | LDD0543 | [9] |
| LDCM0227 | AC110 | HEK-293T | C73(0.92) | LDD0825 | [9] |
| LDCM0228 | AC111 | HEK-293T | C73(1.09) | LDD0826 | [9] |
| LDCM0229 | AC112 | HEK-293T | C73(1.00) | LDD0827 | [9] |
| LDCM0237 | AC12 | HCT 116 | C73(0.69) | LDD0554 | [9] |
| LDCM0246 | AC128 | HEK-293T | C73(1.33) | LDD0844 | [9] |
| LDCM0247 | AC129 | HEK-293T | C73(1.37) | LDD0845 | [9] |
| LDCM0249 | AC130 | HEK-293T | C73(1.65) | LDD0847 | [9] |
| LDCM0250 | AC131 | HEK-293T | C73(1.09) | LDD0848 | [9] |
| LDCM0251 | AC132 | HEK-293T | C73(1.50) | LDD0849 | [9] |
| LDCM0252 | AC133 | HEK-293T | C73(1.11) | LDD0850 | [9] |
| LDCM0253 | AC134 | HEK-293T | C73(1.11) | LDD0851 | [9] |
| LDCM0254 | AC135 | HEK-293T | C73(1.06) | LDD0852 | [9] |
| LDCM0255 | AC136 | HEK-293T | C73(0.84) | LDD0853 | [9] |
| LDCM0256 | AC137 | HEK-293T | C73(1.26) | LDD0854 | [9] |
| LDCM0257 | AC138 | HEK-293T | C73(1.31) | LDD0855 | [9] |
| LDCM0258 | AC139 | HEK-293T | C73(1.39) | LDD0856 | [9] |
| LDCM0259 | AC14 | HCT 116 | C73(1.11) | LDD0576 | [9] |
| LDCM0260 | AC140 | HEK-293T | C73(1.18) | LDD0858 | [9] |
| LDCM0261 | AC141 | HEK-293T | C73(1.20) | LDD0859 | [9] |
| LDCM0262 | AC142 | HEK-293T | C73(1.16) | LDD0860 | [9] |
| LDCM0263 | AC143 | PaTu 8988t | C73(1.16) | LDD1142 | [9] |
| LDCM0264 | AC144 | PaTu 8988t | C73(1.06) | LDD1143 | [9] |
| LDCM0265 | AC145 | PaTu 8988t | C73(1.16) | LDD1144 | [9] |
| LDCM0266 | AC146 | PaTu 8988t | C73(1.50) | LDD1145 | [9] |
| LDCM0267 | AC147 | PaTu 8988t | C73(1.35) | LDD1146 | [9] |
| LDCM0268 | AC148 | PaTu 8988t | C73(1.38) | LDD1147 | [9] |
| LDCM0269 | AC149 | PaTu 8988t | C73(1.24) | LDD1148 | [9] |
| LDCM0270 | AC15 | HCT 116 | C73(1.20) | LDD0587 | [9] |
| LDCM0271 | AC150 | PaTu 8988t | C73(1.53) | LDD1150 | [9] |
| LDCM0272 | AC151 | PaTu 8988t | C73(1.94) | LDD1151 | [9] |
| LDCM0273 | AC152 | PaTu 8988t | C73(1.25) | LDD1152 | [9] |
| LDCM0274 | AC153 | PaTu 8988t | C73(1.97) | LDD1153 | [9] |
| LDCM0621 | AC154 | PaTu 8988t | C73(1.70) | LDD2166 | [9] |
| LDCM0622 | AC155 | PaTu 8988t | C73(1.52) | LDD2167 | [9] |
| LDCM0623 | AC156 | PaTu 8988t | C73(1.35) | LDD2168 | [9] |
| LDCM0624 | AC157 | PaTu 8988t | C73(1.54) | LDD2169 | [9] |
| LDCM0276 | AC17 | HCT 116 | C73(0.97) | LDD0593 | [9] |
| LDCM0277 | AC18 | HCT 116 | C73(0.97) | LDD0594 | [9] |
| LDCM0278 | AC19 | HCT 116 | C73(0.88) | LDD0595 | [9] |
| LDCM0279 | AC2 | HCT 116 | C73(1.13) | LDD0596 | [9] |
| LDCM0280 | AC20 | HCT 116 | C73(1.23) | LDD0597 | [9] |
