Details of the Target
General Information of Target
| Target ID | LDTP13320 | |||||
|---|---|---|---|---|---|---|
| Target Name | Prenylated Rab acceptor protein 1 (RABAC1) | |||||
| Gene Name | RABAC1 | |||||
| Gene ID | 10567 | |||||
| Synonyms |
PRA1; PRAF1; Prenylated Rab acceptor protein 1; PRA1 family protein 1 |
|||||
| 3D Structure | ||||||
| Sequence |
MTKMDIRGAVDAAVPTNIIAAKAAEVRANKVNWQSYLQGQMISAEDCEFIQRFEMKRSPE
EKQEMLQTEGSQCAKTFINLMTHICKEQTVQYILTMVDDMLQENHQRVSIFFDYARCSKN TAWPYFLPMLNRQDPFTVHMAARIIAKLAAWGKELMEGSDLNYYFNWIKTQLSSQKLRGS GVAVETGTVSSSDSSQYVQCVAGCLQLMLRVNEYRFAWVEADGVNCIMGVLSNKCGFQLQ YQMIFSIWLLAFSPQMCEHLRRYNIIPVLSDILQESVKEKVTRIILAAFRNFLEKSTERE TRQEYALAMIQCKVLKQLENLEQQKYDDEDISEDIKFLLEKLGESVQDLSSFDEYSSELK SGRLEWSPVHKSEKFWRENAVRLNEKNYELLKILTKLLEVSDDPQVLAVAAHDVGEYVRH YPRGKRVIEQLGGKQLVMNHMHHEDQQVRYNALLAVQKLMVHNWEYLGKQLQSEQPQTAA ARS |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
PRA1 family
|
|||||
| Subcellular location |
Cell membrane
|
|||||
| Function |
General Rab protein regulator required for vesicle formation from the Golgi complex. May control vesicle docking and fusion by mediating the action of Rab GTPases to the SNARE complexes. In addition it inhibits the removal of Rab GTPases from the membrane by GDI.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
CHEMBL5175495 Probe Info |
 |
6.91 | LDD0196 | [1] | |
|
IA-alkyne Probe Info |
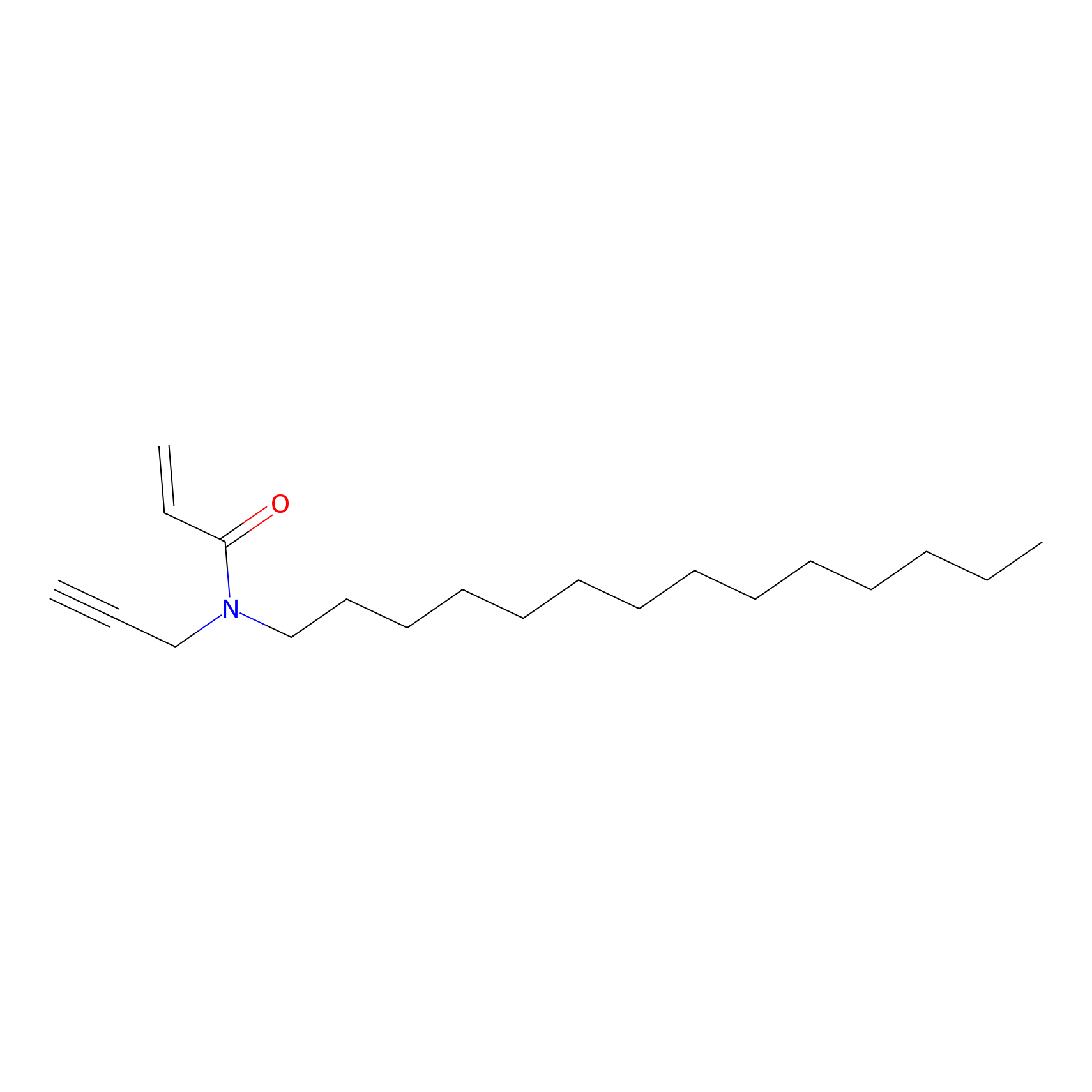
 |
C64(10.00) | LDD2157 | [2] | |
|
Curcusone 37 Probe Info |
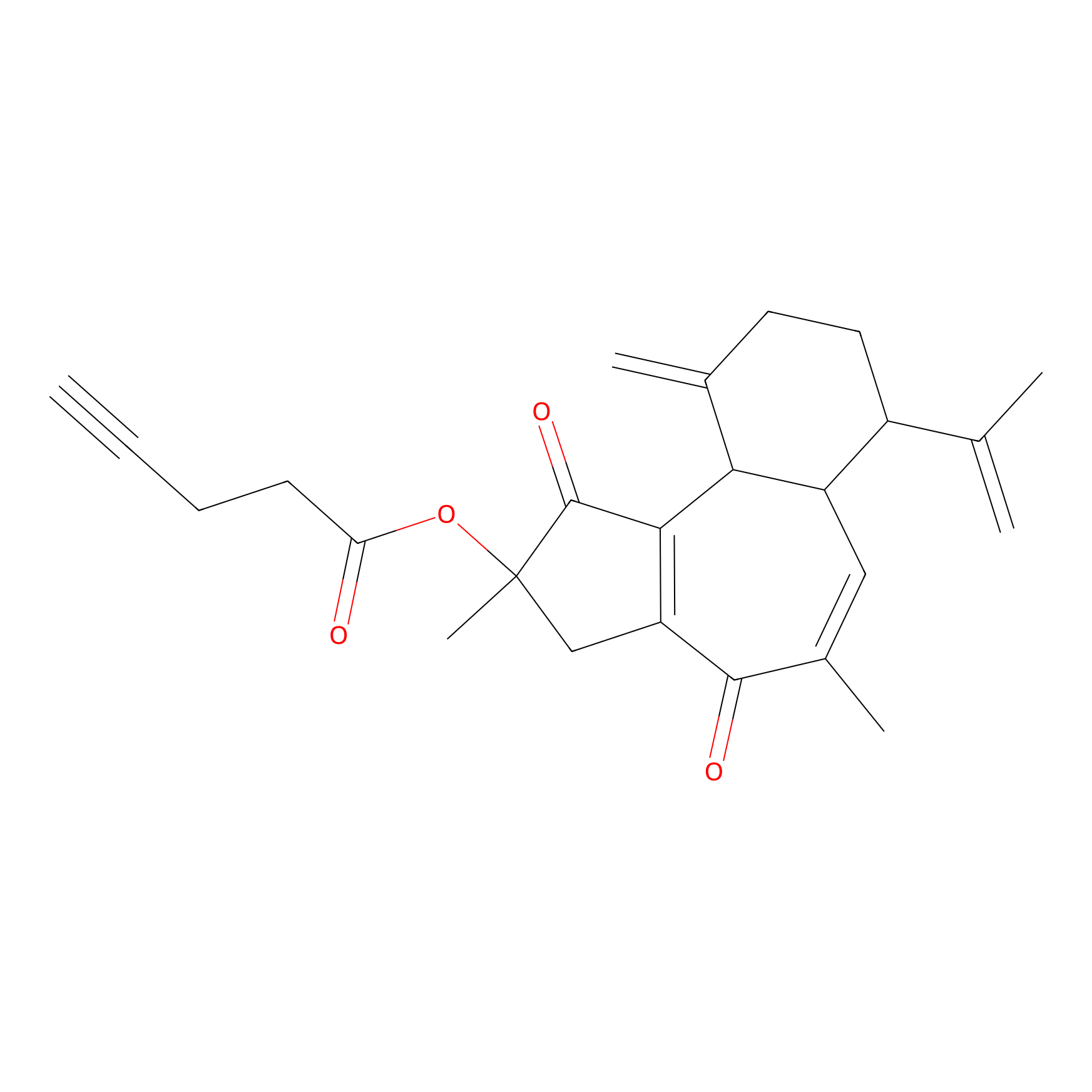 |
4.50 | LDD0188 | [3] | |
|
DBIA Probe Info |
 |
C64(0.97) | LDD0532 | [4] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [5] | |
|
IPM Probe Info |
 |
N.A. | LDD0025 | [6] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [6] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2224 | [7] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [6] | |
|
W1 Probe Info |
 |
N.A. | LDD0236 | [8] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C094 Probe Info |
 |
26.35 | LDD1785 | [9] | |
|
C106 Probe Info |
 |
16.11 | LDD1793 | [9] | |
|
C134 Probe Info |
 |
32.00 | LDD1816 | [9] | |
|
C135 Probe Info |
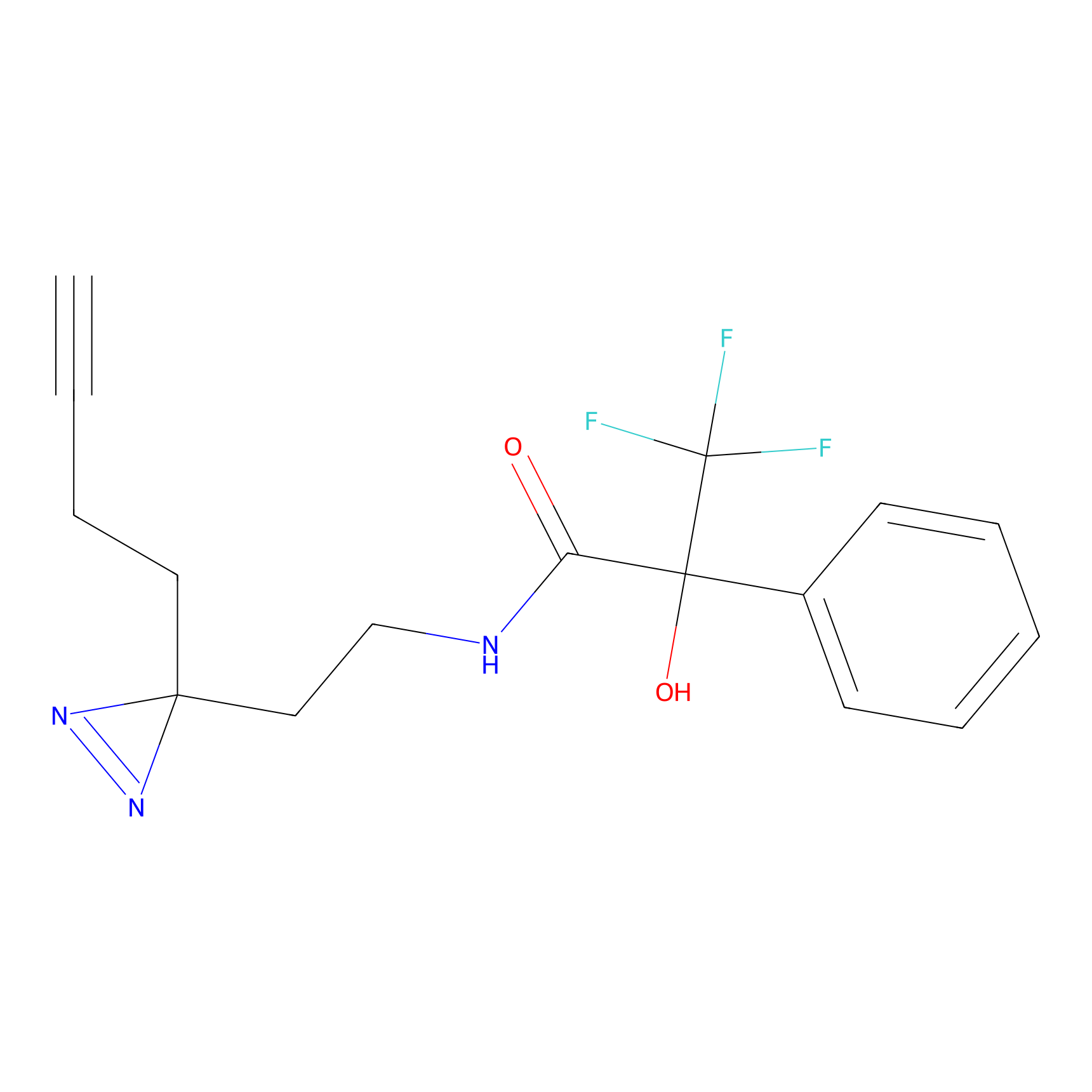 |
10.41 | LDD1817 | [9] | |
|
C177 Probe Info |
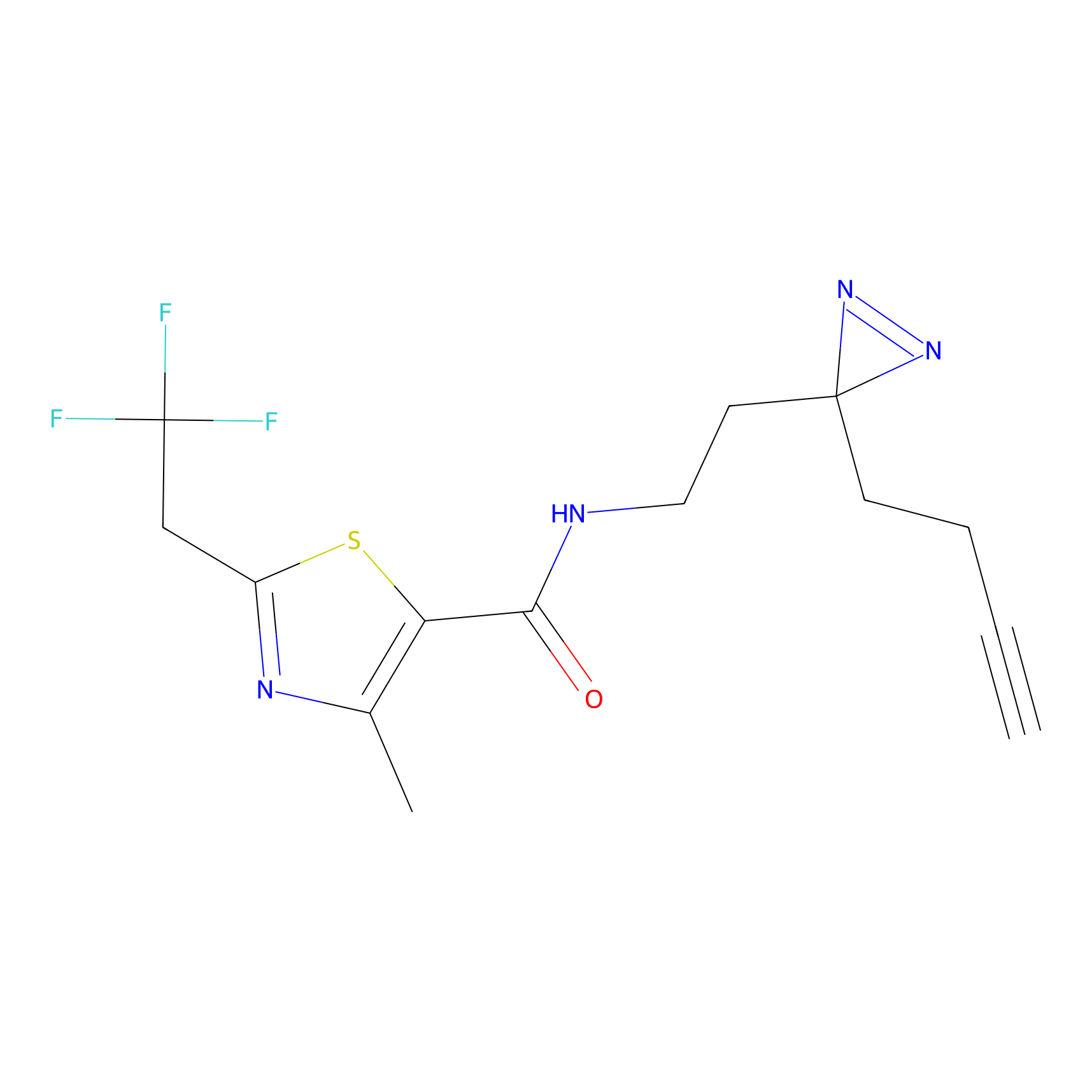 |
8.75 | LDD1856 | [9] | |
|
C178 Probe Info |
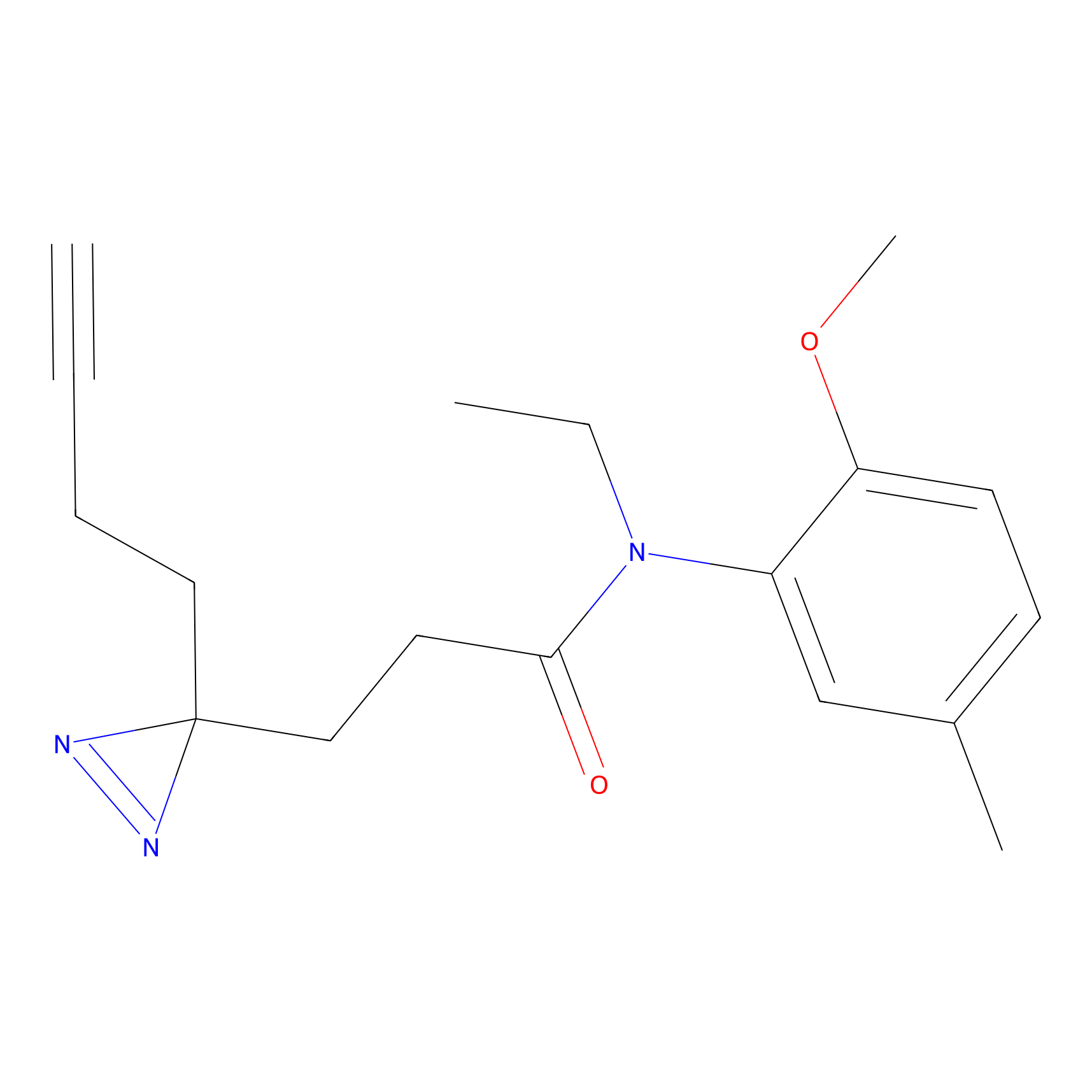 |
18.38 | LDD1857 | [9] | |
