Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
6.47 | LDD0402 | [1] | |
|
Alkylaryl probe 3 Probe Info |
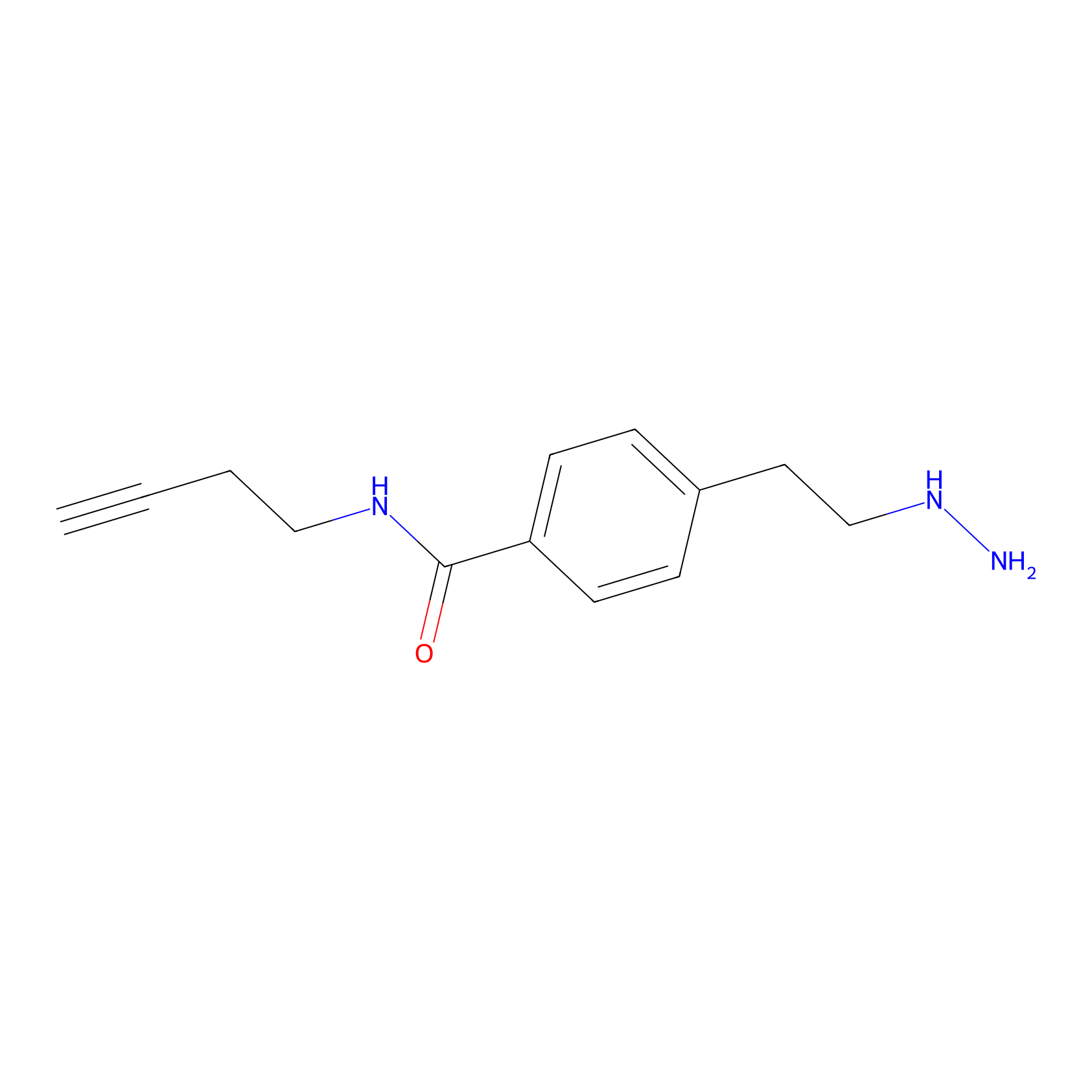 |
20.00 | LDD0381 | [2] | |
|
P1 Probe Info |
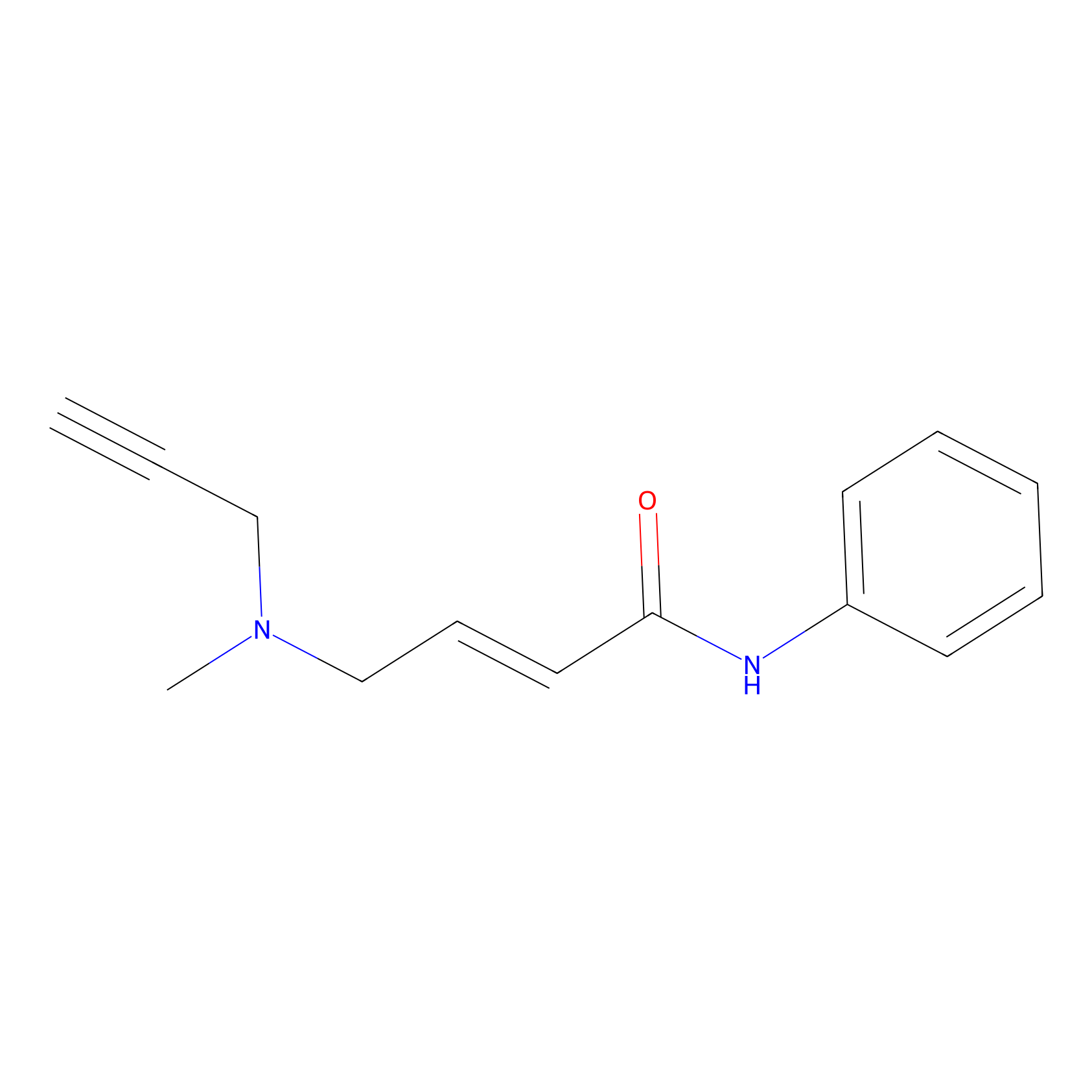 |
1.53 | LDD0452 | [3] | |
|
P2 Probe Info |
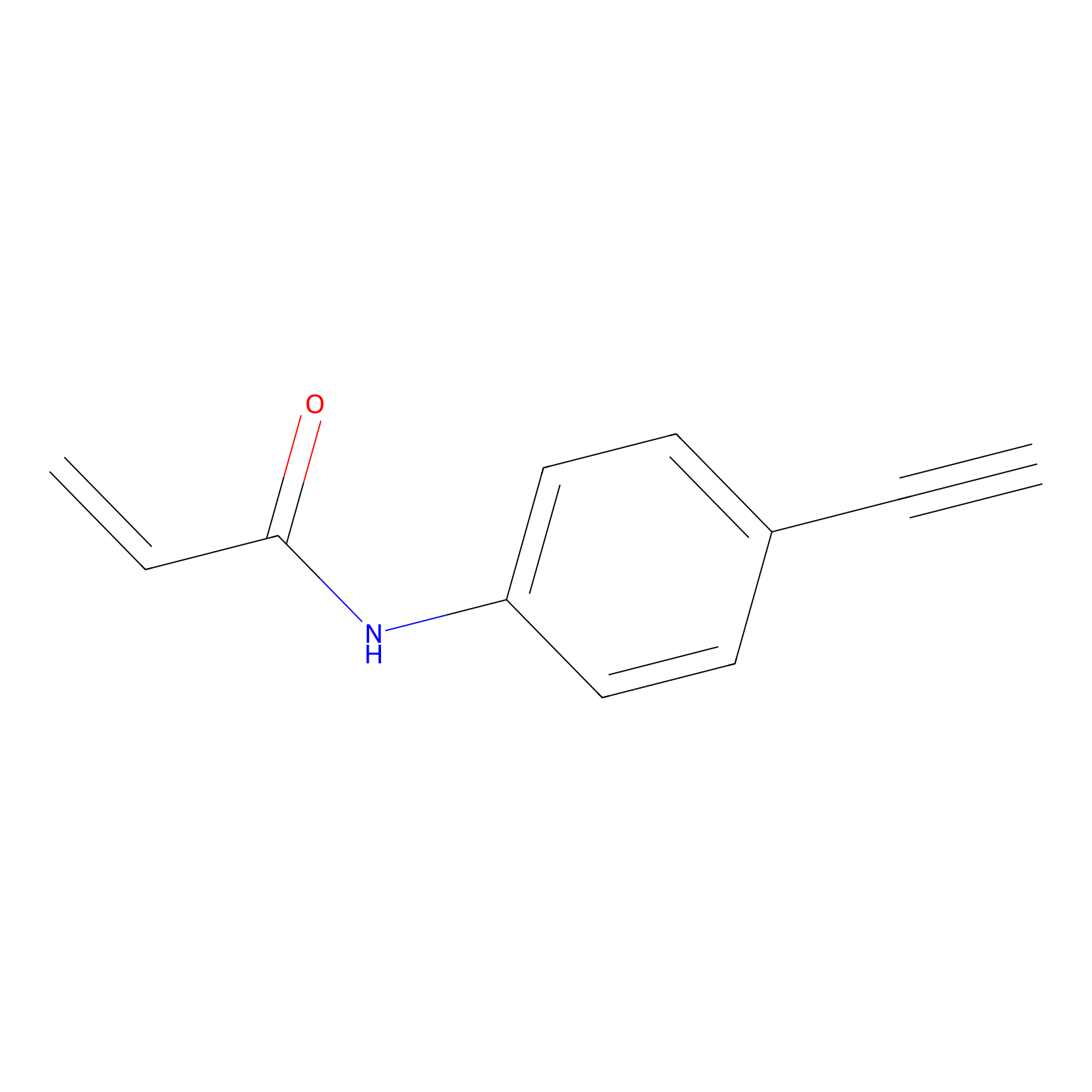 |
2.17 | LDD0449 | [3] | |
|
P3 Probe Info |
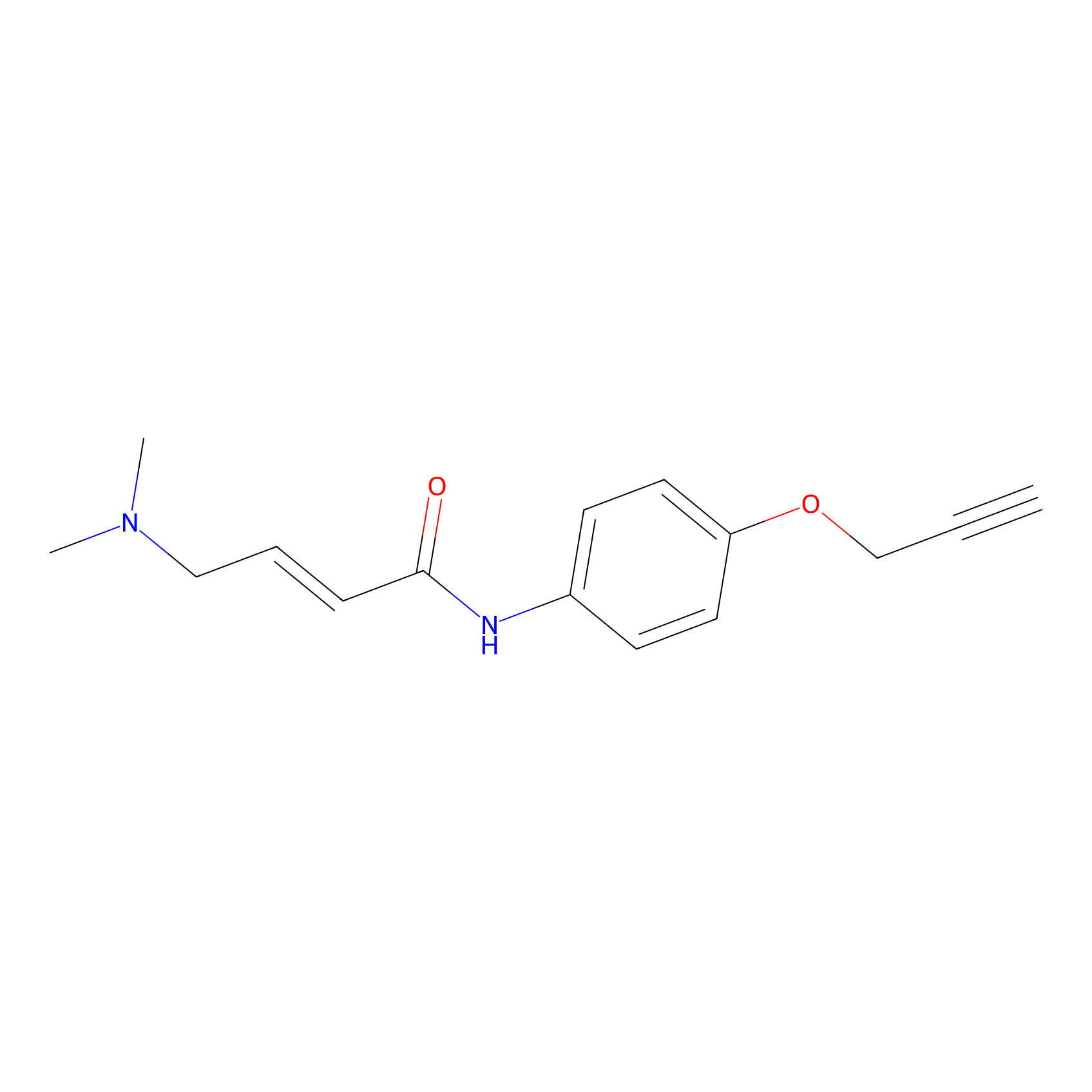 |
1.56 | LDD0450 | [3] | |
|
P8 Probe Info |
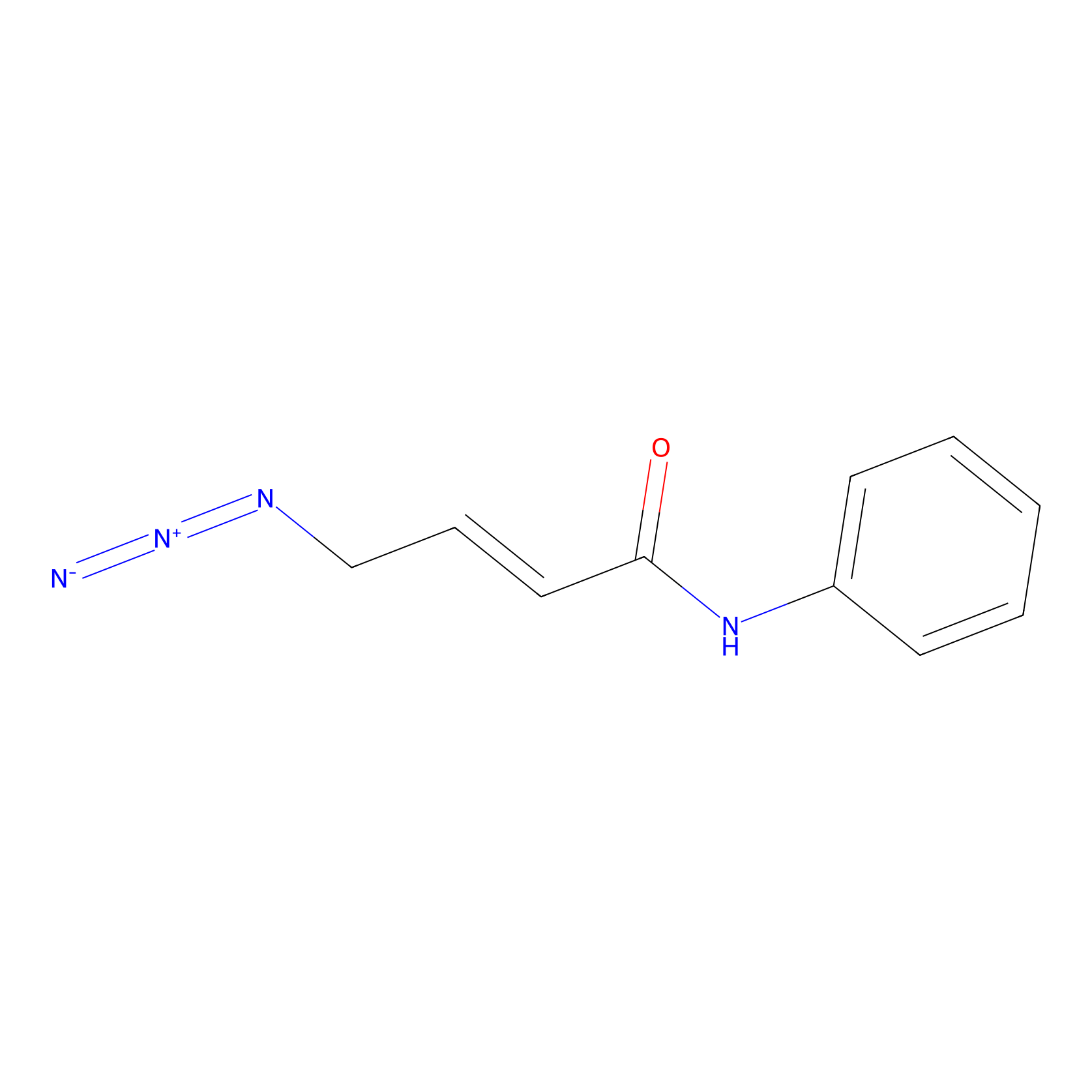 |
1.88 | LDD0451 | [3] | |
|
11RK72 Probe Info |
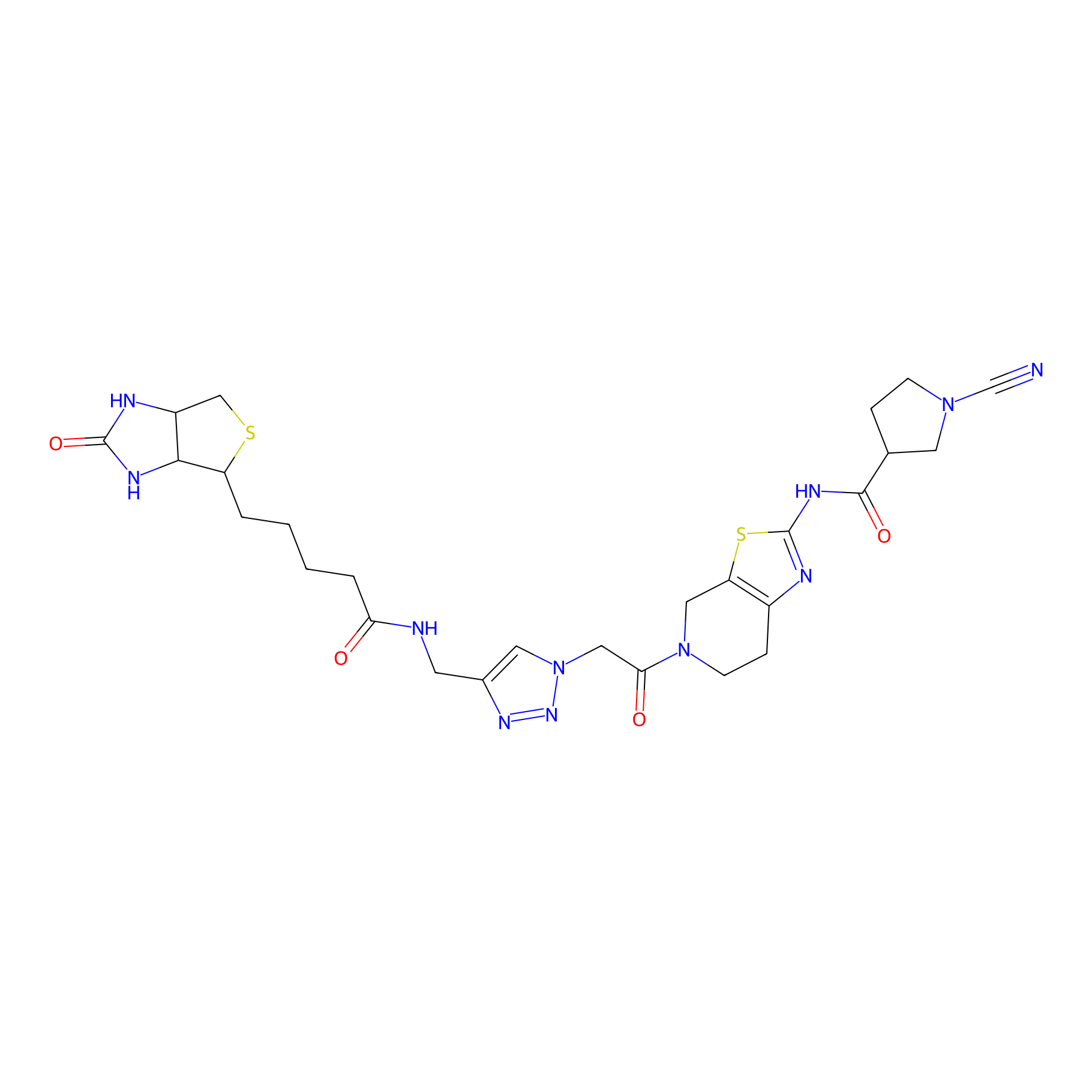 |
5.23 | LDD0327 | [4] | |
|
11RK73 Probe Info |
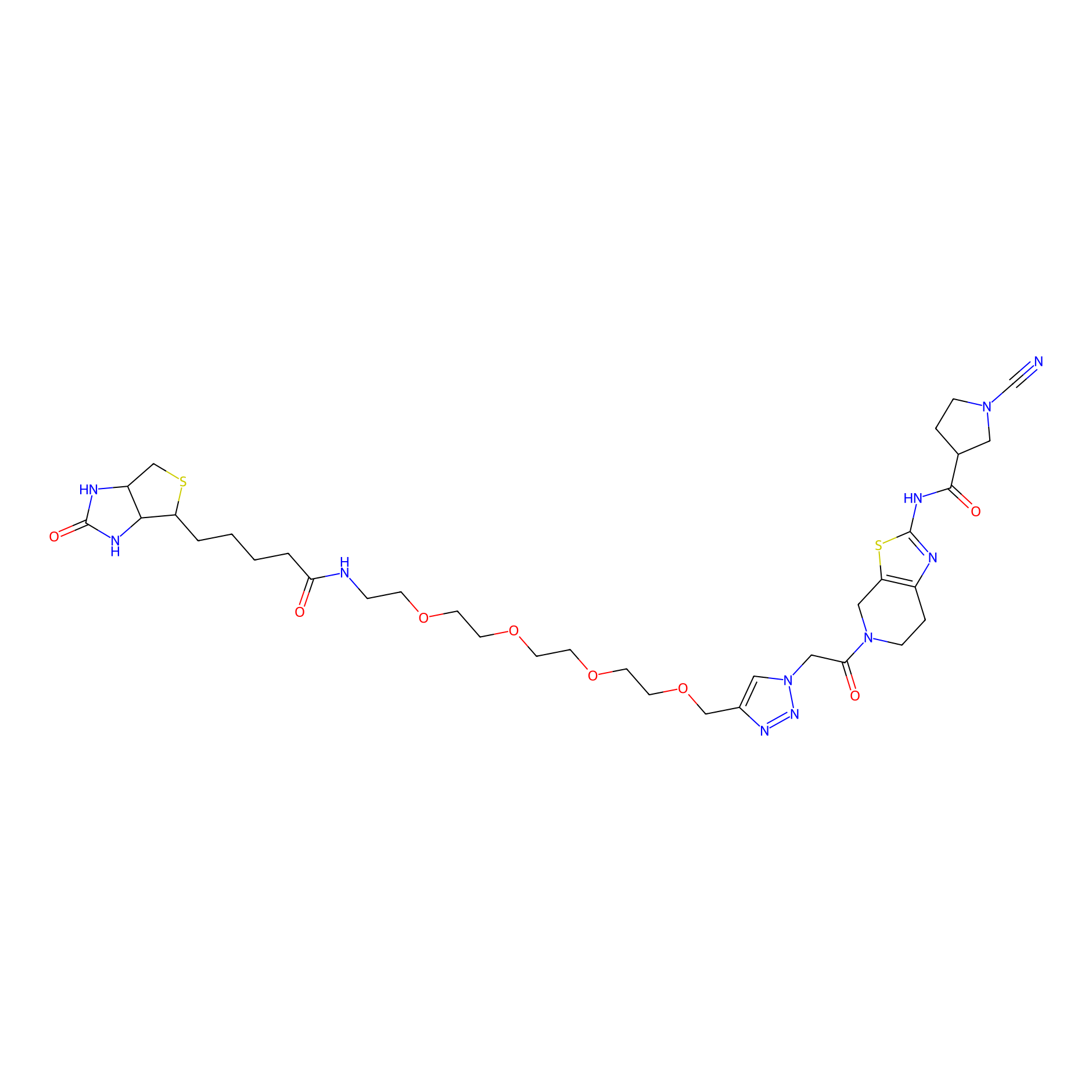 |
4.79 | LDD0328 | [4] | |
|
CHEMBL5175495 Probe Info |
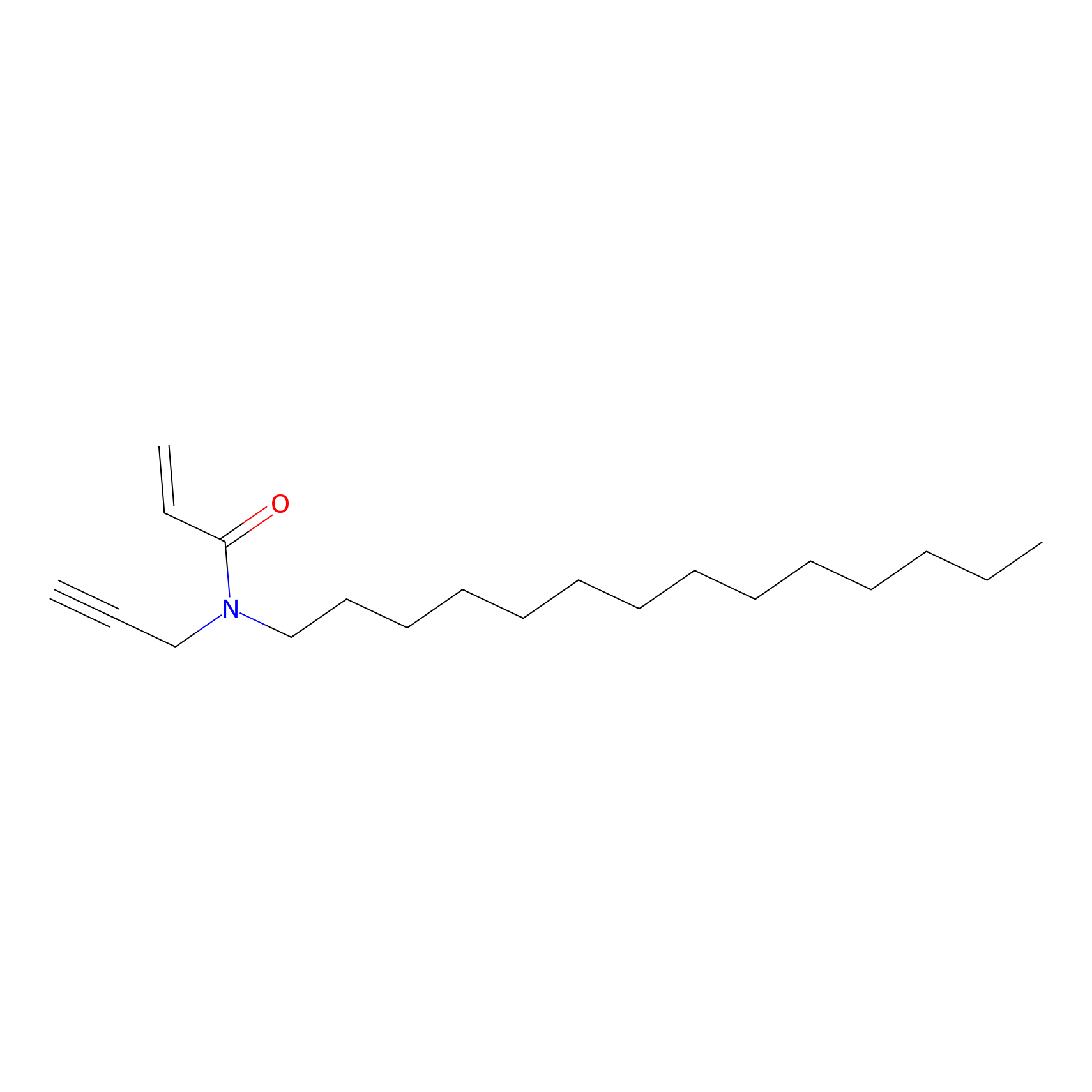 |
11.67 | LDD0196 | [5] | |
|
CY4 Probe Info |
 |
100.00 | LDD0244 | [6] | |
|
N1 Probe Info |
 |
100.00 | LDD0242 | [6] | |