| LDCM0281 | AC21 | HCT 116 | C73(0.91) | LDD0598 | [9] |
| LDCM0282 | AC22 | HCT 116 | C73(1.22) | LDD0599 | [9] |
| LDCM0283 | AC23 | HCT 116 | C73(1.07) | LDD0600 | [9] |
| LDCM0284 | AC24 | HCT 116 | C73(1.20) | LDD0601 | [9] |
| LDCM0285 | AC25 | PaTu 8988t | C73(0.91) | LDD1164 | [9] |
| LDCM0286 | AC26 | PaTu 8988t | C73(1.02) | LDD1165 | [9] |
| LDCM0287 | AC27 | PaTu 8988t | C73(1.15) | LDD1166 | [9] |
| LDCM0288 | AC28 | PaTu 8988t | C73(1.42) | LDD1167 | [9] |
| LDCM0289 | AC29 | PaTu 8988t | C73(1.08) | LDD1168 | [9] |
| LDCM0290 | AC3 | HCT 116 | C73(1.01) | LDD0607 | [9] |
| LDCM0291 | AC30 | PaTu 8988t | C73(1.01) | LDD1170 | [9] |
| LDCM0292 | AC31 | PaTu 8988t | C73(0.84) | LDD1171 | [9] |
| LDCM0293 | AC32 | PaTu 8988t | C73(0.92) | LDD1172 | [9] |
| LDCM0294 | AC33 | PaTu 8988t | C73(1.37) | LDD1173 | [9] |
| LDCM0295 | AC34 | PaTu 8988t | C73(0.96) | LDD1174 | [9] |
| LDCM0296 | AC35 | HCT 116 | C73(0.99) | LDD0613 | [9] |
| LDCM0297 | AC36 | HCT 116 | C73(0.96) | LDD0614 | [9] |
| LDCM0298 | AC37 | HCT 116 | C73(1.05) | LDD0615 | [9] |
| LDCM0299 | AC38 | HCT 116 | C73(1.06) | LDD0616 | [9] |
| LDCM0300 | AC39 | HCT 116 | C73(0.86) | LDD0617 | [9] |
| LDCM0301 | AC4 | HCT 116 | C73(1.08) | LDD0618 | [9] |
| LDCM0302 | AC40 | HCT 116 | C73(0.91) | LDD0619 | [9] |
| LDCM0303 | AC41 | HCT 116 | C73(0.87) | LDD0620 | [9] |
| LDCM0304 | AC42 | HCT 116 | C73(1.00) | LDD0621 | [9] |
| LDCM0305 | AC43 | HCT 116 | C73(0.89) | LDD0622 | [9] |
| LDCM0306 | AC44 | HCT 116 | C73(0.83) | LDD0623 | [9] |
| LDCM0307 | AC45 | HCT 116 | C73(0.83) | LDD0624 | [9] |
| LDCM0308 | AC46 | HCT 116 | C73(1.04) | LDD0625 | [9] |
| LDCM0309 | AC47 | HCT 116 | C73(0.97) | LDD0626 | [9] |
| LDCM0310 | AC48 | HCT 116 | C73(1.22) | LDD0627 | [9] |
| LDCM0311 | AC49 | HCT 116 | C73(1.73) | LDD0628 | [9] |
| LDCM0312 | AC5 | HCT 116 | C73(1.01) | LDD0629 | [9] |
| LDCM0313 | AC50 | HCT 116 | C73(1.28) | LDD0630 | [9] |
| LDCM0314 | AC51 | HCT 116 | C73(1.44) | LDD0631 | [9] |
| LDCM0315 | AC52 | HCT 116 | C73(1.36) | LDD0632 | [9] |
| LDCM0316 | AC53 | HCT 116 | C73(1.14) | LDD0633 | [9] |
| LDCM0317 | AC54 | HCT 116 | C73(1.25) | LDD0634 | [9] |
| LDCM0318 | AC55 | HCT 116 | C73(0.90) | LDD0635 | [9] |
| LDCM0319 | AC56 | HCT 116 | C73(1.37) | LDD0636 | [9] |
| LDCM0320 | AC57 | HEK-293T | C73(1.03) | LDD1559 | [26] |
| LDCM0321 | AC58 | HEK-293T | C73(0.98) | LDD1560 | [26] |
| LDCM0322 | AC59 | HEK-293T | C73(1.01) | LDD1561 | [26] |