|
C246 Probe Info |
 |
12.91 | LDD1919 | [9] | |
|
C278 Probe Info |
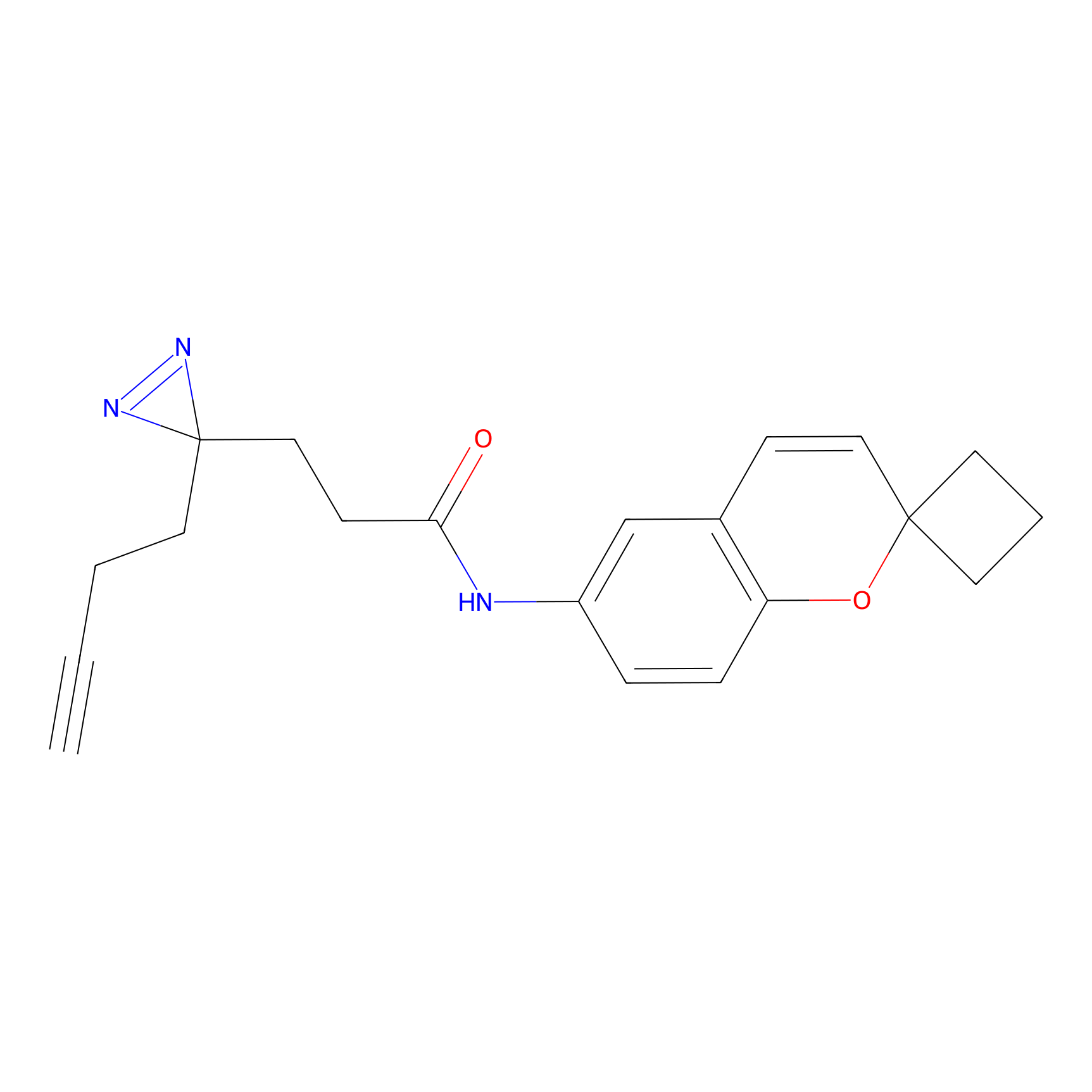 |
49.87 | LDD1948 | [9] | |
|
C289 Probe Info |
 |
37.27 | LDD1959 | [9] | |
|
C350 Probe Info |
 |
25.63 | LDD2011 | [9] | |
|
C355 Probe Info |
 |
24.42 | LDD2016 | [9] | |
|
C362 Probe Info |
 |
83.29 | LDD2023 | [9] | |
|
C363 Probe Info |
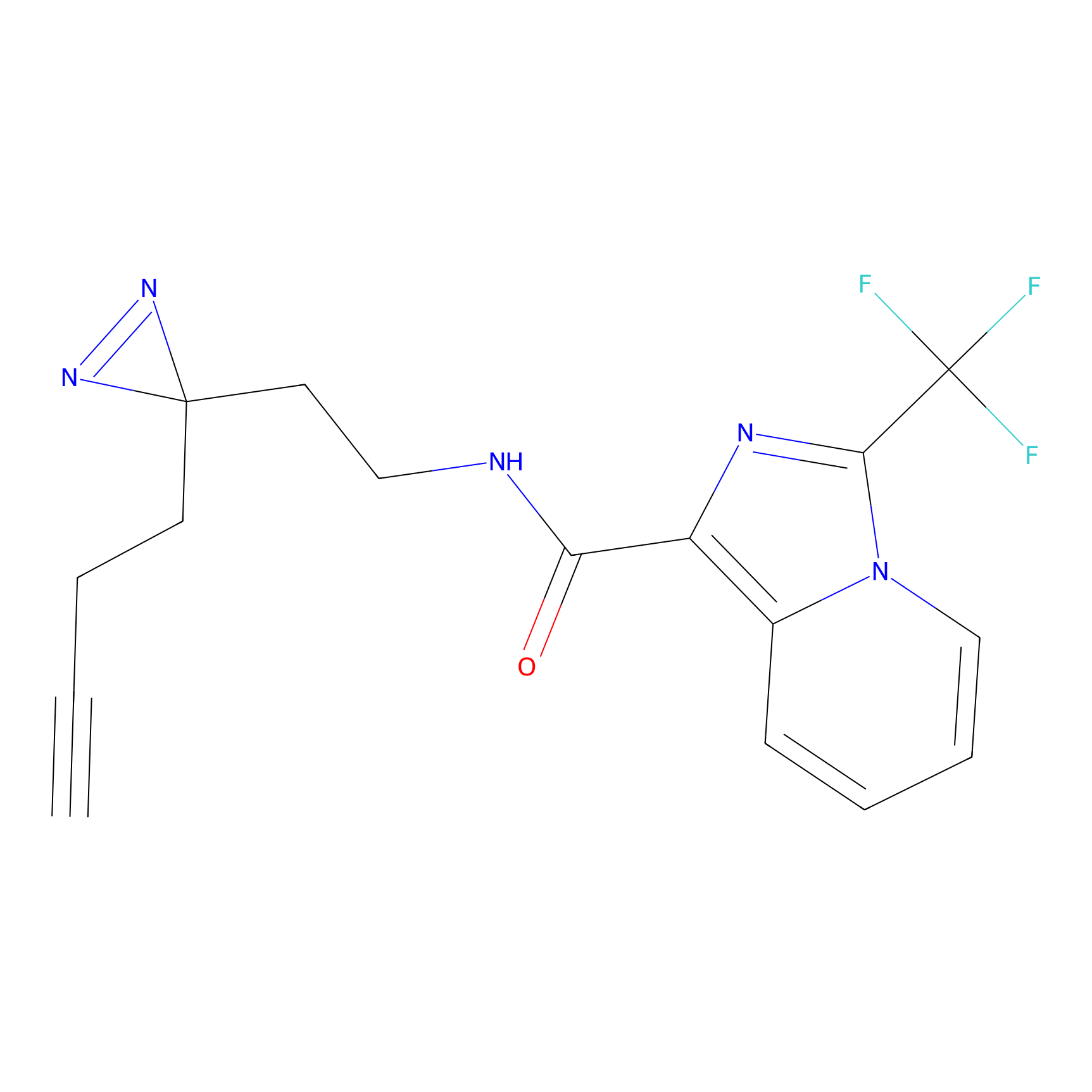 |
20.82 | LDD2024 | [9] | |
|
C364 Probe Info |
 |
24.93 | LDD2025 | [9] | |
|
C366 Probe Info |
 |
7.78 | LDD2027 | [9] | |
|
C367 Probe Info |
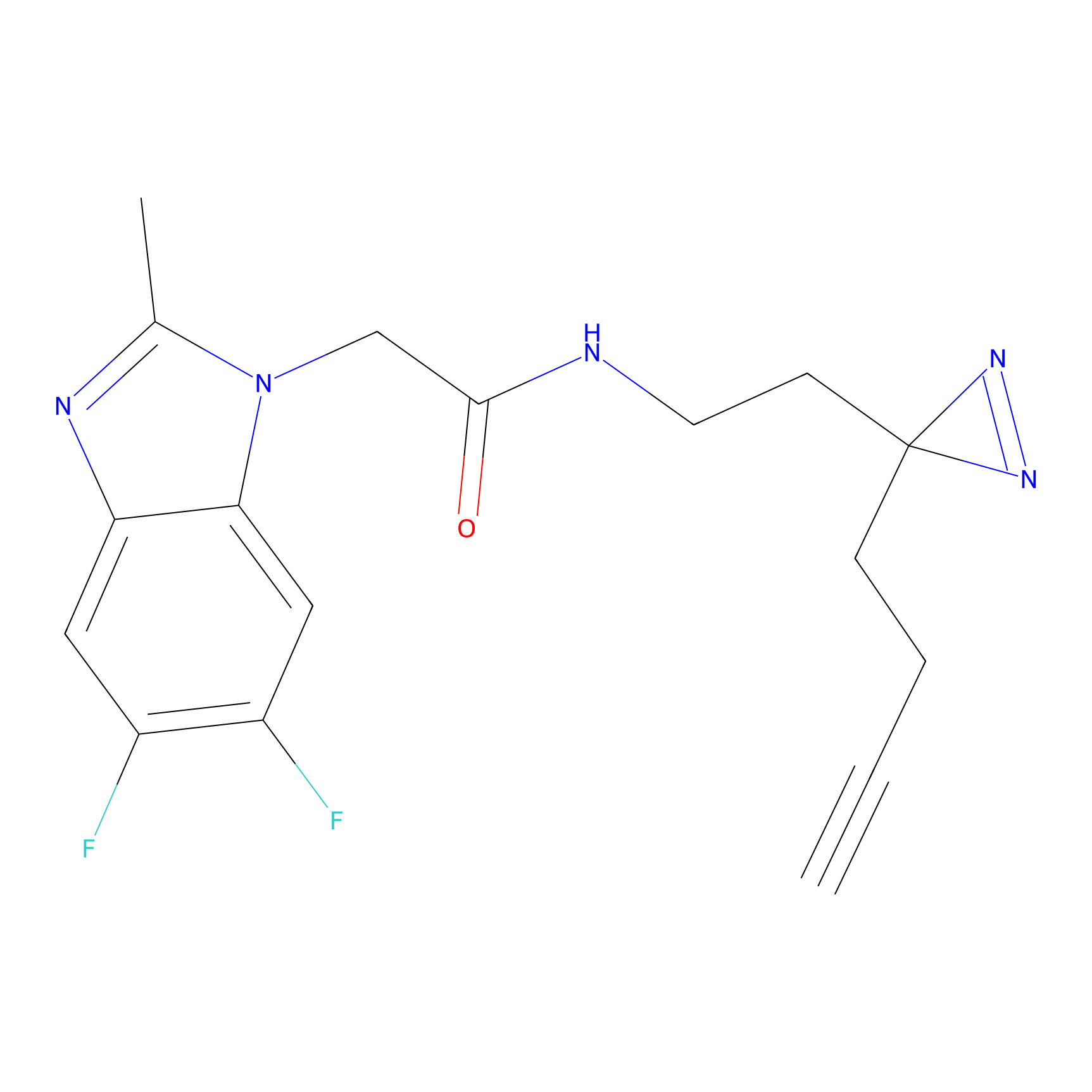 |
6.68 | LDD2028 | [9] | |
|
C390 Probe Info |
 |
41.07 | LDD2049 | [9] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0214 | AC1 | HEK-293T | C64(0.89) | LDD1507 | [10] |
| LDCM0215 | AC10 | HCT 116 | C64(0.97) | LDD0532 | [4] |
| LDCM0216 | AC100 | HCT 116 | C64(1.21) | LDD0533 | [4] |
| LDCM0217 | AC101 | HCT 116 | C64(0.87) | LDD0534 | [4] |
| LDCM0218 | AC102 | HCT 116 | C64(1.22) | LDD0535 | [4] |
| LDCM0219 | AC103 | HCT 116 | C64(1.35) | LDD0536 | [4] |
| LDCM0220 | AC104 | HCT 116 | C64(1.39) | LDD0537 | [4] |
| LDCM0221 | AC105 | HCT 116 | C64(1.33) | LDD0538 | [4] |
| LDCM0222 | AC106 | HCT 116 | C64(1.44) | LDD0539 | [4] |
| LDCM0223 | AC107 | HCT 116 | C64(1.01) | LDD0540 | [4] |
| LDCM0224 | AC108 | HCT 116 | C64(1.26) | LDD0541 | [4] |
| LDCM0225 | AC109 | HCT 116 | C64(1.11) | LDD0542 | [4] |
| LDCM0226 | AC11 | HCT 116 | C64(1.28) | LDD0543 | [4] |
| LDCM0227 | AC110 | HCT 116 | C64(1.23) | LDD0544 | [4] |
| LDCM0228 | AC111 | HCT 116 | C64(1.33) | LDD0545 | [4] |
| LDCM0229 | AC112 | HCT 116 | C64(1.19) | LDD0546 | [4] |
| LDCM0230 | AC113 | HCT 116 | C64(1.09) | LDD0547 | [4] |
| LDCM0231 | AC114 | HCT 116 | C64(1.19) | LDD0548 | [4] |
| LDCM0232 | AC115 | HCT 116 | C64(1.19) | LDD0549 | [4] |
| LDCM0233 | AC116 | HCT 116 | C64(0.95) | LDD0550 | [4] |
| LDCM0234 | AC117 | HCT 116 | C64(0.92) | LDD0551 | [4] |
| LDCM0235 | AC118 | HCT 116 | C64(0.99) | LDD0552 | [4] |
| LDCM0236 | AC119 | HCT 116 | C64(0.95) | LDD0553 | [4] |
| LDCM0237 | AC12 | HCT 116 | C64(1.08) | LDD0554 | [4] |
| LDCM0238 | AC120 | HCT 116 | C64(1.04) | LDD0555 | [4] |
| LDCM0239 | AC121 | HCT 116 | C64(1.06) | LDD0556 | [4] |
| LDCM0240 | AC122 | HCT 116 | C64(1.06) | LDD0557 | [4] |
| LDCM0241 | AC123 | HCT 116 | C64(1.34) | LDD0558 | [4] |
| LDCM0242 | AC124 | HCT 116 | C64(1.06) | LDD0559 | [4] |