|
W1 Probe Info |
 |
20.19 | LDD0235 | [7] | |
|
C-Sul Probe Info |
 |
2.24 | LDD0066 | [8] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [9] | |
|
ONAyne Probe Info |
 |
K166(0.00); K615(0.00) | LDD0273 | [10] | |
|
IPM Probe Info |
 |
N.A. | LDD0241 | [7] | |
|
OPA-S-S-alkyne Probe Info |
 |
K257(1.64); K255(1.64); K615(3.54); K245(5.72) | LDD3494 | [11] | |
|
Alkylaryl probe 2 Probe Info |
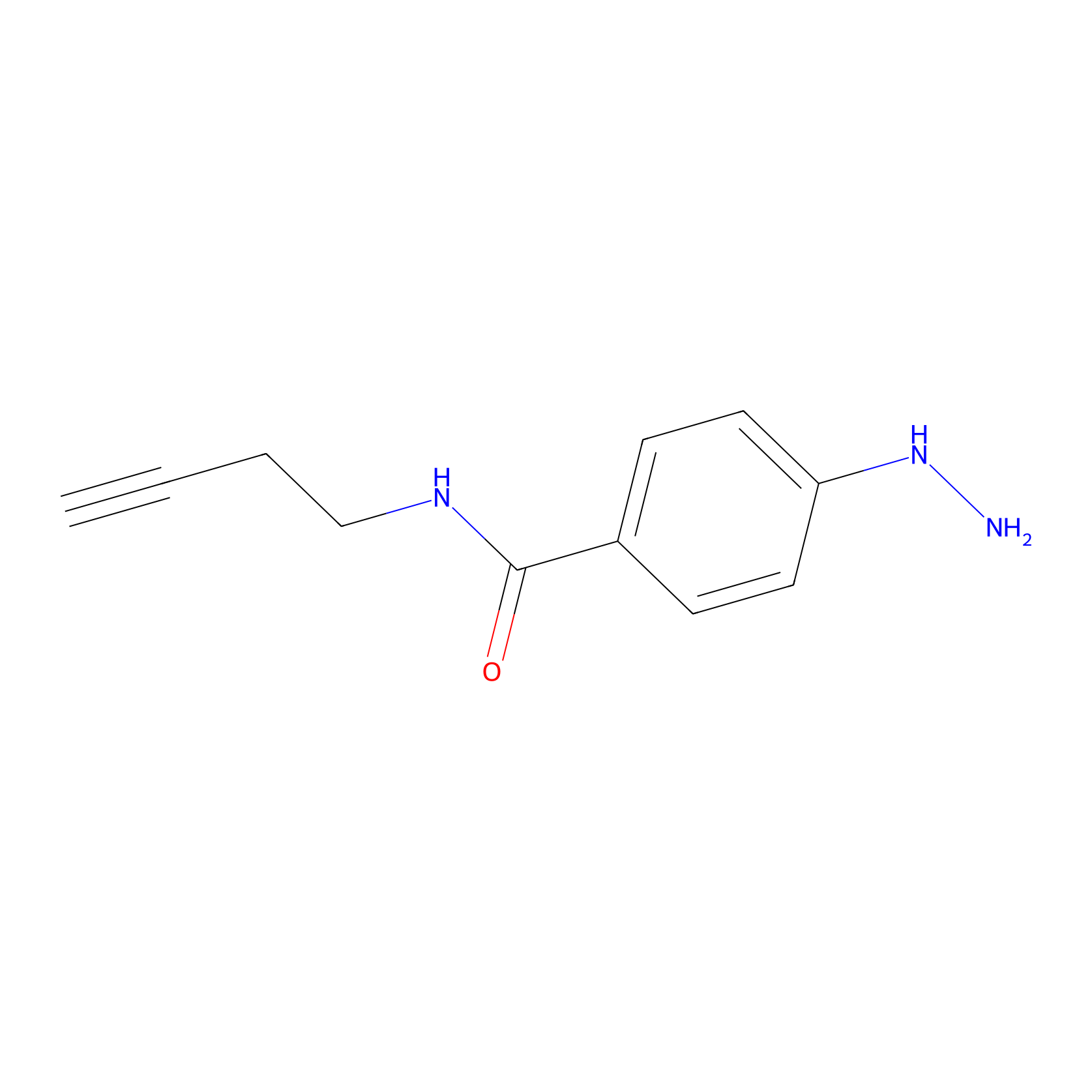 |
3.00 | LDD0390 | [2] | |
|
DBIA Probe Info |
 |
C462(2.57) | LDD3312 | [12] | |
|
AZ-9 Probe Info |
 |
10.00 | LDD2154 | [13] | |
|
5E-2FA Probe Info |
 |
H414(0.00); H236(0.00); H619(0.00); H542(0.00) | LDD2235 | [14] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [15] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [16] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [16] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [16] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [17] | |
