Details of the Target
General Information of Target
| Target ID | LDTP03987 | |||||
|---|---|---|---|---|---|---|
| Target Name | Small ribosomal subunit protein eS27 (RPS27) | |||||
| Gene Name | RPS27 | |||||
| Gene ID | 6232 | |||||
| Synonyms |
MPS1; Small ribosomal subunit protein eS27; 40S ribosomal protein S27; Metallopan-stimulin 1; MPS-1 |
|||||
| 3D Structure | ||||||
| Sequence |
MPLAKDLLHPSPEEEKRKHKKKRLVQSPNSYFMDVKCPGCYKITTVFSHAQTVVLCVGCS
TVLCQPTGGKARLTEGCSFRRKQH |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
Eukaryotic ribosomal protein eS27 family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Component of the small ribosomal subunit. The ribosome is a large ribonucleoprotein complex responsible for the synthesis of proteins in the cell. Required for proper rRNA processing and maturation of 18S rRNAs. Part of the small subunit (SSU) processome, first precursor of the small eukaryotic ribosomal subunit. During the assembly of the SSU processome in the nucleolus, many ribosome biogenesis factors, an RNA chaperone and ribosomal proteins associate with the nascent pre-rRNA and work in concert to generate RNA folding, modifications, rearrangements and cleavage as well as targeted degradation of pre-ribosomal RNA by the RNA exosome.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
14.82 | LDD0402 | [1] | |
|
P1 Probe Info |
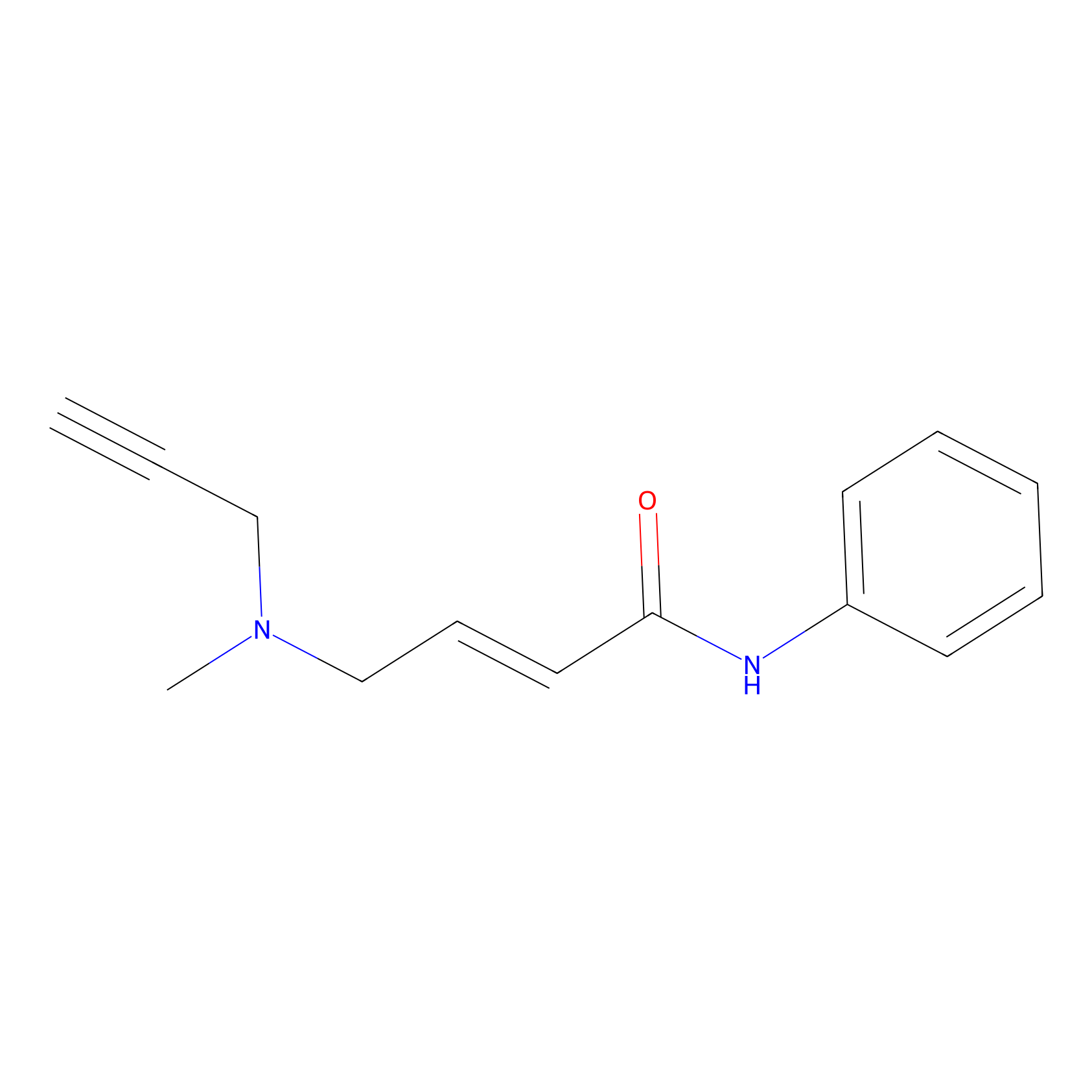 |
1.81 | LDD0448 | [2] | |
|
P2 Probe Info |
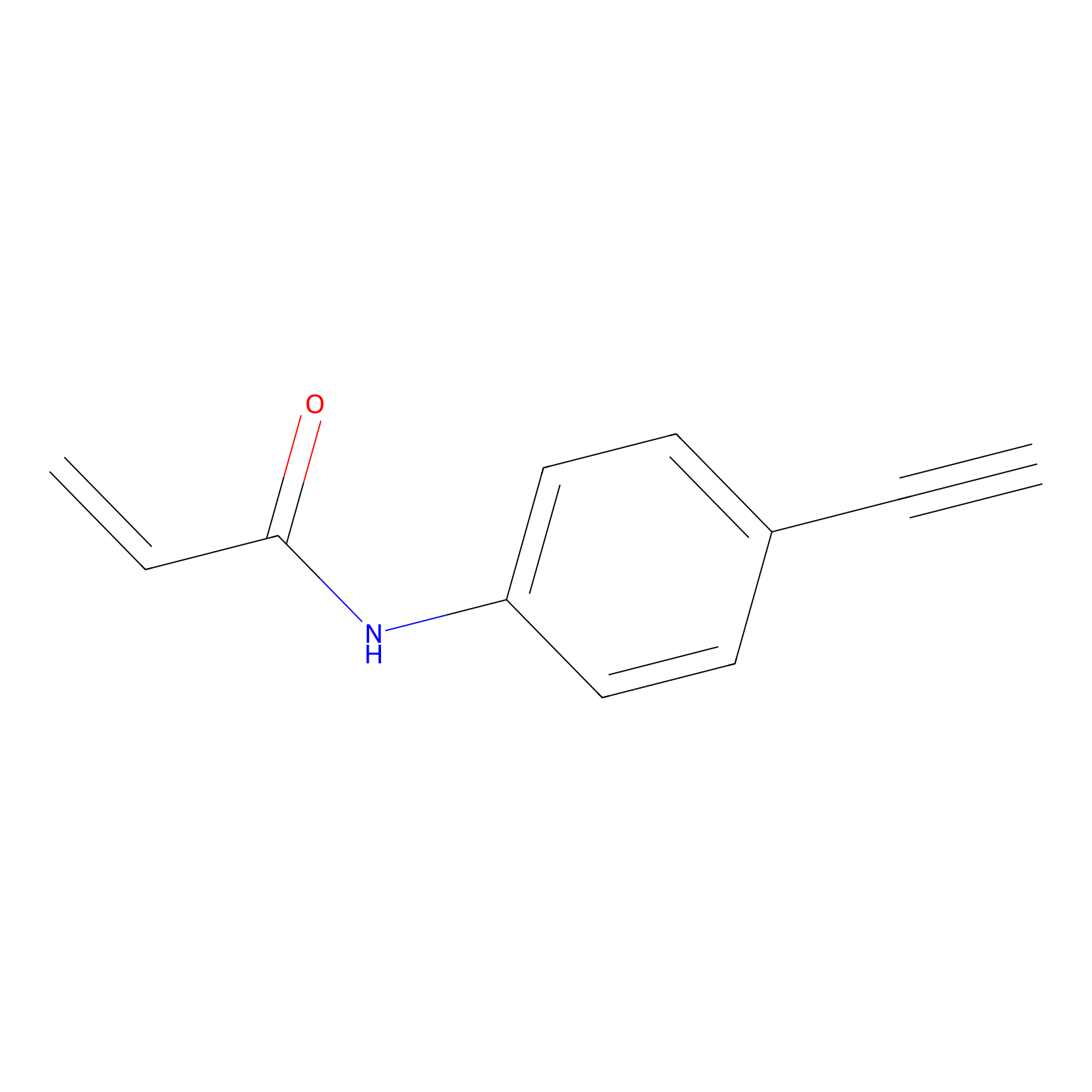 |
1.66 | LDD0449 | [2] | |
|
P8 Probe Info |
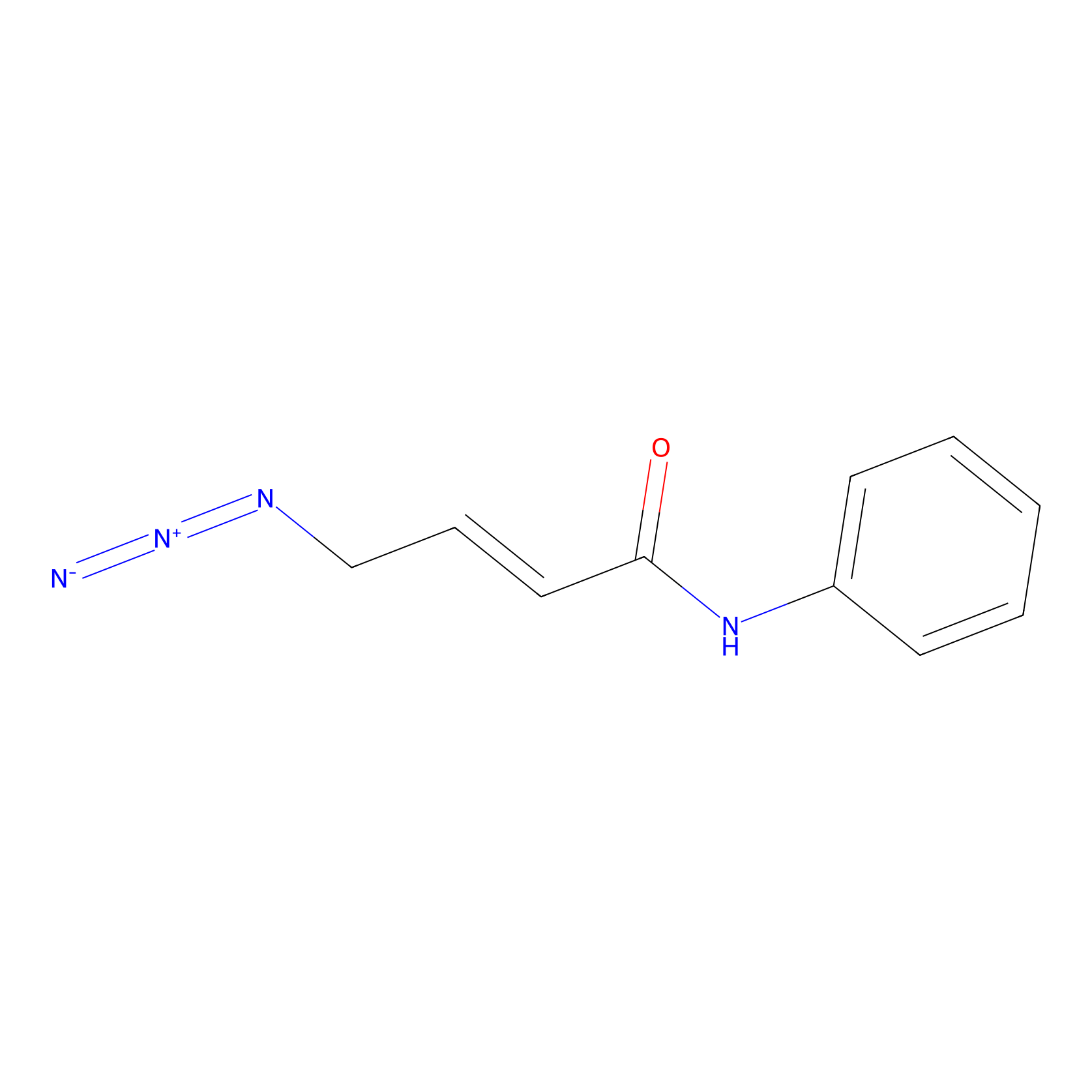 |
1.87 | LDD0451 | [2] | |
|
CY-1 Probe Info |
 |
6.12 | LDD0243 | [3] | |
|
CY4 Probe Info |
 |
4.17 | LDD0244 | [3] | |
|
N1 Probe Info |
 |
6.28 | LDD0242 | [3] | |
|
TH211 Probe Info |
 |
Y31(13.36) | LDD0260 | [4] | |
|
TH214 Probe Info |
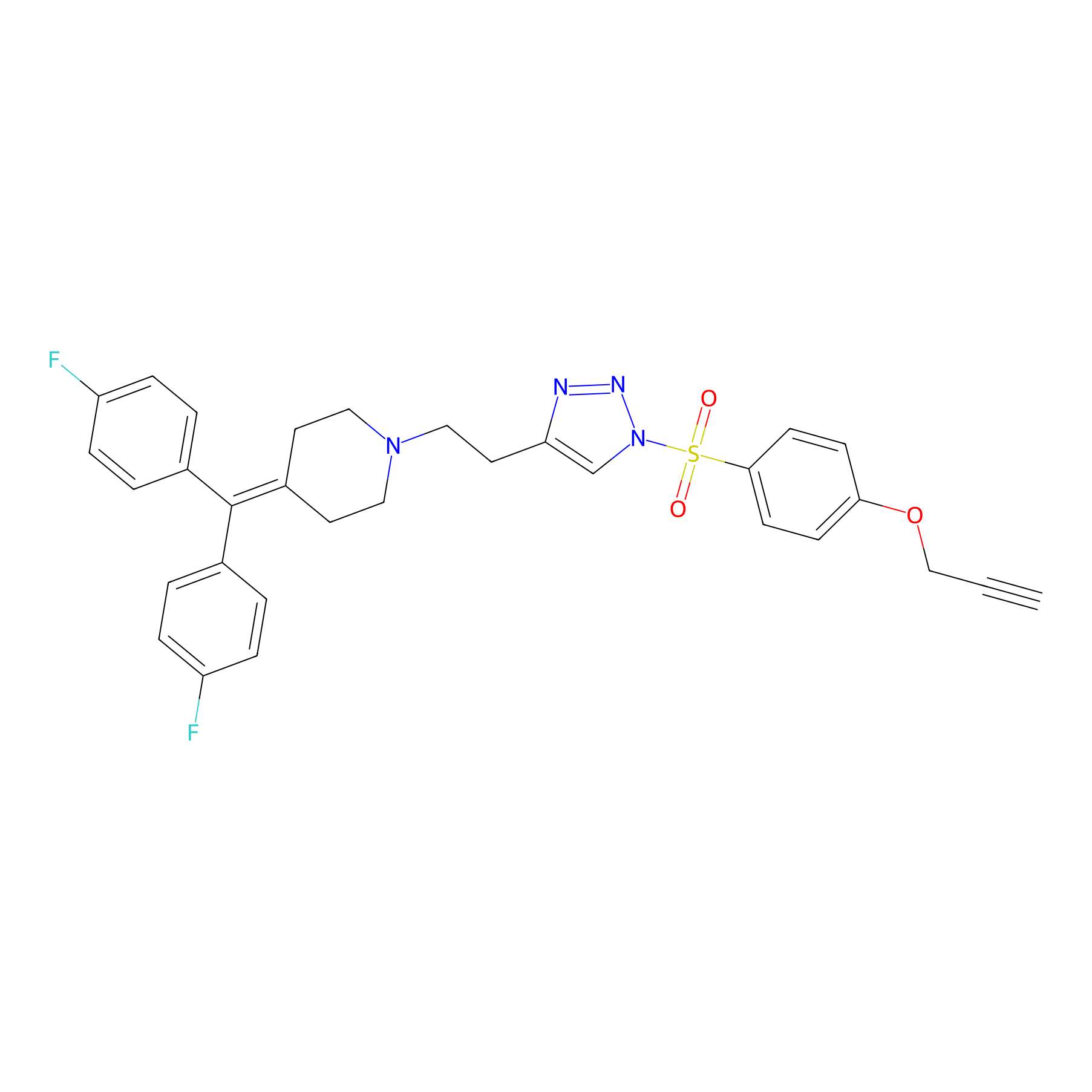 |
Y31(13.92) | LDD0258 | [4] | |
|
TH216 Probe Info |
 |
Y31(15.16) | LDD0259 | [4] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [5] | |
|
STPyne Probe Info |
 |
K16(6.92) | LDD0277 | [6] | |