| LDCM0243 | AC125 | HCT 116 | C64(1.09) | LDD0560 | [4] |
| LDCM0244 | AC126 | HCT 116 | C64(1.01) | LDD0561 | [4] |
| LDCM0245 | AC127 | HCT 116 | C64(1.00) | LDD0562 | [4] |
| LDCM0259 | AC14 | HCT 116 | C64(1.13) | LDD0576 | [4] |
| LDCM0270 | AC15 | HCT 116 | C64(0.96) | LDD0587 | [4] |
| LDCM0276 | AC17 | HCT 116 | C64(1.20) | LDD0593 | [4] |
| LDCM0277 | AC18 | HCT 116 | C64(1.02) | LDD0594 | [4] |
| LDCM0278 | AC19 | HCT 116 | C64(0.99) | LDD0595 | [4] |
| LDCM0279 | AC2 | HEK-293T | C64(0.99) | LDD1518 | [10] |
| LDCM0280 | AC20 | HCT 116 | C64(1.10) | LDD0597 | [4] |
| LDCM0281 | AC21 | HCT 116 | C64(1.04) | LDD0598 | [4] |
| LDCM0282 | AC22 | HCT 116 | C64(0.87) | LDD0599 | [4] |
| LDCM0283 | AC23 | HCT 116 | C64(0.94) | LDD0600 | [4] |
| LDCM0284 | AC24 | HCT 116 | C64(1.01) | LDD0601 | [4] |
| LDCM0285 | AC25 | HEK-293T | C64(0.98) | LDD1524 | [10] |
| LDCM0286 | AC26 | HEK-293T | C64(0.99) | LDD1525 | [10] |
| LDCM0287 | AC27 | HEK-293T | C64(0.94) | LDD1526 | [10] |
| LDCM0288 | AC28 | HEK-293T | C64(1.02) | LDD1527 | [10] |
| LDCM0289 | AC29 | HEK-293T | C64(0.94) | LDD1528 | [10] |
| LDCM0290 | AC3 | HEK-293T | C64(0.91) | LDD1529 | [10] |
| LDCM0291 | AC30 | HEK-293T | C64(0.93) | LDD1530 | [10] |
| LDCM0292 | AC31 | HEK-293T | C64(0.97) | LDD1531 | [10] |
| LDCM0293 | AC32 | HEK-293T | C64(1.01) | LDD1532 | [10] |
| LDCM0294 | AC33 | HEK-293T | C64(0.93) | LDD1533 | [10] |
| LDCM0295 | AC34 | HEK-293T | C64(0.94) | LDD1534 | [10] |
| LDCM0296 | AC35 | HCT 116 | C64(0.93) | LDD0613 | [4] |
| LDCM0297 | AC36 | HCT 116 | C64(0.79) | LDD0614 | [4] |
| LDCM0298 | AC37 | HCT 116 | C64(0.80) | LDD0615 | [4] |
| LDCM0299 | AC38 | HCT 116 | C64(0.93) | LDD0616 | [4] |
| LDCM0300 | AC39 | HCT 116 | C64(0.87) | LDD0617 | [4] |
| LDCM0301 | AC4 | HEK-293T | C64(0.93) | LDD1540 | [10] |
| LDCM0302 | AC40 | HCT 116 | C64(0.86) | LDD0619 | [4] |
| LDCM0303 | AC41 | HCT 116 | C64(1.04) | LDD0620 | [4] |
| LDCM0304 | AC42 | HCT 116 | C64(0.80) | LDD0621 | [4] |
| LDCM0305 | AC43 | HCT 116 | C64(0.99) | LDD0622 | [4] |
| LDCM0306 | AC44 | HCT 116 | C64(0.81) | LDD0623 | [4] |
| LDCM0307 | AC45 | HCT 116 | C64(0.70) | LDD0624 | [4] |
| LDCM0308 | AC46 | HEK-293T | C64(0.94) | LDD1547 | [10] |
| LDCM0309 | AC47 | HEK-293T | C64(0.96) | LDD1548 | [10] |
| LDCM0310 | AC48 | HEK-293T | C64(0.93) | LDD1549 | [10] |
| LDCM0311 | AC49 | HEK-293T | C64(0.94) | LDD1550 | [10] |