| LDCM0323 | AC6 | HCT 116 | C73(0.91) | LDD0640 | [9] |
| LDCM0324 | AC60 | HEK-293T | C73(1.00) | LDD1563 | [26] |
| LDCM0325 | AC61 | HEK-293T | C73(1.01) | LDD1564 | [26] |
| LDCM0326 | AC62 | HEK-293T | C73(0.96) | LDD1565 | [26] |
| LDCM0327 | AC63 | HEK-293T | C73(0.97) | LDD1566 | [26] |
| LDCM0328 | AC64 | HEK-293T | C73(1.00) | LDD1567 | [26] |
| LDCM0332 | AC68 | HCT 116 | C73(0.81) | LDD0649 | [9] |
| LDCM0333 | AC69 | HCT 116 | C73(0.90) | LDD0650 | [9] |
| LDCM0334 | AC7 | HCT 116 | C73(0.72) | LDD0651 | [9] |
| LDCM0335 | AC70 | HCT 116 | C73(0.91) | LDD0652 | [9] |
| LDCM0336 | AC71 | HCT 116 | C73(0.85) | LDD0653 | [9] |
| LDCM0337 | AC72 | HCT 116 | C73(1.02) | LDD0654 | [9] |
| LDCM0338 | AC73 | HCT 116 | C73(0.95) | LDD0655 | [9] |
| LDCM0339 | AC74 | HCT 116 | C73(0.85) | LDD0656 | [9] |
| LDCM0340 | AC75 | HCT 116 | C73(0.95) | LDD0657 | [9] |
| LDCM0341 | AC76 | HCT 116 | C73(0.87) | LDD0658 | [9] |
| LDCM0342 | AC77 | HCT 116 | C73(0.96) | LDD0659 | [9] |
| LDCM0343 | AC78 | HCT 116 | C73(0.93) | LDD0660 | [9] |
| LDCM0344 | AC79 | HCT 116 | C73(0.97) | LDD0661 | [9] |
| LDCM0345 | AC8 | HCT 116 | C73(1.01) | LDD0662 | [9] |
| LDCM0346 | AC80 | HCT 116 | C73(0.82) | LDD0663 | [9] |
| LDCM0347 | AC81 | HCT 116 | C73(0.85) | LDD0664 | [9] |
| LDCM0348 | AC82 | HCT 116 | C73(0.84) | LDD0665 | [9] |
| LDCM0349 | AC83 | HCT 116 | C73(1.20) | LDD0666 | [9] |
| LDCM0350 | AC84 | HCT 116 | C73(1.16) | LDD0667 | [9] |
| LDCM0351 | AC85 | HCT 116 | C73(1.21) | LDD0668 | [9] |
| LDCM0352 | AC86 | HCT 116 | C73(1.37) | LDD0669 | [9] |
| LDCM0353 | AC87 | HCT 116 | C73(1.02) | LDD0670 | [9] |
| LDCM0354 | AC88 | HCT 116 | C73(1.02) | LDD0671 | [9] |
| LDCM0355 | AC89 | HCT 116 | C73(1.21) | LDD0672 | [9] |
| LDCM0357 | AC90 | HCT 116 | C73(0.96) | LDD0674 | [9] |
| LDCM0358 | AC91 | HCT 116 | C73(1.20) | LDD0675 | [9] |
| LDCM0359 | AC92 | HCT 116 | C73(0.99) | LDD0676 | [9] |
| LDCM0360 | AC93 | HCT 116 | C73(0.89) | LDD0677 | [9] |
| LDCM0361 | AC94 | HCT 116 | C73(1.10) | LDD0678 | [9] |
| LDCM0362 | AC95 | HCT 116 | C73(1.05) | LDD0679 | [9] |
| LDCM0363 | AC96 | HCT 116 | C73(1.00) | LDD0680 | [9] |
| LDCM0364 | AC97 | HCT 116 | C73(1.08) | LDD0681 | [9] |
| LDCM0365 | AC98 | HEK-293T | C73(0.93) | LDD0963 | [9] |
| LDCM0366 | AC99 | HEK-293T | C73(0.97) | LDD0964 | [9] |
| LDCM0248 | AKOS034007472 | HCT 116 | C73(0.78) | LDD0565 | [9] |
| LDCM0356 | AKOS034007680 | HCT 116 | C73(0.70) | LDD0673 | [9] |