|
BTD Probe Info |
 |
C77(4.75) | LDD1699 | [7] | |
|
ONAyne Probe Info |
 |
N.A. | LDD0273 | [6] | |
|
AF-1 Probe Info |
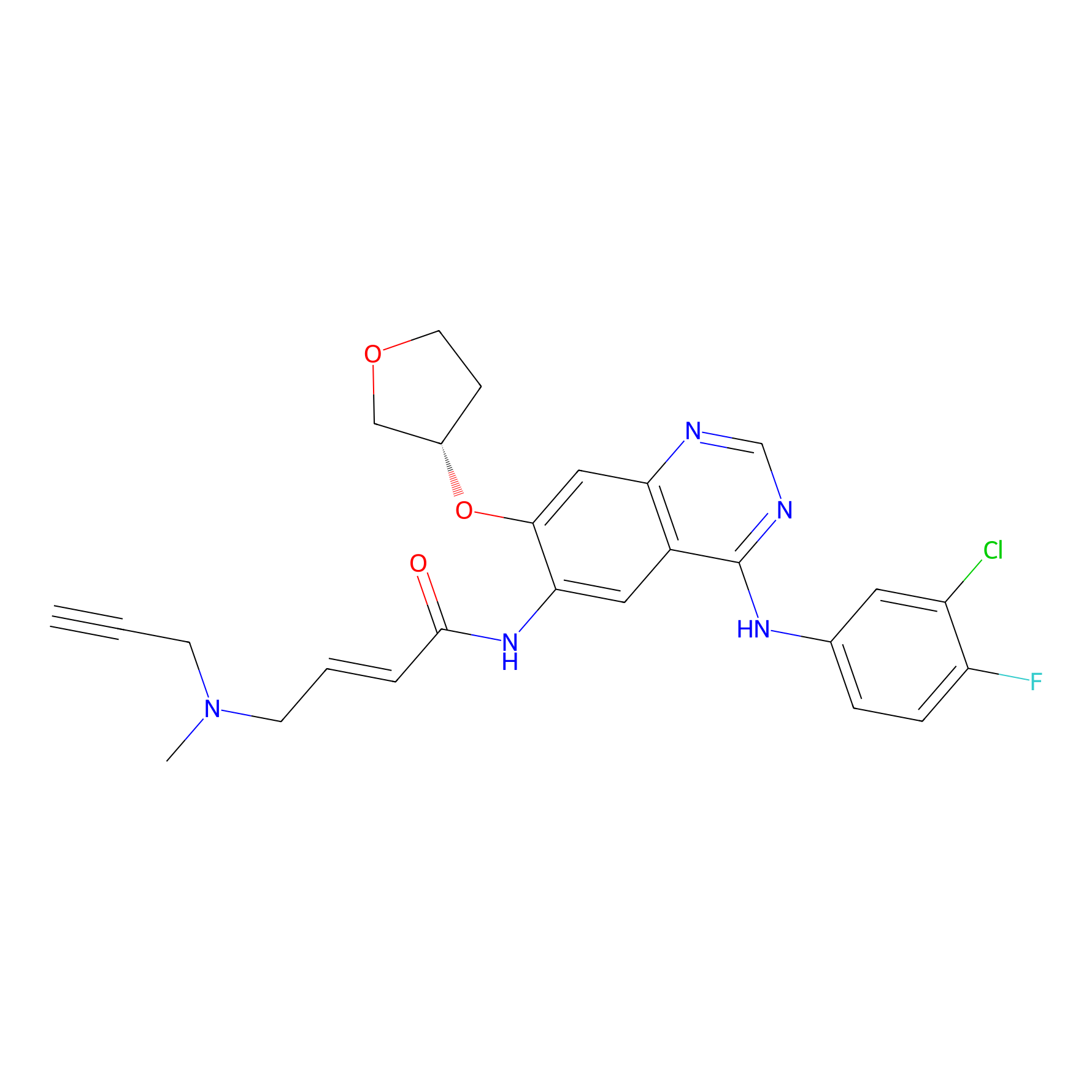 |
1.64 | LDD0421 | [8] | |
|
HPAP Probe Info |
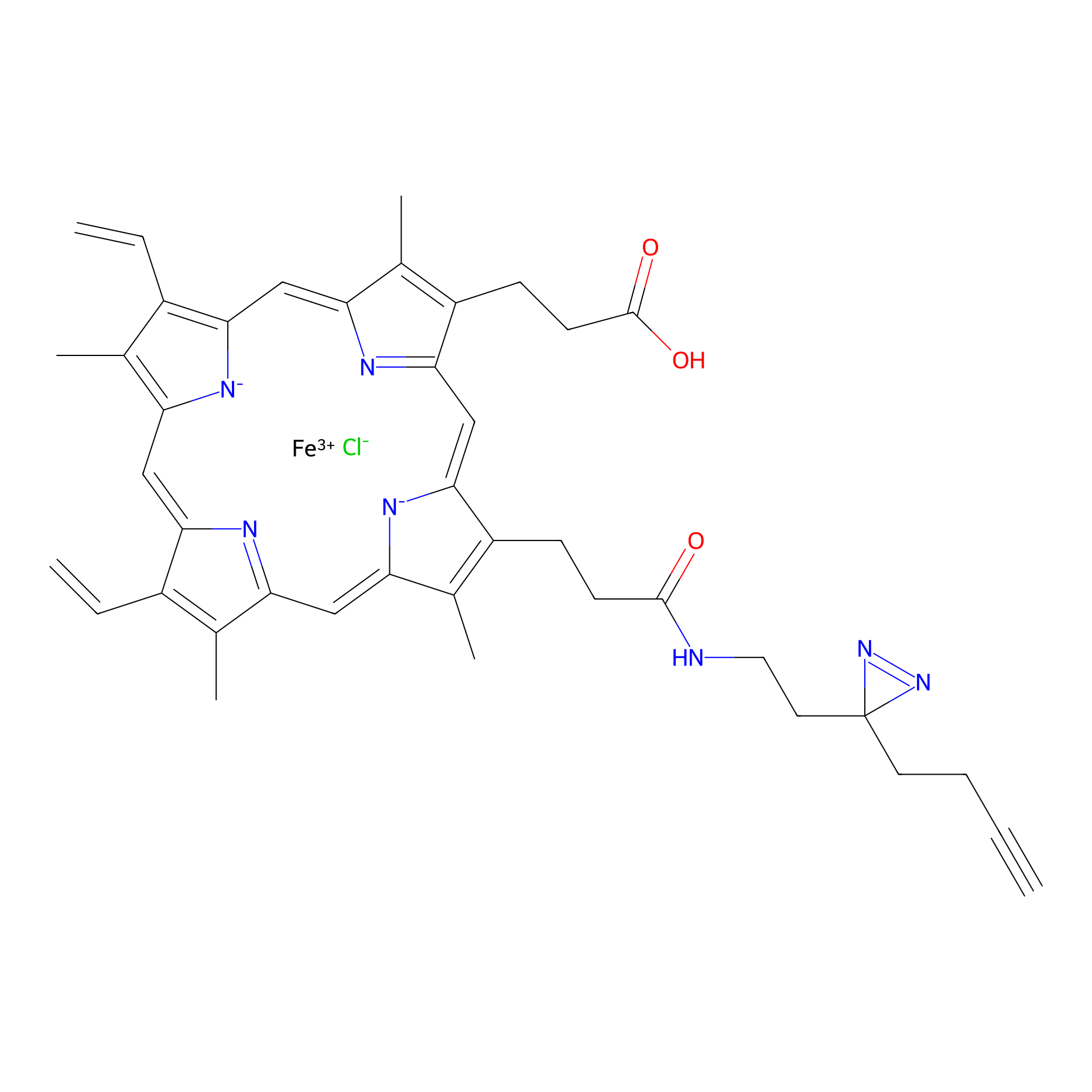 |
3.87 | LDD0062 | [9] | |
|
HHS-482 Probe Info |
 |
Y31(0.07) | LDD0289 | [10] | |
|
DBIA Probe Info |
 |
C77(1.51) | LDD0078 | [11] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [12] | |
|
ATP probe Probe Info |
 |
K5(0.00); K16(0.00) | LDD0199 | [13] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [14] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0032 | [15] | |
|
IPIAA_H Probe Info |
 |
N.A. | LDD0030 | [16] | |
|
IPIAA_L Probe Info |
 |
N.A. | LDD0031 | [16] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [14] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [17] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [18] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [17] | |
|
Compound 10 Probe Info |
 |
N.A. | LDD2216 | [19] | |