| LDCM0312 | AC5 | HEK-293T | C64(1.01) | LDD1551 | [10] |
| LDCM0313 | AC50 | HEK-293T | C64(1.05) | LDD1552 | [10] |
| LDCM0314 | AC51 | HEK-293T | C64(1.01) | LDD1553 | [10] |
| LDCM0315 | AC52 | HEK-293T | C64(0.97) | LDD1554 | [10] |
| LDCM0316 | AC53 | HEK-293T | C64(1.00) | LDD1555 | [10] |
| LDCM0317 | AC54 | HEK-293T | C64(0.99) | LDD1556 | [10] |
| LDCM0318 | AC55 | HEK-293T | C64(1.02) | LDD1557 | [10] |
| LDCM0319 | AC56 | HEK-293T | C64(1.00) | LDD1558 | [10] |
| LDCM0320 | AC57 | HCT 116 | C64(0.92) | LDD0637 | [4] |
| LDCM0321 | AC58 | HCT 116 | C64(1.00) | LDD0638 | [4] |
| LDCM0322 | AC59 | HCT 116 | C64(0.96) | LDD0639 | [4] |
| LDCM0323 | AC6 | HCT 116 | C64(1.25) | LDD0640 | [4] |
| LDCM0324 | AC60 | HCT 116 | C64(1.03) | LDD0641 | [4] |
| LDCM0325 | AC61 | HCT 116 | C64(0.91) | LDD0642 | [4] |
| LDCM0326 | AC62 | HCT 116 | C64(1.03) | LDD0643 | [4] |
| LDCM0327 | AC63 | HCT 116 | C64(1.20) | LDD0644 | [4] |
| LDCM0328 | AC64 | HCT 116 | C64(1.17) | LDD0645 | [4] |
| LDCM0329 | AC65 | HCT 116 | C64(1.07) | LDD0646 | [4] |
| LDCM0330 | AC66 | HCT 116 | C64(1.03) | LDD0647 | [4] |
| LDCM0331 | AC67 | HCT 116 | C64(1.06) | LDD0648 | [4] |
| LDCM0334 | AC7 | HCT 116 | C64(1.18) | LDD0651 | [4] |
| LDCM0345 | AC8 | HCT 116 | C64(1.13) | LDD0662 | [4] |
| LDCM0349 | AC83 | HCT 116 | C64(1.09) | LDD0666 | [4] |
| LDCM0350 | AC84 | HCT 116 | C64(0.90) | LDD0667 | [4] |
| LDCM0351 | AC85 | HCT 116 | C64(1.09) | LDD0668 | [4] |
| LDCM0352 | AC86 | HCT 116 | C64(1.03) | LDD0669 | [4] |
| LDCM0353 | AC87 | HCT 116 | C64(1.13) | LDD0670 | [4] |
| LDCM0354 | AC88 | HCT 116 | C64(1.02) | LDD0671 | [4] |
| LDCM0355 | AC89 | HCT 116 | C64(0.99) | LDD0672 | [4] |
| LDCM0357 | AC90 | HCT 116 | C64(1.13) | LDD0674 | [4] |
| LDCM0358 | AC91 | HCT 116 | C64(1.01) | LDD0675 | [4] |
| LDCM0359 | AC92 | HCT 116 | C64(0.79) | LDD0676 | [4] |
| LDCM0360 | AC93 | HCT 116 | C64(1.18) | LDD0677 | [4] |
| LDCM0361 | AC94 | HCT 116 | C64(1.10) | LDD0678 | [4] |
| LDCM0362 | AC95 | HCT 116 | C64(1.00) | LDD0679 | [4] |
| LDCM0363 | AC96 | HCT 116 | C64(1.06) | LDD0680 | [4] |
| LDCM0364 | AC97 | HCT 116 | C64(1.16) | LDD0681 | [4] |
| LDCM0365 | AC98 | HCT 116 | C64(1.36) | LDD0682 | [4] |
| LDCM0366 | AC99 | HCT 116 | C64(1.41) | LDD0683 | [4] |
| LDCM0248 | AKOS034007472 | HCT 116 | C64(1.02) | LDD0565 | [4] |