| LDCM0275 | AKOS034007705 | HCT 116 | C73(0.97) | LDD0592 | [9] |
| LDCM0156 | Aniline | NCI-H1299 | 12.74 | LDD0403 | [1] |
| LDCM0087 | Capsaicin | HEK-293T | 5.98 | LDD0185 | [14] |
| LDCM0632 | CL-Sc | Hep-G2 | C73(0.75) | LDD2227 | [13] |
| LDCM0367 | CL1 | HEK-293T | C73(0.89) | LDD0965 | [9] |
| LDCM0368 | CL10 | HEK-293T | C73(0.72) | LDD0966 | [9] |
| LDCM0369 | CL100 | HCT 116 | C73(1.09) | LDD0686 | [9] |
| LDCM0370 | CL101 | HCT 116 | C73(0.99) | LDD0687 | [9] |
| LDCM0371 | CL102 | HCT 116 | C73(0.57) | LDD0688 | [9] |
| LDCM0372 | CL103 | HCT 116 | C73(0.57) | LDD0689 | [9] |
| LDCM0373 | CL104 | HCT 116 | C73(0.69) | LDD0690 | [9] |
| LDCM0374 | CL105 | HCT 116 | C73(0.80) | LDD0691 | [9] |
| LDCM0375 | CL106 | HCT 116 | C73(0.80) | LDD0692 | [9] |
| LDCM0376 | CL107 | HCT 116 | C73(0.98) | LDD0693 | [9] |
| LDCM0377 | CL108 | HCT 116 | C73(0.78) | LDD0694 | [9] |
| LDCM0378 | CL109 | HCT 116 | C73(1.11) | LDD0695 | [9] |
| LDCM0379 | CL11 | HEK-293T | C73(0.80) | LDD0977 | [9] |
| LDCM0380 | CL110 | HCT 116 | C73(0.84) | LDD0697 | [9] |
| LDCM0381 | CL111 | HCT 116 | C73(0.84) | LDD0698 | [9] |
| LDCM0382 | CL112 | PaTu 8988t | C73(0.99) | LDD1261 | [9] |
| LDCM0383 | CL113 | PaTu 8988t | C73(1.04) | LDD1262 | [9] |
| LDCM0384 | CL114 | PaTu 8988t | C73(0.99) | LDD1263 | [9] |
| LDCM0385 | CL115 | PaTu 8988t | C73(0.97) | LDD1264 | [9] |
| LDCM0386 | CL116 | PaTu 8988t | C73(1.17) | LDD1265 | [9] |
| LDCM0387 | CL117 | HCT 116 | C73(0.73) | LDD0704 | [9] |
| LDCM0388 | CL118 | HCT 116 | C73(1.14) | LDD0705 | [9] |
| LDCM0389 | CL119 | HCT 116 | C73(1.24) | LDD0706 | [9] |
| LDCM0390 | CL12 | HEK-293T | C73(0.92) | LDD0988 | [9] |
| LDCM0391 | CL120 | HCT 116 | C73(1.03) | LDD0708 | [9] |
| LDCM0392 | CL121 | HCT 116 | C73(1.25) | LDD0709 | [9] |
| LDCM0393 | CL122 | HCT 116 | C73(1.19) | LDD0710 | [9] |
| LDCM0394 | CL123 | HCT 116 | C73(0.98) | LDD0711 | [9] |
| LDCM0395 | CL124 | HCT 116 | C73(1.14) | LDD0712 | [9] |
| LDCM0396 | CL125 | HEK-293T | C73(1.09) | LDD1600 | [26] |
| LDCM0397 | CL126 | HEK-293T | C73(1.01) | LDD1601 | [26] |
| LDCM0398 | CL127 | HEK-293T | C73(1.01) | LDD1602 | [26] |
| LDCM0399 | CL128 | HEK-293T | C73(1.05) | LDD1603 | [26] |
| LDCM0400 | CL13 | HEK-293T | C73(0.85) | LDD0998 | [9] |
| LDCM0401 | CL14 | HEK-293T | C73(0.92) | LDD0999 | [9] |
| LDCM0402 | CL15 | HEK-293T | C73(0.87) | LDD1000 | [9] |
| LDCM0403 | CL16 | PaTu 8988t | C73(1.08) | LDD1282 | [9] |
| LDCM0404 | CL17 | PaTu 8988t | C73(0.97) | LDD1283 | [9] |