|
IPM Probe Info |
 |
N.A. | LDD2156 | [20] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [21] | |
|
OSF Probe Info |
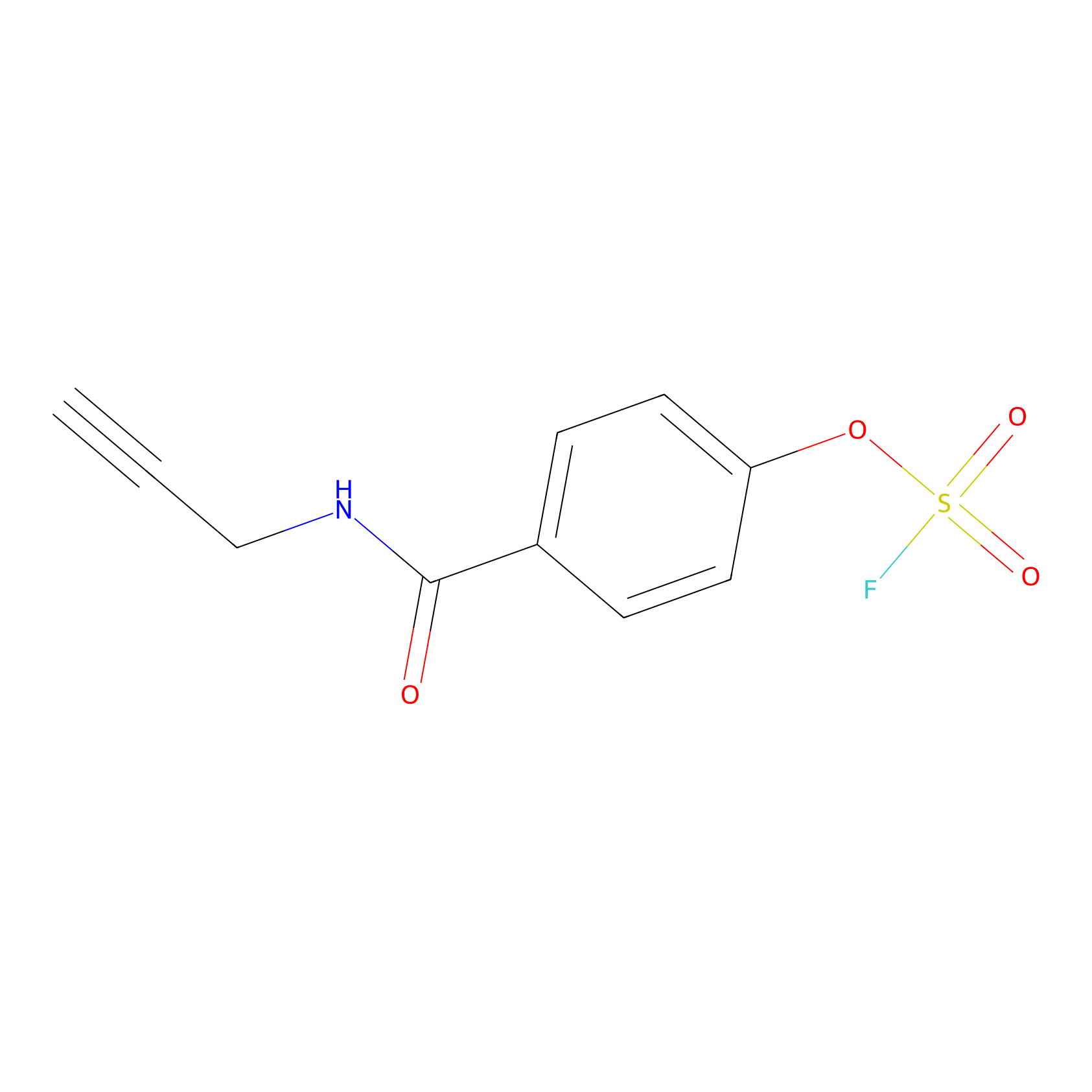 |
N.A. | LDD0029 | [22] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [22] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [23] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [24] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [18] | |
|
HHS-475 Probe Info |
 |
Y31(1.03) | LDD2238 | [25] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0166 | Afatinib | A431 | 1.64 | LDD0421 | [8] |
| LDCM0156 | Aniline | NCI-H1299 | 11.63 | LDD0403 | [1] |
| LDCM0020 | ARS-1620 | HCC44 | C77(1.51) | LDD0078 | [11] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [23] |
| LDCM0632 | CL-Sc | Hep-G2 | C77(1.74); C77(1.16); C77(0.93) | LDD2227 | [18] |
| LDCM0634 | CY-0357 | Hep-G2 | C77(0.63) | LDD2228 | [18] |
| LDCM0625 | F8 | Ramos | C77(1.43) | LDD2187 | [26] |
| LDCM0572 | Fragment10 | MDA-MB-231 | C77(3.17) | LDD1389 | [27] |