| LDCM0356 | AKOS034007680 | HCT 116 | C64(1.12) | LDD0673 | [4] |
| LDCM0275 | AKOS034007705 | HCT 116 | C64(1.08) | LDD0592 | [4] |
| LDCM0367 | CL1 | HEK-293T | C64(1.11) | LDD1571 | [10] |
| LDCM0368 | CL10 | HEK-293T | C64(0.82) | LDD1572 | [10] |
| LDCM0369 | CL100 | HEK-293T | C64(1.04) | LDD1573 | [10] |
| LDCM0370 | CL101 | HCT 116 | C64(0.94) | LDD0687 | [4] |
| LDCM0371 | CL102 | HCT 116 | C64(0.90) | LDD0688 | [4] |
| LDCM0372 | CL103 | HCT 116 | C64(1.04) | LDD0689 | [4] |
| LDCM0373 | CL104 | HCT 116 | C64(1.09) | LDD0690 | [4] |
| LDCM0374 | CL105 | HCT 116 | C64(1.22) | LDD0691 | [4] |
| LDCM0375 | CL106 | HCT 116 | C64(1.08) | LDD0692 | [4] |
| LDCM0376 | CL107 | HCT 116 | C64(0.99) | LDD0693 | [4] |
| LDCM0377 | CL108 | HCT 116 | C64(1.18) | LDD0694 | [4] |
| LDCM0378 | CL109 | HCT 116 | C64(0.95) | LDD0695 | [4] |
| LDCM0379 | CL11 | HEK-293T | C64(0.81) | LDD1583 | [10] |
| LDCM0380 | CL110 | HCT 116 | C64(1.17) | LDD0697 | [4] |
| LDCM0381 | CL111 | HCT 116 | C64(1.01) | LDD0698 | [4] |
| LDCM0382 | CL112 | HEK-293T | C64(1.04) | LDD1586 | [10] |
| LDCM0383 | CL113 | HEK-293T | C64(1.10) | LDD1587 | [10] |
| LDCM0384 | CL114 | HEK-293T | C64(0.92) | LDD1588 | [10] |
| LDCM0385 | CL115 | HEK-293T | C64(0.93) | LDD1589 | [10] |
| LDCM0386 | CL116 | HEK-293T | C64(1.02) | LDD1590 | [10] |
| LDCM0387 | CL117 | HCT 116 | C64(0.94) | LDD0704 | [4] |
| LDCM0388 | CL118 | HCT 116 | C64(1.21) | LDD0705 | [4] |
| LDCM0389 | CL119 | HCT 116 | C64(0.93) | LDD0706 | [4] |
| LDCM0390 | CL12 | HEK-293T | C64(0.97) | LDD1594 | [10] |
| LDCM0391 | CL120 | HCT 116 | C64(1.09) | LDD0708 | [4] |
| LDCM0392 | CL121 | HEK-293T | C64(0.96) | LDD1596 | [10] |
| LDCM0393 | CL122 | HEK-293T | C64(0.94) | LDD1597 | [10] |
| LDCM0394 | CL123 | HEK-293T | C64(0.94) | LDD1598 | [10] |
| LDCM0395 | CL124 | HEK-293T | C64(1.15) | LDD1599 | [10] |
| LDCM0396 | CL125 | HCT 116 | C64(1.12) | LDD0713 | [4] |
| LDCM0397 | CL126 | HCT 116 | C64(0.95) | LDD0714 | [4] |
| LDCM0398 | CL127 | HCT 116 | C64(0.83) | LDD0715 | [4] |
| LDCM0399 | CL128 | HCT 116 | C64(0.80) | LDD0716 | [4] |
| LDCM0400 | CL13 | HEK-293T | C64(1.03) | LDD1604 | [10] |
| LDCM0401 | CL14 | HEK-293T | C64(0.91) | LDD1605 | [10] |
| LDCM0402 | CL15 | HEK-293T | C64(0.92) | LDD1606 | [10] |
| LDCM0403 | CL16 | HEK-293T | C64(1.21) | LDD1607 | [10] |
| LDCM0404 | CL17 | HEK-293T | C64(0.81) | LDD1608 | [10] |