| LDCM0405 | CL18 | PaTu 8988t | C73(1.19) | LDD1284 | [9] |
| LDCM0406 | CL19 | PaTu 8988t | C73(0.96) | LDD1285 | [9] |
| LDCM0407 | CL2 | HEK-293T | C73(1.01) | LDD1005 | [9] |
| LDCM0408 | CL20 | PaTu 8988t | C73(1.09) | LDD1287 | [9] |
| LDCM0409 | CL21 | PaTu 8988t | C73(1.31) | LDD1288 | [9] |
| LDCM0410 | CL22 | PaTu 8988t | C73(1.61) | LDD1289 | [9] |
| LDCM0411 | CL23 | PaTu 8988t | C73(1.43) | LDD1290 | [9] |
| LDCM0412 | CL24 | PaTu 8988t | C73(0.95) | LDD1291 | [9] |
| LDCM0413 | CL25 | PaTu 8988t | C73(1.37) | LDD1292 | [9] |
| LDCM0414 | CL26 | PaTu 8988t | C73(1.14) | LDD1293 | [9] |
| LDCM0415 | CL27 | PaTu 8988t | C73(1.40) | LDD1294 | [9] |
| LDCM0416 | CL28 | PaTu 8988t | C73(1.06) | LDD1295 | [9] |
| LDCM0417 | CL29 | PaTu 8988t | C73(1.03) | LDD1296 | [9] |
| LDCM0418 | CL3 | HEK-293T | C73(1.03) | LDD1016 | [9] |
| LDCM0419 | CL30 | PaTu 8988t | C73(1.16) | LDD1298 | [9] |
| LDCM0420 | CL31 | HCT 116 | C73(1.00) | LDD0737 | [9] |
| LDCM0421 | CL32 | HCT 116 | C73(0.94) | LDD0738 | [9] |
| LDCM0422 | CL33 | HCT 116 | C73(1.27) | LDD0739 | [9] |
| LDCM0423 | CL34 | HCT 116 | C73(1.01) | LDD0740 | [9] |
| LDCM0424 | CL35 | HCT 116 | C73(0.87) | LDD0741 | [9] |
| LDCM0425 | CL36 | HCT 116 | C73(0.95) | LDD0742 | [9] |
| LDCM0426 | CL37 | HCT 116 | C73(0.77) | LDD0743 | [9] |
| LDCM0428 | CL39 | HCT 116 | C73(0.87) | LDD0745 | [9] |
| LDCM0429 | CL4 | HEK-293T | C73(1.17) | LDD1027 | [9] |
| LDCM0430 | CL40 | HCT 116 | C73(0.97) | LDD0747 | [9] |
| LDCM0431 | CL41 | HCT 116 | C73(0.82) | LDD0748 | [9] |
| LDCM0432 | CL42 | HCT 116 | C73(1.06) | LDD0749 | [9] |
| LDCM0433 | CL43 | HCT 116 | C73(1.04) | LDD0750 | [9] |
| LDCM0434 | CL44 | HCT 116 | C73(0.99) | LDD0751 | [9] |
| LDCM0435 | CL45 | HCT 116 | C73(0.98) | LDD0752 | [9] |
| LDCM0436 | CL46 | HCT 116 | C73(0.79) | LDD0753 | [9] |
| LDCM0437 | CL47 | HCT 116 | C73(0.96) | LDD0754 | [9] |
| LDCM0438 | CL48 | HCT 116 | C73(0.77) | LDD0755 | [9] |
| LDCM0439 | CL49 | HCT 116 | C73(0.72) | LDD0756 | [9] |
| LDCM0440 | CL5 | HEK-293T | C73(0.92) | LDD1038 | [9] |
| LDCM0441 | CL50 | HCT 116 | C73(0.82) | LDD0758 | [9] |
| LDCM0442 | CL51 | HCT 116 | C73(0.81) | LDD0759 | [9] |
| LDCM0443 | CL52 | HCT 116 | C73(0.81) | LDD0760 | [9] |
| LDCM0444 | CL53 | HCT 116 | C73(1.17) | LDD0761 | [9] |
| LDCM0445 | CL54 | HCT 116 | C73(0.84) | LDD0762 | [9] |
| LDCM0446 | CL55 | HCT 116 | C73(0.77) | LDD0763 | [9] |
| LDCM0447 | CL56 | HCT 116 | C73(1.09) | LDD0764 | [9] |