|
BTD Probe Info |
 |
N.A. | LDD0004 | [18] | |
|
Compound 10 Probe Info |
 |
N.A. | LDD2216 | [19] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [19] | |
|
NHS Probe Info |
 |
K255(0.00); K166(0.00); K147(0.00); K160(0.00) | LDD0010 | [18] | |
|
STPyne Probe Info |
 |
K166(0.00); K160(0.00) | LDD0009 | [18] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [20] | |
|
1c-yne Probe Info |
 |
K160(0.00); K140(0.00) | LDD0228 | [21] | |
|
1d-yne Probe Info |
 |
K269(0.00); K615(0.00) | LDD0357 | [21] | |
|
Acrolein Probe Info |
 |
H414(0.00); H236(0.00); H556(0.00); H265(0.00) | LDD0217 | [22] | |
|
Crotonaldehyde Probe Info |
 |
H556(0.00); H236(0.00); H619(0.00); H265(0.00) | LDD0219 | [22] | |
|
Methacrolein Probe Info |
 |
H236(0.00); H556(0.00); H220(0.00); H619(0.00) | LDD0218 | [22] | |
|
AOyne Probe Info |
 |
5.10 | LDD0443 | [23] | |
|
NAIA_5 Probe Info |
 |
C210(0.00); C431(0.00) | LDD2223 | [24] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C055 Probe Info |
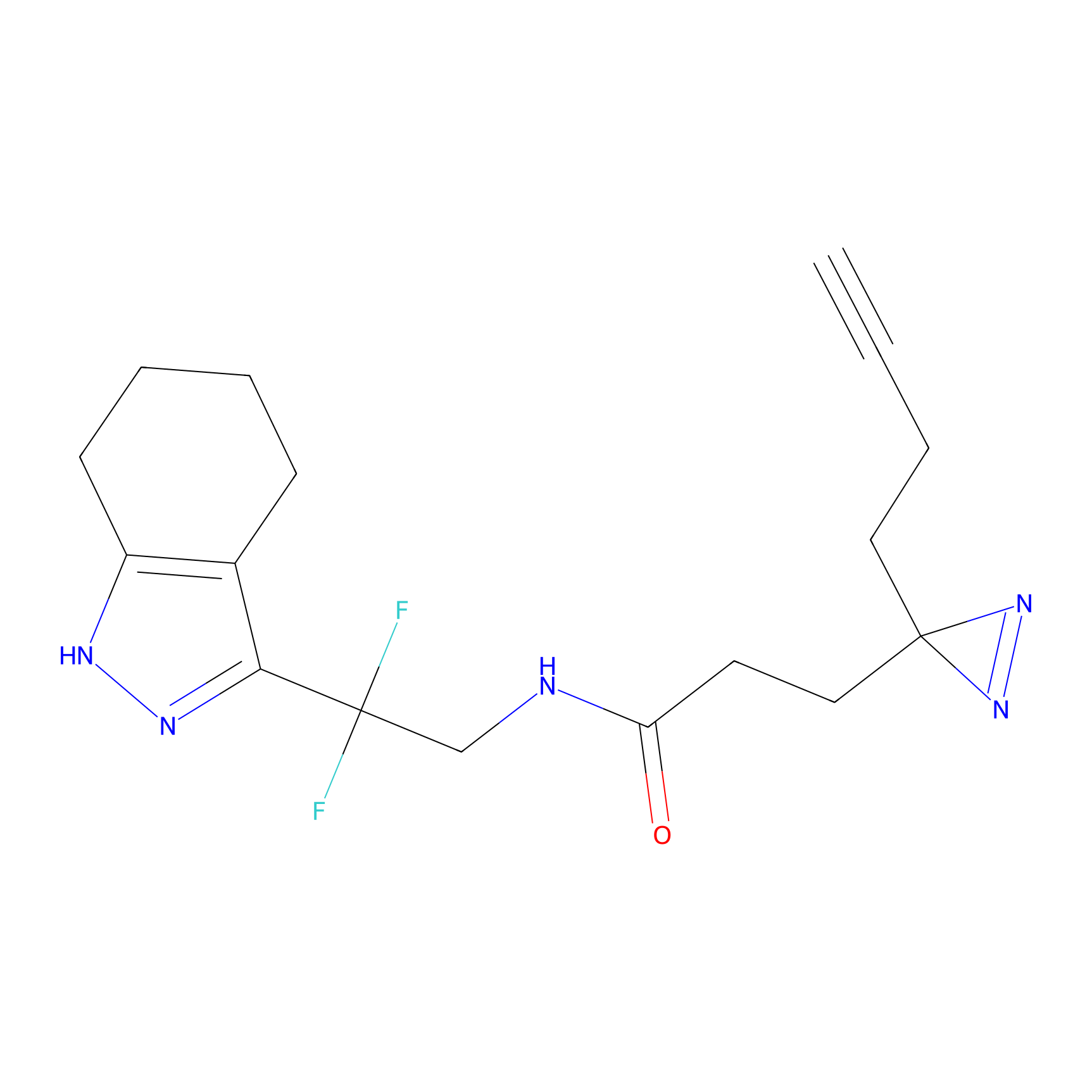 |
19.29 | LDD1752 | [25] | |
|
C158 Probe Info |
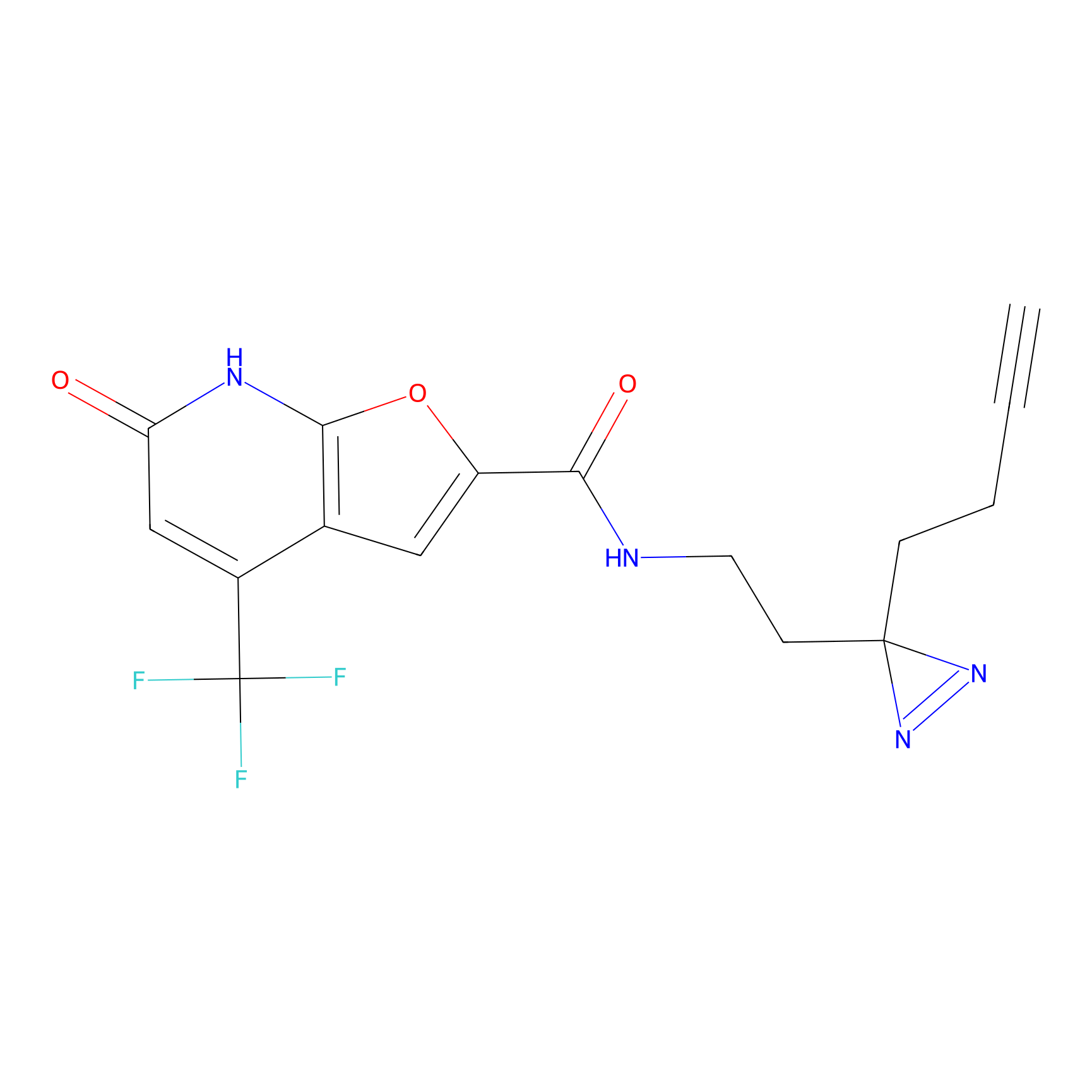 |
9.25 | LDD1838 | [25] | |
|
C160 Probe Info |
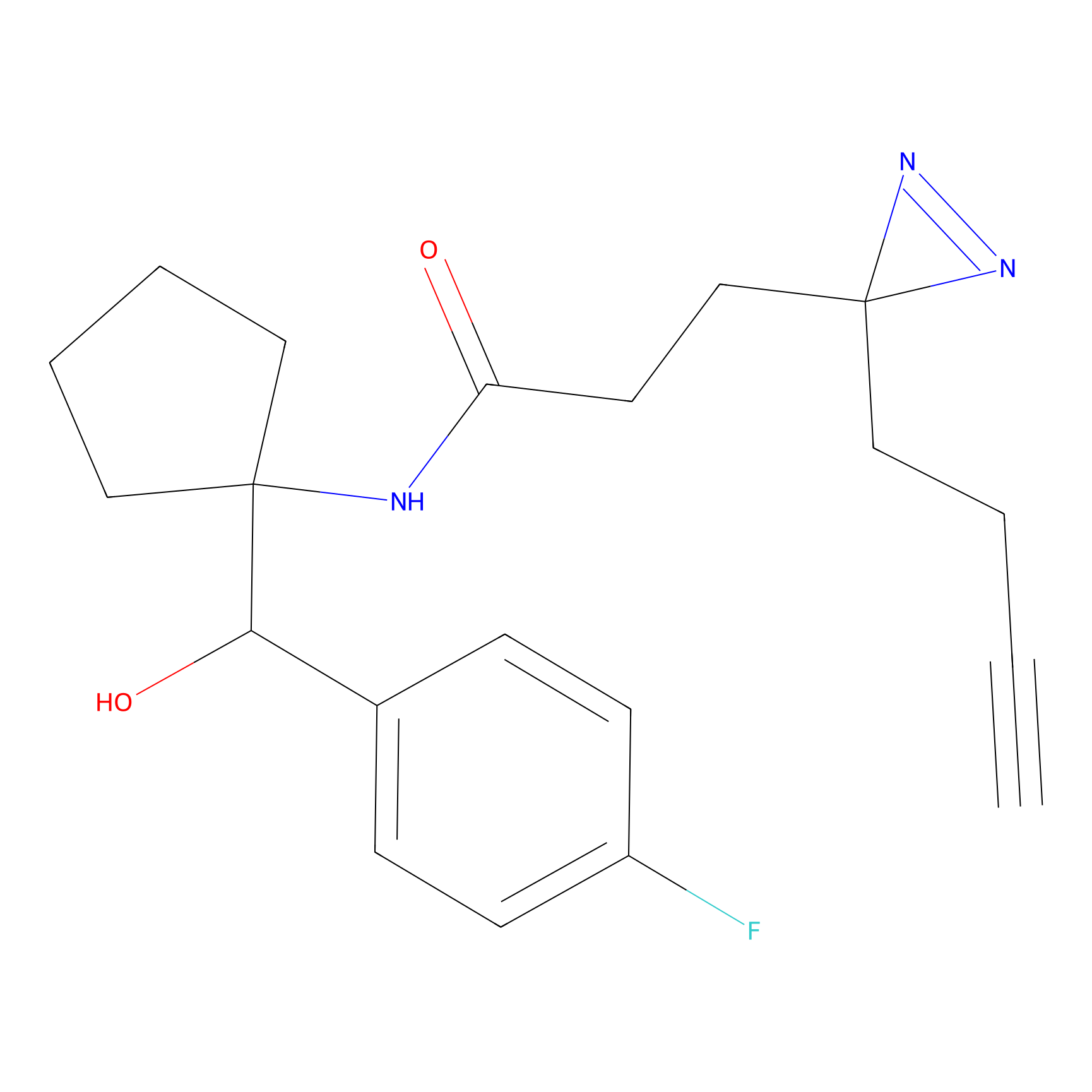 |
20.97 | LDD1840 | [25] | |
|
C170 Probe Info |
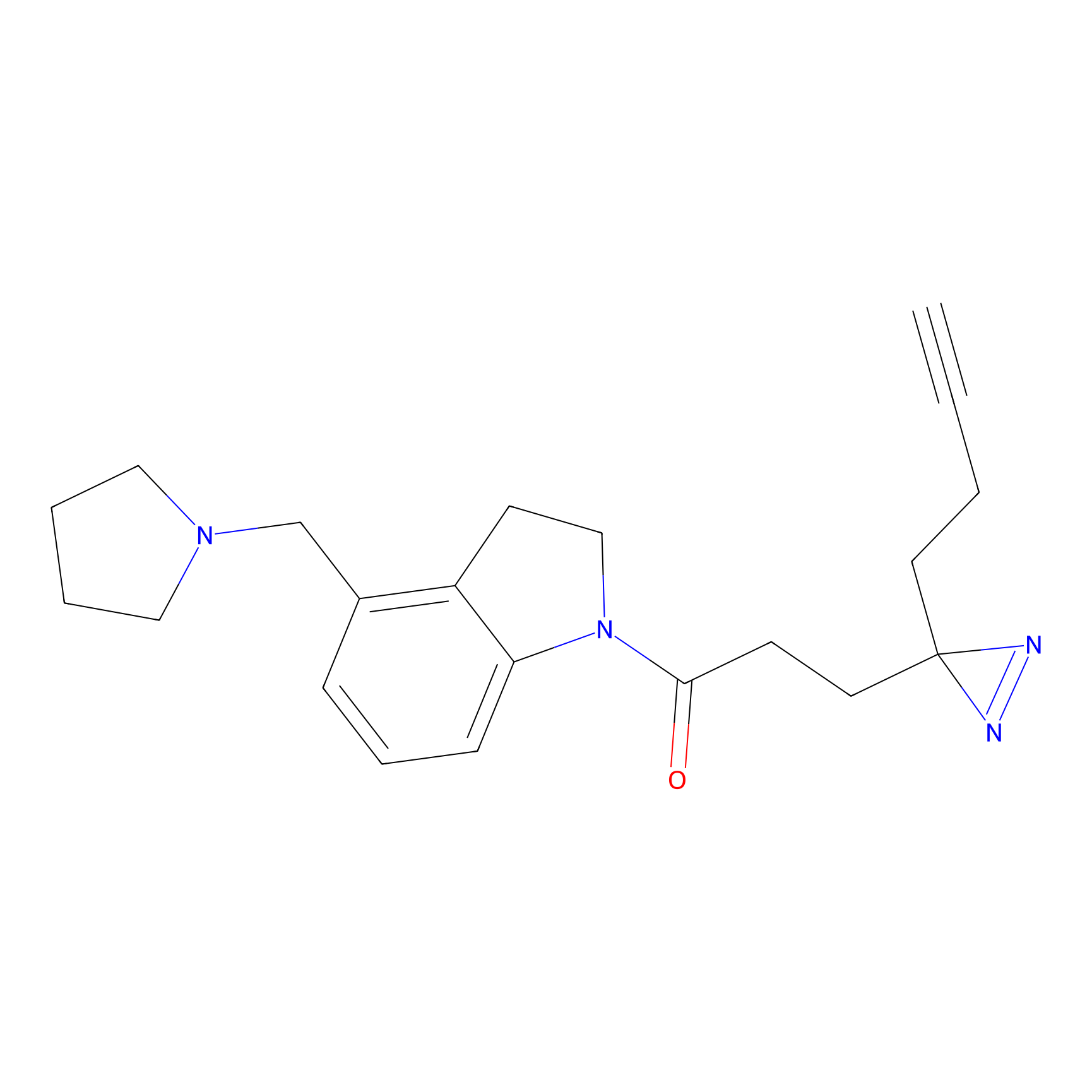 |
7.89 | LDD1850 | [25] | |
|
C193 Probe Info |
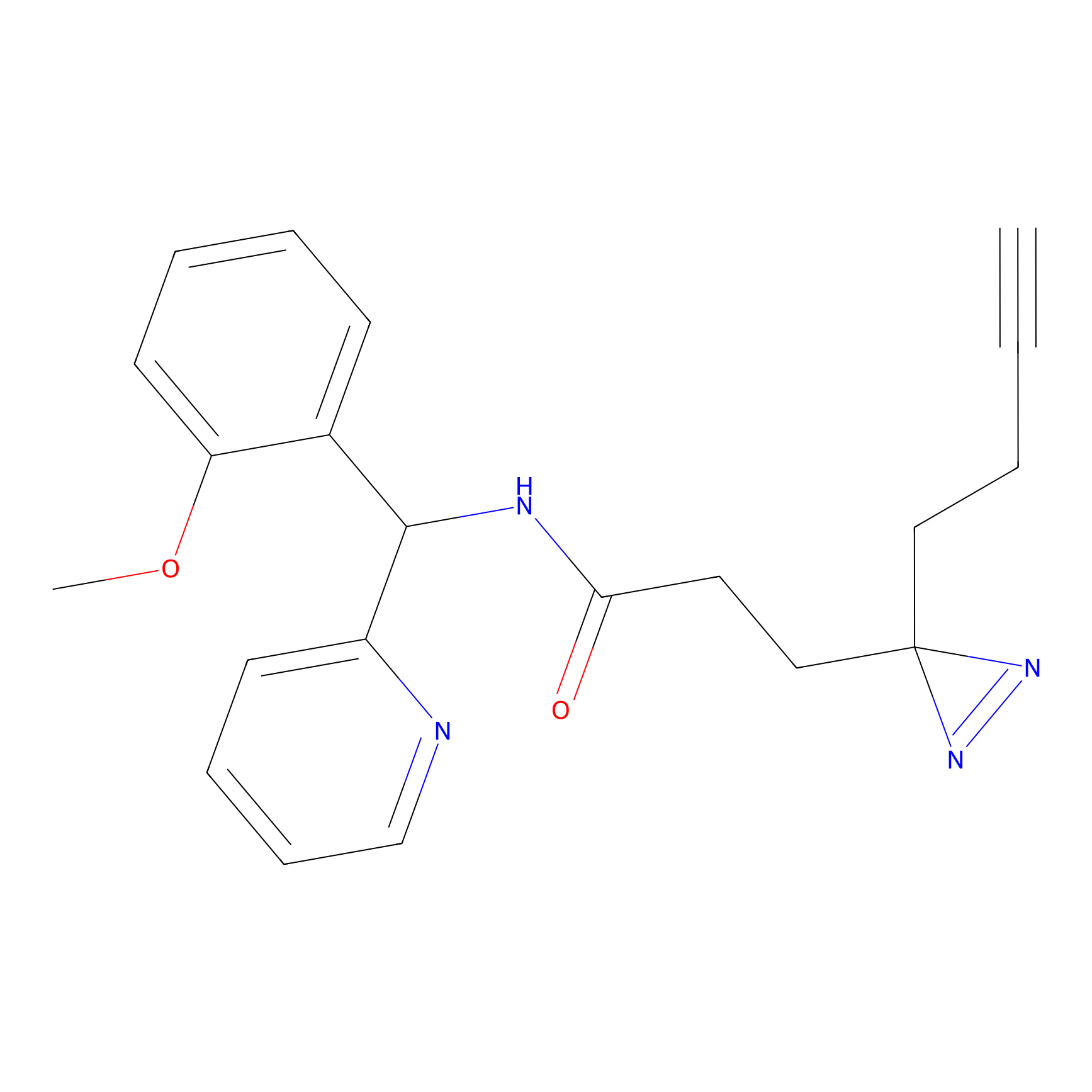 |
7.06 | LDD1869 | [25] | |
|
C194 Probe Info |
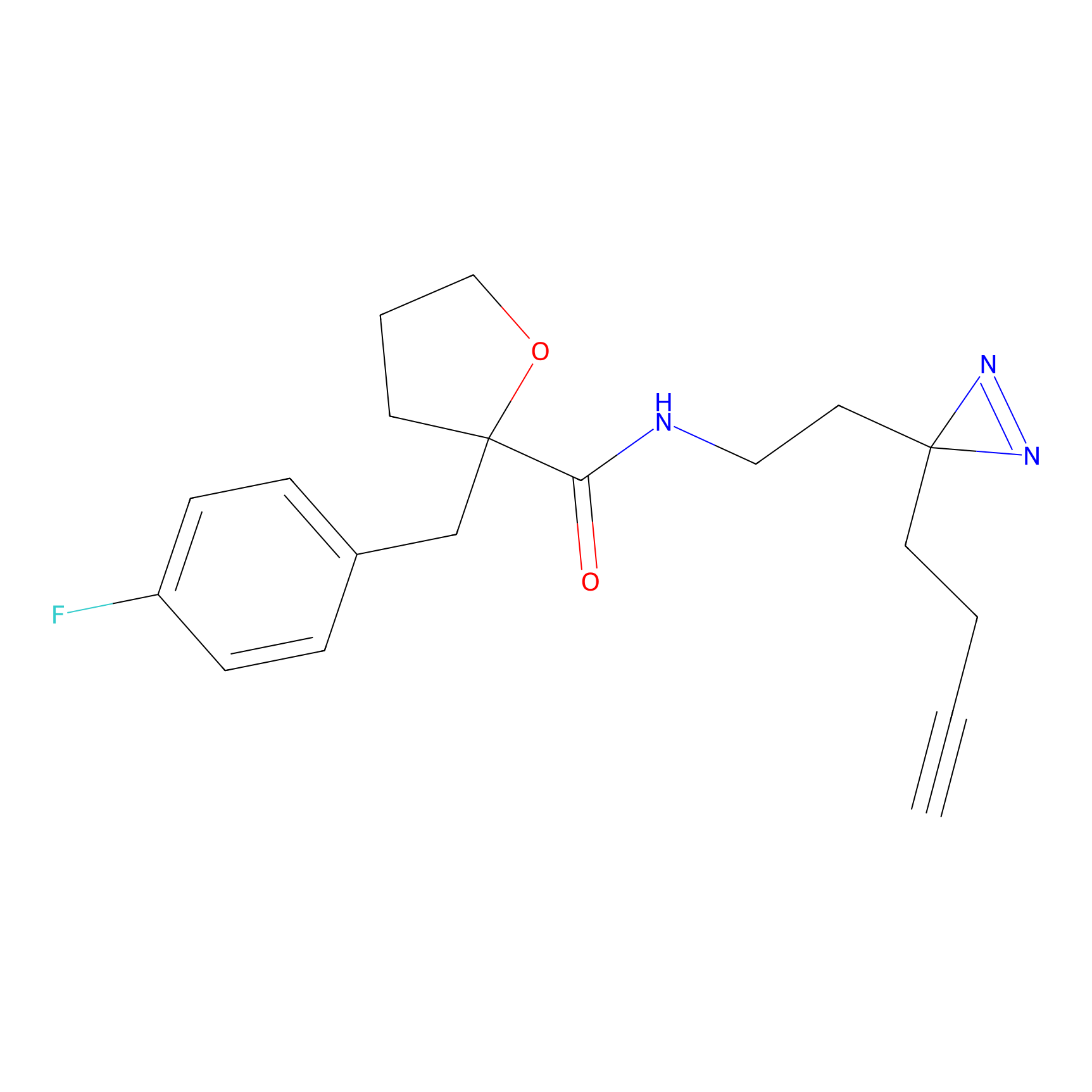 |
9.71 | LDD1870 | [25] | |
|
C198 Probe Info |
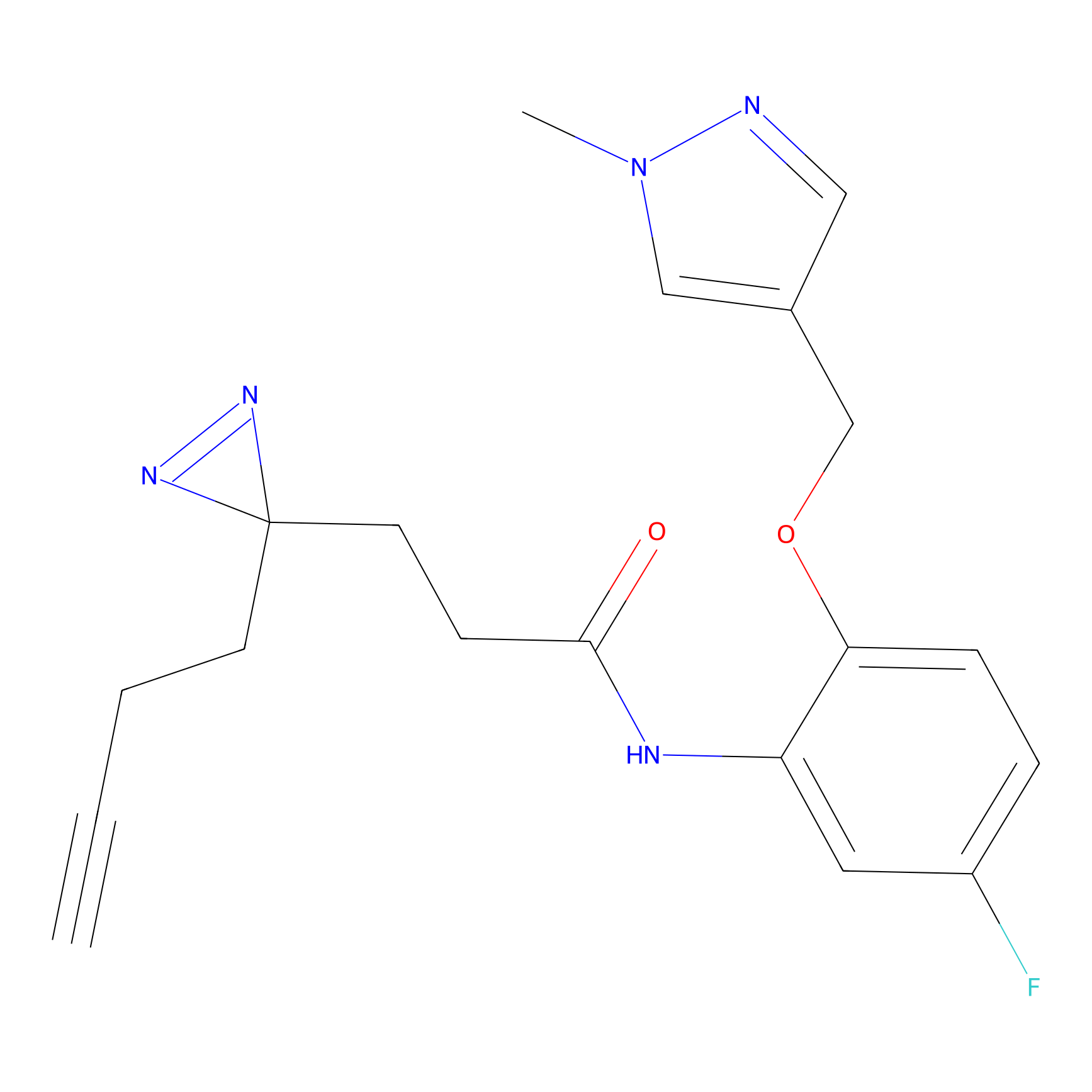 |
10.78 | LDD1874 | [25] | |
|
C275 Probe Info |
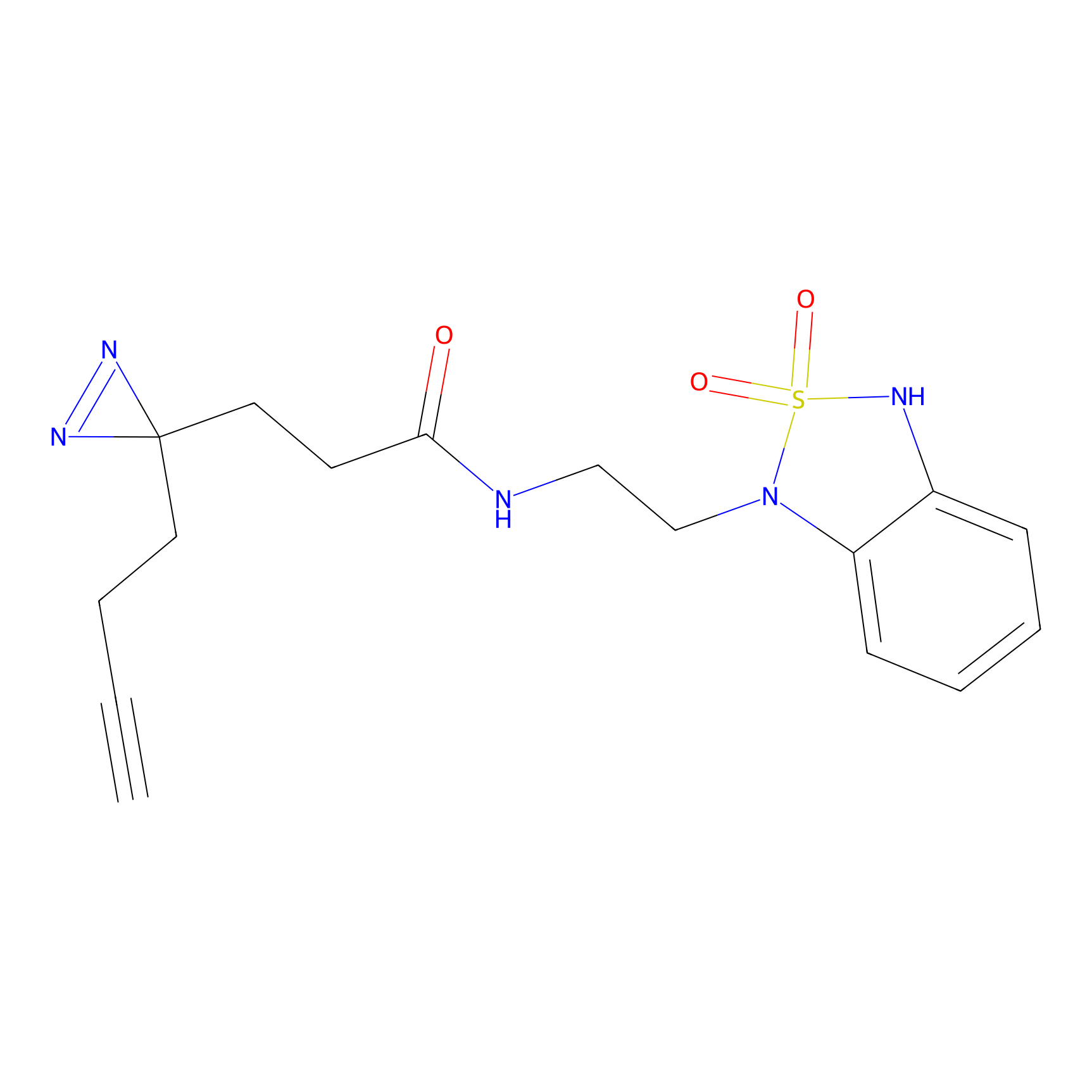 |
6.36 | LDD1945 | [25] | |
|
C284 Probe Info |