| LDCM0405 | CL18 | HEK-293T | C64(1.01) | LDD1609 | [10] |
| LDCM0406 | CL19 | HEK-293T | C64(0.90) | LDD1610 | [10] |
| LDCM0407 | CL2 | HEK-293T | C64(0.93) | LDD1611 | [10] |
| LDCM0408 | CL20 | HEK-293T | C64(1.00) | LDD1612 | [10] |
| LDCM0409 | CL21 | HEK-293T | C64(1.02) | LDD1613 | [10] |
| LDCM0410 | CL22 | HEK-293T | C64(0.97) | LDD1614 | [10] |
| LDCM0411 | CL23 | HEK-293T | C64(0.93) | LDD1615 | [10] |
| LDCM0412 | CL24 | HEK-293T | C64(1.06) | LDD1616 | [10] |
| LDCM0413 | CL25 | HEK-293T | C64(1.06) | LDD1617 | [10] |
| LDCM0414 | CL26 | HEK-293T | C64(0.98) | LDD1618 | [10] |
| LDCM0415 | CL27 | HEK-293T | C64(1.02) | LDD1619 | [10] |
| LDCM0416 | CL28 | HEK-293T | C64(1.03) | LDD1620 | [10] |
| LDCM0417 | CL29 | HEK-293T | C64(1.01) | LDD1621 | [10] |
| LDCM0418 | CL3 | HEK-293T | C64(0.98) | LDD1622 | [10] |
| LDCM0419 | CL30 | HEK-293T | C64(1.02) | LDD1623 | [10] |
| LDCM0420 | CL31 | HCT 116 | C64(0.89) | LDD0737 | [4] |
| LDCM0421 | CL32 | HCT 116 | C64(0.76) | LDD0738 | [4] |
| LDCM0422 | CL33 | HCT 116 | C64(1.00) | LDD0739 | [4] |
| LDCM0423 | CL34 | HCT 116 | C64(1.01) | LDD0740 | [4] |
| LDCM0424 | CL35 | HCT 116 | C64(1.43) | LDD0741 | [4] |
| LDCM0425 | CL36 | HCT 116 | C64(1.11) | LDD0742 | [4] |
| LDCM0426 | CL37 | HCT 116 | C64(1.10) | LDD0743 | [4] |
| LDCM0428 | CL39 | HCT 116 | C64(1.05) | LDD0745 | [4] |
| LDCM0429 | CL4 | HEK-293T | C64(0.97) | LDD1633 | [10] |
| LDCM0430 | CL40 | HCT 116 | C64(1.06) | LDD0747 | [4] |
| LDCM0431 | CL41 | HCT 116 | C64(1.24) | LDD0748 | [4] |
| LDCM0432 | CL42 | HCT 116 | C64(0.84) | LDD0749 | [4] |
| LDCM0433 | CL43 | HCT 116 | C64(1.06) | LDD0750 | [4] |
| LDCM0434 | CL44 | HCT 116 | C64(0.86) | LDD0751 | [4] |
| LDCM0435 | CL45 | HCT 116 | C64(1.32) | LDD0752 | [4] |
| LDCM0436 | CL46 | HEK-293T | C64(0.98) | LDD1640 | [10] |
| LDCM0437 | CL47 | HEK-293T | C64(0.96) | LDD1641 | [10] |
| LDCM0438 | CL48 | HEK-293T | C64(1.06) | LDD1642 | [10] |
| LDCM0439 | CL49 | HEK-293T | C64(1.10) | LDD1643 | [10] |
| LDCM0440 | CL5 | HEK-293T | C64(0.90) | LDD1644 | [10] |
| LDCM0441 | CL50 | HEK-293T | C64(1.00) | LDD1645 | [10] |
| LDCM0443 | CL52 | HEK-293T | C64(1.13) | LDD1646 | [10] |
| LDCM0444 | CL53 | HEK-293T | C64(0.89) | LDD1647 | [10] |
| LDCM0445 | CL54 | HEK-293T | C64(0.94) | LDD1648 | [10] |
| LDCM0446 | CL55 | HEK-293T | C64(0.98) | LDD1649 | [10] |
| LDCM0447 | CL56 | HEK-293T | C64(0.92) | LDD1650 | [10] |