| LDCM0448 | CL57 | HCT 116 | C73(1.22) | LDD0765 | [9] |
| LDCM0449 | CL58 | HCT 116 | C73(0.93) | LDD0766 | [9] |
| LDCM0450 | CL59 | HCT 116 | C73(0.93) | LDD0767 | [9] |
| LDCM0451 | CL6 | HEK-293T | C73(1.03) | LDD1049 | [9] |
| LDCM0452 | CL60 | HCT 116 | C73(0.77) | LDD0769 | [9] |
| LDCM0453 | CL61 | HCT 116 | C73(0.99) | LDD0770 | [9] |
| LDCM0454 | CL62 | HCT 116 | C73(1.00) | LDD0771 | [9] |
| LDCM0455 | CL63 | HCT 116 | C73(1.12) | LDD0772 | [9] |
| LDCM0456 | CL64 | HCT 116 | C73(1.05) | LDD0773 | [9] |
| LDCM0457 | CL65 | HCT 116 | C73(1.10) | LDD0774 | [9] |
| LDCM0458 | CL66 | HCT 116 | C73(0.99) | LDD0775 | [9] |
| LDCM0459 | CL67 | HCT 116 | C73(1.24) | LDD0776 | [9] |
| LDCM0460 | CL68 | HCT 116 | C73(1.30) | LDD0777 | [9] |
| LDCM0461 | CL69 | HCT 116 | C73(1.14) | LDD0778 | [9] |
| LDCM0462 | CL7 | HEK-293T | C73(0.71) | LDD1060 | [9] |
| LDCM0463 | CL70 | HCT 116 | C73(1.04) | LDD0780 | [9] |
| LDCM0464 | CL71 | HCT 116 | C73(1.15) | LDD0781 | [9] |
| LDCM0465 | CL72 | HCT 116 | C73(1.36) | LDD0782 | [9] |
| LDCM0466 | CL73 | HCT 116 | C73(1.39) | LDD0783 | [9] |
| LDCM0467 | CL74 | HCT 116 | C73(1.40) | LDD0784 | [9] |
| LDCM0469 | CL76 | HCT 116 | C73(1.18) | LDD0786 | [9] |
| LDCM0470 | CL77 | HCT 116 | C73(1.04) | LDD0787 | [9] |
| LDCM0471 | CL78 | HCT 116 | C73(1.09) | LDD0788 | [9] |
| LDCM0472 | CL79 | HCT 116 | C73(1.05) | LDD0789 | [9] |
| LDCM0473 | CL8 | HEK-293T | C73(2.21) | LDD1071 | [9] |
| LDCM0474 | CL80 | HCT 116 | C73(0.97) | LDD0791 | [9] |
| LDCM0475 | CL81 | HCT 116 | C73(1.14) | LDD0792 | [9] |
| LDCM0476 | CL82 | HCT 116 | C73(0.99) | LDD0793 | [9] |
| LDCM0477 | CL83 | HCT 116 | C73(0.80) | LDD0794 | [9] |
| LDCM0478 | CL84 | HCT 116 | C73(0.74) | LDD0795 | [9] |
| LDCM0479 | CL85 | HCT 116 | C73(0.98) | LDD0796 | [9] |
| LDCM0480 | CL86 | HCT 116 | C73(1.14) | LDD0797 | [9] |
| LDCM0481 | CL87 | HCT 116 | C73(1.00) | LDD0798 | [9] |
| LDCM0482 | CL88 | HCT 116 | C73(1.09) | LDD0799 | [9] |
| LDCM0483 | CL89 | HCT 116 | C73(0.93) | LDD0800 | [9] |
| LDCM0484 | CL9 | HEK-293T | C73(1.33) | LDD1082 | [9] |
| LDCM0485 | CL90 | HCT 116 | C73(1.08) | LDD0802 | [9] |
| LDCM0486 | CL91 | HCT 116 | C73(0.88) | LDD0803 | [9] |
| LDCM0487 | CL92 | HCT 116 | C73(0.85) | LDD0804 | [9] |
| LDCM0488 | CL93 | HCT 116 | C73(0.87) | LDD0805 | [9] |
| LDCM0489 | CL94 | HCT 116 | C73(0.99) | LDD0806 | [9] |
| LDCM0490 | CL95 | HCT 116 | C73(0.99) | LDD0807 | [9] |
| LDCM0491 | CL96 | HCT 116 | C73(1.02) | LDD0808 | [9] |