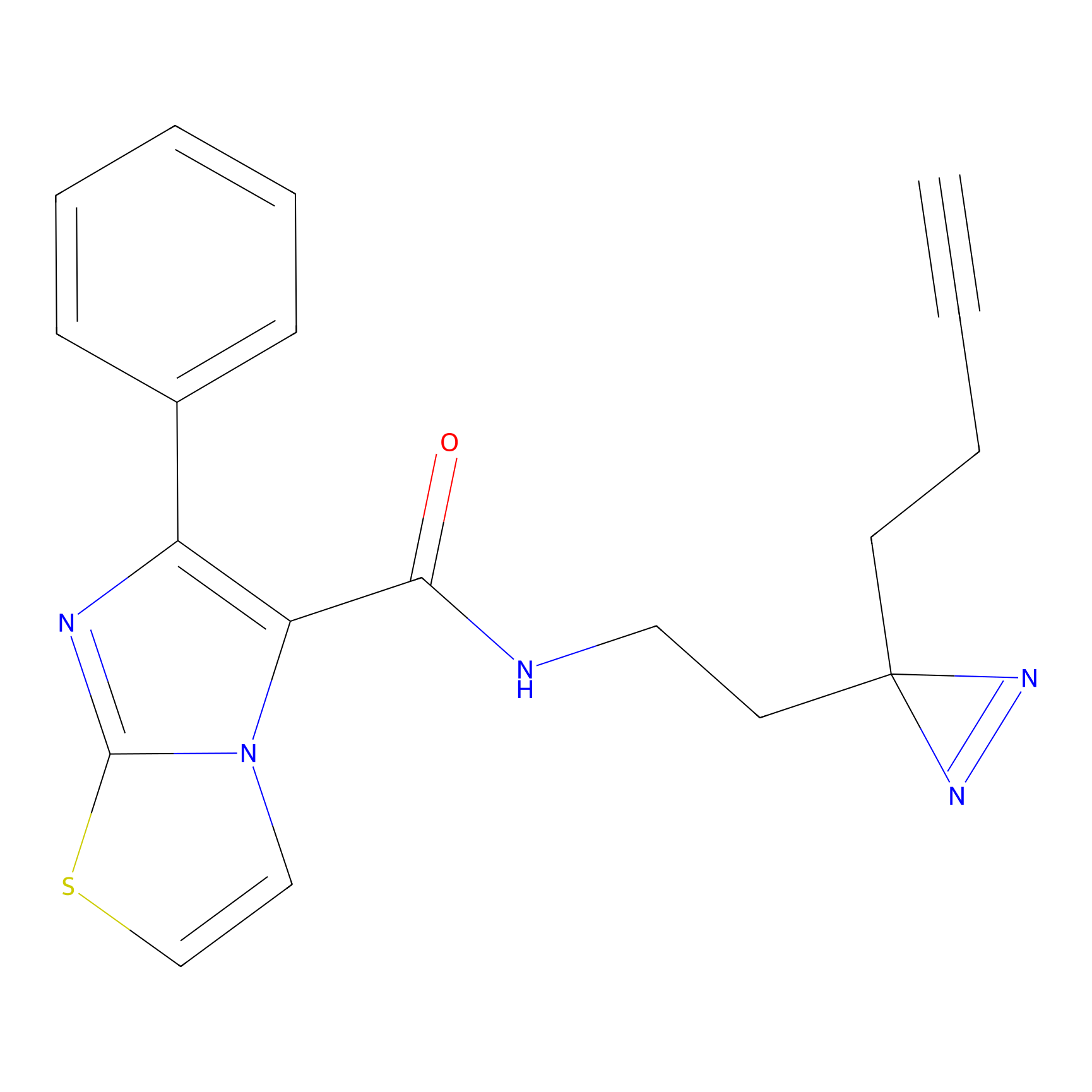 |
38.59 | LDD1954 | [25] | |
|
C310 Probe Info |
 |
8.75 | LDD1977 | [25] | |
|
FFF probe11 Probe Info |
 |
5.95 | LDD0471 | [26] | |
|
FFF probe12 Probe Info |
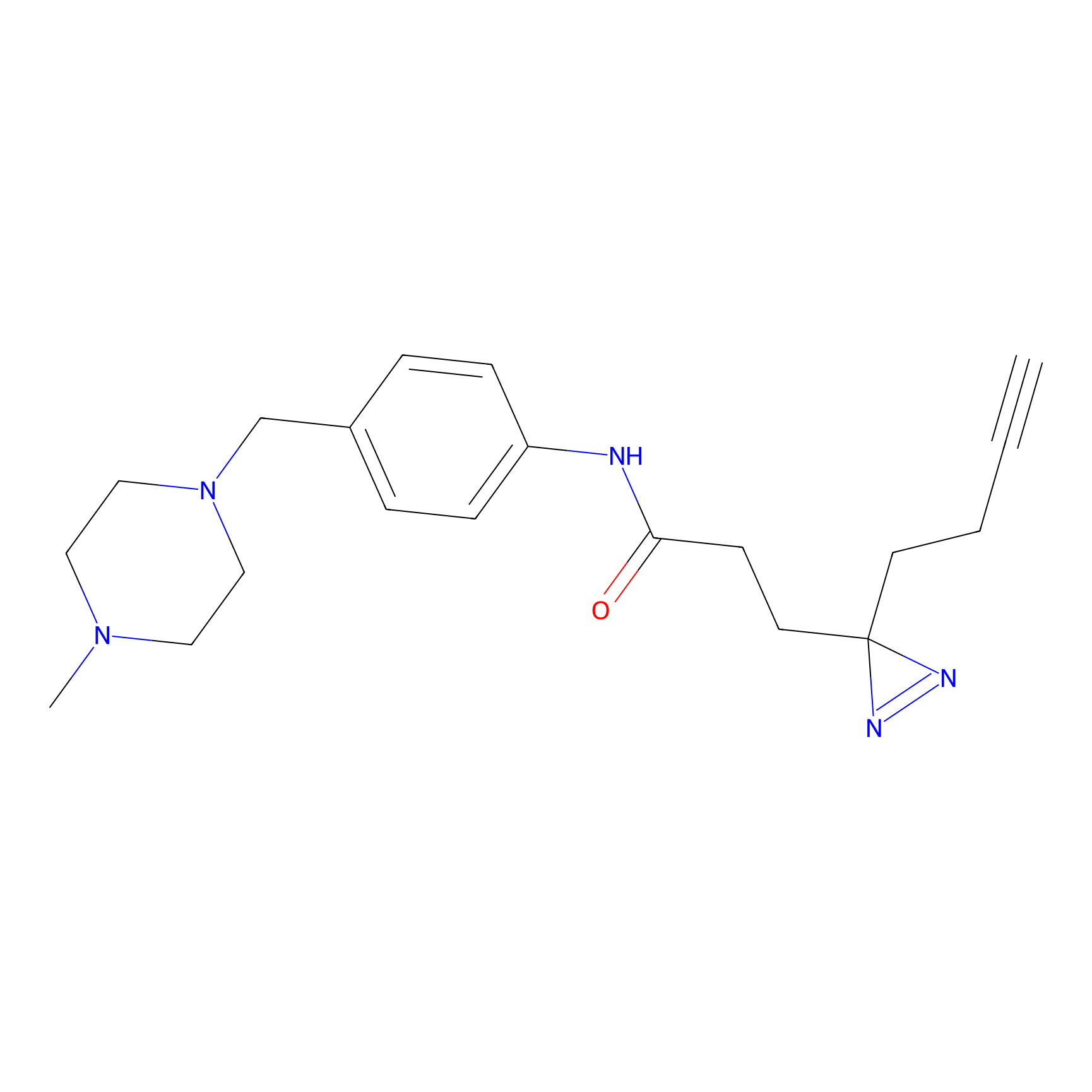 |
11.16 | LDD0473 | [26] | |
|
FFF probe3 Probe Info |
 |
5.23 | LDD0464 | [26] | |
|
FFF probe9 Probe Info |
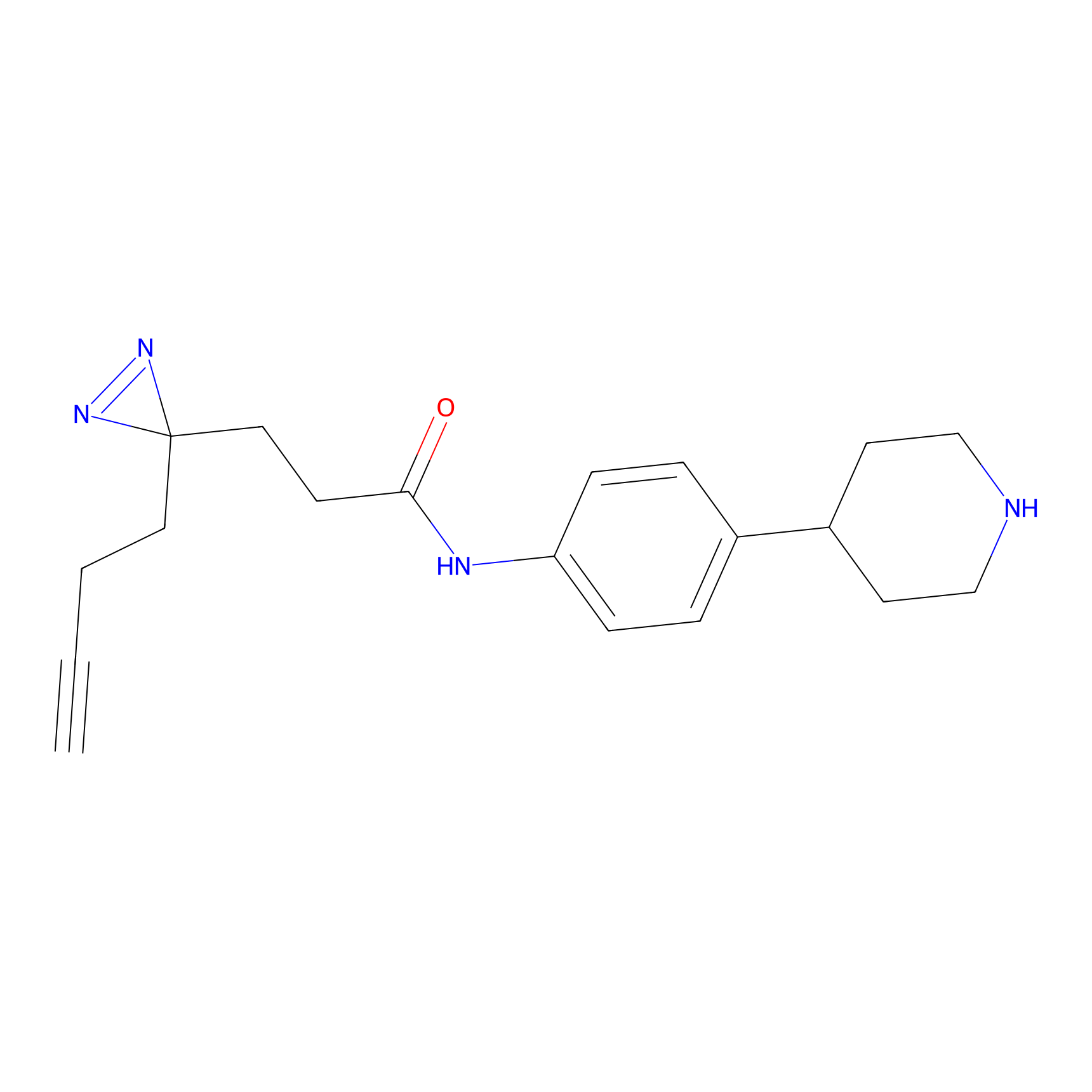 |
5.30 | LDD0470 | [26] | |
|
STS-2 Probe Info |
 |
3.77 | LDD0139 | [27] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C431(0.53) | LDD2117 | [28] |
| LDCM0151 | AZ-11 | HeLa | 10.00 | LDD2154 | [13] |
| LDCM0108 | Chloroacetamide | HeLa | H220(0.00); H556(0.00); H236(0.00); H265(0.00) | LDD0222 | [22] |
| LDCM0625 | F8 | Ramos | C431(0.65); C210(1.66) | LDD2187 | [29] |
| LDCM0572 | Fragment10 | Ramos | C431(0.83) | LDD2189 | [29] |
| LDCM0573 | Fragment11 | Ramos | C431(0.22); C210(13.80) | LDD2190 | [29] |