| LDCM0448 | CL57 | HEK-293T | C64(1.02) | LDD1651 | [10] |
| LDCM0449 | CL58 | HEK-293T | C64(1.11) | LDD1652 | [10] |
| LDCM0450 | CL59 | HEK-293T | C64(1.02) | LDD1653 | [10] |
| LDCM0451 | CL6 | HEK-293T | C64(1.01) | LDD1654 | [10] |
| LDCM0452 | CL60 | HEK-293T | C64(0.98) | LDD1655 | [10] |
| LDCM0453 | CL61 | HEK-293T | C64(1.02) | LDD1656 | [10] |
| LDCM0454 | CL62 | HEK-293T | C64(0.99) | LDD1657 | [10] |
| LDCM0455 | CL63 | HEK-293T | C64(0.96) | LDD1658 | [10] |
| LDCM0456 | CL64 | HEK-293T | C64(0.94) | LDD1659 | [10] |
| LDCM0457 | CL65 | HEK-293T | C64(0.94) | LDD1660 | [10] |
| LDCM0458 | CL66 | HEK-293T | C64(1.05) | LDD1661 | [10] |
| LDCM0459 | CL67 | HEK-293T | C64(0.92) | LDD1662 | [10] |
| LDCM0460 | CL68 | HEK-293T | C64(1.19) | LDD1663 | [10] |
| LDCM0461 | CL69 | HEK-293T | C64(1.10) | LDD1664 | [10] |
| LDCM0462 | CL7 | HEK-293T | C64(0.91) | LDD1665 | [10] |
| LDCM0463 | CL70 | HEK-293T | C64(1.03) | LDD1666 | [10] |
| LDCM0464 | CL71 | HEK-293T | C64(1.00) | LDD1667 | [10] |
| LDCM0465 | CL72 | HEK-293T | C64(1.02) | LDD1668 | [10] |
| LDCM0466 | CL73 | HEK-293T | C64(1.04) | LDD1669 | [10] |
| LDCM0467 | CL74 | HEK-293T | C64(0.98) | LDD1670 | [10] |
| LDCM0469 | CL76 | HEK-293T | C64(1.24) | LDD1672 | [10] |
| LDCM0470 | CL77 | HEK-293T | C64(0.82) | LDD1673 | [10] |
| LDCM0471 | CL78 | HEK-293T | C64(1.09) | LDD1674 | [10] |
| LDCM0472 | CL79 | HEK-293T | C64(1.03) | LDD1675 | [10] |
| LDCM0473 | CL8 | HEK-293T | C64(0.95) | LDD1676 | [10] |
| LDCM0474 | CL80 | HEK-293T | C64(1.02) | LDD1677 | [10] |
| LDCM0475 | CL81 | HEK-293T | C64(1.15) | LDD1678 | [10] |
| LDCM0476 | CL82 | HEK-293T | C64(1.02) | LDD1679 | [10] |
| LDCM0477 | CL83 | HEK-293T | C64(0.94) | LDD1680 | [10] |
| LDCM0478 | CL84 | HEK-293T | C64(1.00) | LDD1681 | [10] |
| LDCM0479 | CL85 | HEK-293T | C64(0.98) | LDD1682 | [10] |
| LDCM0480 | CL86 | HEK-293T | C64(1.00) | LDD1683 | [10] |
| LDCM0481 | CL87 | HEK-293T | C64(1.08) | LDD1684 | [10] |
| LDCM0482 | CL88 | HEK-293T | C64(1.21) | LDD1685 | [10] |
| LDCM0483 | CL89 | HEK-293T | C64(0.97) | LDD1686 | [10] |
| LDCM0484 | CL9 | HEK-293T | C64(1.06) | LDD1687 | [10] |
| LDCM0485 | CL90 | HEK-293T | C64(0.76) | LDD1688 | [10] |
| LDCM0486 | CL91 | HEK-293T | C64(0.94) | LDD1689 | [10] |
| LDCM0487 | CL92 | HEK-293T | C64(0.92) | LDD1690 | [10] |
| LDCM0488 | CL93 | HEK-293T | C64(1.03) | LDD1691 | [10] |