| LDCM0492 | CL97 | HCT 116 | C73(1.08) | LDD0809 | [9] |
| LDCM0493 | CL98 | HCT 116 | C73(0.98) | LDD0810 | [9] |
| LDCM0494 | CL99 | HCT 116 | C73(0.88) | LDD0811 | [9] |
| LDCM0193 | Compound 36 | HEK-293T | 4.43 | LDD0511 | [24] |
| LDCM0495 | E2913 | HEK-293T | C73(1.00) | LDD1698 | [26] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C73(1.18) | LDD1702 | [4] |
| LDCM0031 | Epigallocatechin gallate | HEK-293T | 9.20 | LDD0183 | [14] |
| LDCM0625 | F8 | Ramos | C73(2.65) | LDD2187 | [27] |
| LDCM0573 | Fragment11 | Ramos | C73(0.34) | LDD2190 | [27] |
| LDCM0576 | Fragment14 | Ramos | C73(0.57) | LDD2193 | [27] |
| LDCM0586 | Fragment28 | Ramos | C73(1.08) | LDD2198 | [27] |
| LDCM0468 | Fragment33 | HCT 116 | C73(1.13) | LDD0785 | [9] |
| LDCM0566 | Fragment4 | Ramos | C73(0.65) | LDD2184 | [27] |
| LDCM0427 | Fragment51 | HCT 116 | C73(0.94) | LDD0744 | [9] |
| LDCM0569 | Fragment7 | Ramos | C73(0.61) | LDD2186 | [27] |
| LDCM0116 | HHS-0101 | DM93 | Y200(1.04) | LDD0264 | [6] |
| LDCM0117 | HHS-0201 | DM93 | Y200(0.82) | LDD0265 | [6] |
| LDCM0118 | HHS-0301 | DM93 | Y200(1.36) | LDD0266 | [6] |
| LDCM0119 | HHS-0401 | DM93 | Y200(1.14) | LDD0267 | [6] |
| LDCM0120 | HHS-0701 | DM93 | Y200(0.82) | LDD0268 | [6] |
| LDCM0022 | KB02 | HCT 116 | C73(1.29) | LDD0080 | [9] |
| LDCM0023 | KB03 | HCT 116 | C73(0.99) | LDD0081 | [9] |
| LDCM0024 | KB05 | HCT 116 | C73(1.52) | LDD0082 | [9] |
| LDCM0030 | Luteolin | HEK-293T | 6.22 | LDD0182 | [14] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [8] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C73(0.97) | LDD2092 | [4] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C73(1.14) | LDD2094 | [4] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C73(0.90) | LDD2098 | [4] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C73(0.77) | LDD2109 | [4] |
| LDCM0559 | Nucleophilic fragment 9b | MDA-MB-231 | C73(2.69) | LDD2153 | [4] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C73(1.33) | LDD2206 | [28] |
| LDCM0032 | Oleacein | HEK-293T | 4.69 | LDD0184 | [14] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C73(1.03) | LDD2207 | [28] |
| LDCM0131 | RA190 | MM1.R | C73(1.03) | LDD0304 | [29] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Protein arginine N-methyltransferase 5 (PRMT5) | Protein arginine N-methyltransferase family | O14744 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| FERM domain-containing protein 7 (FRMD7) | . | Q6ZUT3 | |||
Other
References