| LDCM0574 | Fragment12 | Ramos | C431(0.62) | LDD2191 | [29] |
| LDCM0575 | Fragment13 | Ramos | C431(1.19); C210(1.88) | LDD2192 | [29] |
| LDCM0576 | Fragment14 | Ramos | C431(1.18); C210(2.72) | LDD2193 | [29] |
| LDCM0579 | Fragment20 | Ramos | C431(0.68) | LDD2194 | [29] |
| LDCM0580 | Fragment21 | Ramos | C431(0.81); C210(0.67) | LDD2195 | [29] |
| LDCM0582 | Fragment23 | Ramos | C431(1.24); C210(1.52) | LDD2196 | [29] |
| LDCM0578 | Fragment27 | Ramos | C431(1.02); C210(1.01) | LDD2197 | [29] |
| LDCM0586 | Fragment28 | Ramos | C431(1.20); C210(0.59) | LDD2198 | [29] |
| LDCM0588 | Fragment30 | Ramos | C431(0.76); C210(1.65) | LDD2199 | [29] |
| LDCM0589 | Fragment31 | Ramos | C431(0.93); C210(1.03) | LDD2200 | [29] |
| LDCM0590 | Fragment32 | Ramos | C431(0.56) | LDD2201 | [29] |
| LDCM0468 | Fragment33 | Ramos | C431(0.65); C210(1.48) | LDD2202 | [29] |