| LDCM0489 | CL94 | HEK-293T | C64(0.96) | LDD1692 | [10] |
| LDCM0490 | CL95 | HEK-293T | C64(0.89) | LDD1693 | [10] |
| LDCM0491 | CL96 | HEK-293T | C64(0.93) | LDD1694 | [10] |
| LDCM0492 | CL97 | HEK-293T | C64(0.99) | LDD1695 | [10] |
| LDCM0493 | CL98 | HEK-293T | C64(0.99) | LDD1696 | [10] |
| LDCM0494 | CL99 | HEK-293T | C64(1.07) | LDD1697 | [10] |
| LDCM0033 | Curcusone1d | MCF-7 | 4.50 | LDD0188 | [3] |
| LDCM0495 | E2913 | HEK-293T | C64(1.00) | LDD1698 | [10] |
| LDCM0625 | F8 | Ramos | C64(0.83) | LDD2187 | [11] |
| LDCM0572 | Fragment10 | Ramos | C64(0.28) | LDD2189 | [11] |
| LDCM0573 | Fragment11 | Ramos | C64(1.64) | LDD2190 | [11] |
| LDCM0574 | Fragment12 | Ramos | C64(0.29) | LDD2191 | [11] |
| LDCM0575 | Fragment13 | Ramos | C64(1.41) | LDD2192 | [11] |
| LDCM0576 | Fragment14 | Ramos | C64(0.75) | LDD2193 | [11] |
| LDCM0579 | Fragment20 | Ramos | C64(0.49) | LDD2194 | [11] |
| LDCM0582 | Fragment23 | Ramos | C64(0.54) | LDD2196 | [11] |
| LDCM0588 | Fragment30 | Ramos | C64(3.76) | LDD2199 | [11] |
| LDCM0590 | Fragment32 | Ramos | C64(0.21) | LDD2201 | [11] |
| LDCM0468 | Fragment33 | HEK-293T | C64(1.13) | LDD1671 | [10] |
| LDCM0596 | Fragment38 | Ramos | C64(1.13) | LDD2203 | [11] |
| LDCM0566 | Fragment4 | Ramos | C64(0.69) | LDD2184 | [11] |
| LDCM0427 | Fragment51 | HCT 116 | C64(1.92) | LDD0744 | [4] |
| LDCM0610 | Fragment52 | Ramos | C64(0.44) | LDD2204 | [11] |
| LDCM0569 | Fragment7 | Ramos | C64(0.81) | LDD2186 | [11] |
| LDCM0571 | Fragment9 | Ramos | C64(0.54) | LDD2188 | [11] |
| LDCM0022 | KB02 | HEK-293T | C64(0.93) | LDD1492 | [10] |
| LDCM0023 | KB03 | HEK-293T | C64(1.01) | LDD1497 | [10] |
| LDCM0024 | KB05 | HEK-293T | C64(0.87) | LDD1502 | [10] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C64(0.58) | LDD2207 | [12] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
GPCR
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Probable G-protein coupled receptor 152 (GPR152) | G-protein coupled receptor 1 family | Q8TDT2 | |||
Immunoglobulin
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Junctional adhesion molecule A (F11R) | Immunoglobulin superfamily | Q9Y624 | |||
| SLAM family member 6 (SLAMF6) | . | Q96DU3 | |||
Cytokine and receptor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Tumor necrosis factor receptor superfamily member 3 (LTBR) | . | P36941 | |||
Other
References