| LDCM0573 | Fragment11 | MDA-MB-231 | C77(3.27) | LDD1391 | [27] |
| LDCM0574 | Fragment12 | Ramos | C77(2.19) | LDD1394 | [27] |
| LDCM0575 | Fragment13 | MDA-MB-231 | C77(1.45) | LDD1395 | [27] |
| LDCM0576 | Fragment14 | MDA-MB-231 | C77(1.42) | LDD1397 | [27] |
| LDCM0577 | Fragment15 | Ramos | C77(1.09) | LDD1400 | [27] |
| LDCM0579 | Fragment20 | MDA-MB-231 | C77(1.26) | LDD1402 | [27] |
| LDCM0580 | Fragment21 | MDA-MB-231 | C77(1.66) | LDD1404 | [27] |
| LDCM0581 | Fragment22 | MDA-MB-231 | C77(1.54) | LDD1406 | [27] |
| LDCM0582 | Fragment23 | MDA-MB-231 | C77(1.31) | LDD1408 | [27] |
| LDCM0584 | Fragment25 | MDA-MB-231 | C77(0.96) | LDD1411 | [27] |
| LDCM0585 | Fragment26 | Ramos | C77(0.96) | LDD1412 | [27] |
| LDCM0578 | Fragment27 | MDA-MB-231 | C77(0.86) | LDD1401 | [27] |
| LDCM0586 | Fragment28 | MDA-MB-231 | C77(1.14) | LDD1415 | [27] |
| LDCM0587 | Fragment29 | MDA-MB-231 | C77(1.29) | LDD1417 | [27] |
| LDCM0588 | Fragment30 | MDA-MB-231 | C77(1.34) | LDD1419 | [27] |