| LDCM0596 | Fragment38 | Ramos | C431(0.99); C210(1.77) | LDD2203 | [29] |
| LDCM0566 | Fragment4 | Ramos | C431(1.07) | LDD2184 | [29] |
| LDCM0610 | Fragment52 | Ramos | C431(0.64) | LDD2204 | [29] |
| LDCM0614 | Fragment56 | Ramos | C431(0.77); C210(1.45) | LDD2205 | [29] |
| LDCM0569 | Fragment7 | Ramos | C431(0.87) | LDD2186 | [29] |
| LDCM0571 | Fragment9 | Ramos | C431(0.57) | LDD2188 | [29] |
| LDCM0107 | IAA | HeLa | H414(0.00); H236(0.00); H619(0.00); H265(0.00) | LDD0221 | [22] |
| LDCM0022 | KB02 | HEK-293T | C431(0.90) | LDD1492 | [30] |
| LDCM0023 | KB03 | HEK-293T | C431(0.94) | LDD1497 | [30] |
| LDCM0024 | KB05 | HMCB | C462(2.57) | LDD3312 | [12] |
| LDCM0109 | NEM | HeLa | H236(0.00); H414(0.00); H619(0.00); H556(0.00) | LDD0223 | [22] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C431(0.76) | LDD2109 | [28] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C431(0.78) | LDD2123 | [28] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C431(0.98) | LDD2125 | [28] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C431(0.70) | LDD2127 | [28] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C431(0.90) | LDD2136 | [28] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C431(1.00) | LDD2137 | [28] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C431(0.97) | LDD2146 | [28] |
| LDCM0099 | Phenelzine | HEK-293T | 3.00 | LDD0390 | [2] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| S-adenosylmethionine synthase isoform type-2 (MAT2A) | AdoMet synthase family | P31153 | |||
| 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase (EBP) | EBP family | Q15125 | |||
Transporter and channel
GPCR
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Prosaposin receptor GPR37 (GPR37) | G-protein coupled receptor 1 family | O15354 | |||
Other
References