| LDCM0589 | Fragment31 | Ramos | C77(0.86) | LDD2200 | [26] |
| LDCM0590 | Fragment32 | MDA-MB-231 | C77(4.53) | LDD1423 | [27] |
| LDCM0468 | Fragment33 | MDA-MB-231 | C77(1.27) | LDD1425 | [27] |
| LDCM0592 | Fragment34 | MDA-MB-231 | C77(1.29) | LDD1427 | [27] |
| LDCM0593 | Fragment35 | Ramos | C77(0.86) | LDD1430 | [27] |
| LDCM0594 | Fragment36 | MDA-MB-231 | C77(1.78) | LDD1431 | [27] |
| LDCM0595 | Fragment37 | Ramos | C77(0.85) | LDD1432 | [27] |
| LDCM0596 | Fragment38 | MDA-MB-231 | C77(1.00) | LDD1433 | [27] |
| LDCM0566 | Fragment4 | MDA-MB-231 | C77(4.42) | LDD1378 | [27] |
| LDCM0598 | Fragment40 | MDA-MB-231 | C77(0.83) | LDD1436 | [27] |
| LDCM0599 | Fragment41 | MDA-MB-231 | C77(1.24) | LDD1438 | [27] |
| LDCM0600 | Fragment42 | Ramos | C77(1.14) | LDD1440 | [27] |
| LDCM0601 | Fragment43 | MDA-MB-231 | C77(2.49) | LDD1441 | [27] |
| LDCM0602 | Fragment44 | MDA-MB-231 | C77(0.77) | LDD1443 | [27] |
| LDCM0603 | Fragment45 | MDA-MB-231 | C77(6.20) | LDD1444 | [27] |
| LDCM0604 | Fragment46 | MDA-MB-231 | C77(0.89) | LDD1445 | [27] |
| LDCM0605 | Fragment47 | MDA-MB-231 | C77(1.02) | LDD1446 | [27] |
| LDCM0606 | Fragment48 | MDA-MB-231 | C77(1.16) | LDD1447 | [27] |
| LDCM0608 | Fragment50 | MDA-MB-231 | C77(5.06) | LDD1449 | [27] |
| LDCM0427 | Fragment51 | MDA-MB-231 | C77(0.98) | LDD1450 | [27] |
| LDCM0610 | Fragment52 | MDA-MB-231 | C77(1.43) | LDD1452 | [27] |
| LDCM0611 | Fragment53 | MDA-MB-231 | C77(1.07) | LDD1454 | [27] |
| LDCM0612 | Fragment54 | MDA-MB-231 | C77(0.96) | LDD1456 | [27] |
| LDCM0613 | Fragment55 | MDA-MB-231 | C77(1.12) | LDD1457 | [27] |
| LDCM0614 | Fragment56 | MDA-MB-231 | C77(1.13) | LDD1458 | [27] |
| LDCM0569 | Fragment7 | Ramos | C77(1.34) | LDD2186 | [26] |
| LDCM0570 | Fragment8 | MDA-MB-231 | C77(3.07) | LDD1385 | [27] |
| LDCM0571 | Fragment9 | MDA-MB-231 | C77(1.22) | LDD1387 | [27] |
| LDCM0015 | HNE | MDA-MB-231 | C77(0.90) | LDD0346 | [26] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [23] |
| LDCM0127 | JWB152 | DM93 | Y31(0.07) | LDD0289 | [10] |
| LDCM0128 | JWB198 | DM93 | Y31(0.17) | LDD0290 | [10] |
| LDCM0022 | KB02 | MDA-MB-231 | C77(8.33) | LDD1374 | [27] |
| LDCM0023 | KB03 | MDA-MB-231 | C77(8.51) | LDD1376 | [27] |
| LDCM0024 | KB05 | Ramos | C77(2.70) | LDD1381 | [27] |
| LDCM0109 | NEM | HeLa | H9(0.00); K5(0.00) | LDD0223 | [23] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C77(1.26) | LDD2090 | [7] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C77(0.98) | LDD2111 | [7] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C77(0.85) | LDD2125 | [7] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C77(1.16) | LDD2136 | [7] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C77(0.91) | LDD2137 | [7] |
| LDCM0551 | Nucleophilic fragment 5b | MDA-MB-231 | C77(6.46) | LDD2145 | [7] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C77(0.65) | LDD2148 | [7] |
| LDCM0014 | Panhematin | HEK-293T | 3.87 | LDD0062 | [9] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Proteasome activator complex subunit 3 (PSME3) | PA28 family | P61289 | |||
References
