Details of the Target
General Information of Target
| Target ID | LDTP10255 | |||||
|---|---|---|---|---|---|---|
| Target Name | Protein Hook homolog 2 (HOOK2) | |||||
| Gene Name | HOOK2 | |||||
| Gene ID | 29911 | |||||
| Synonyms |
Protein Hook homolog 2; h-hook2; hHK2 |
|||||
| 3D Structure | ||||||
| Sequence |
MAEAEESPGDPGTASPRPLFAGLSDISISQDIPVEGEITIPMRSRIREFDSSTLNESVRN
TIMRDLKAVGKKFMHVLYPRKSNTLLRDWDLWGPLILCVTLALMLQRDSADSEKDGGPQF AEVFVIVWFGAVTITLNSKLLGGNISFFQSLCVLGYCILPLTVAMLICRLVLLADPGPVN FMVRLFVVIVMFAWSIVASTAFLADSQPPNRRALAVYPVFLFYFVISWMILTFTPQ |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
Hook family
|
|||||
| Subcellular location |
Cytoplasm, cytoskeleton, microtubule organizing center, centrosome
|
|||||
| Function |
Component of the FTS/Hook/FHIP complex (FHF complex). The FHF complex may function to promote vesicle trafficking and/or fusion via the homotypic vesicular protein sorting complex (the HOPS complex). Contributes to the establishment and maintenance of centrosome function. May function in the positioning or formation of aggresomes, which are pericentriolar accumulations of misfolded proteins, proteasomes and chaperones. FHF complex promotes the distribution of AP-4 complex to the perinuclear area of the cell.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| 22RV1 | Deletion: p.E371NfsTer11 | . | |||
| CAMA1 | SNV: p.E660Q | . | |||
| CHL1 | SNV: p.G120C | . | |||
| HCT15 | SNV: p.A274T | DBIA Probe Info | |||
| HEC1 | SNV: p.R193H | DBIA Probe Info | |||
| HEC1B | SNV: p.R193H | . | |||
| JURKAT | SNV: p.Q29Ter | . | |||
| KMCH1 | SNV: p.D554H | DBIA Probe Info | |||
| KURAMOCHI | SNV: p.I537V | . | |||
| LN229 | SNV: p.C25F | . | |||
| MCC26 | SNV: p.E182K | . | |||
| MDAMB231 | SNV: p.P108R | . | |||
| P31FUJ | SNV: p.Q43L | . | |||
| RCC10RGB | SNV: p.F179I | . | |||
| SKMM2 | SNV: p.E483G | . | |||
| SNU1079 | SNV: p.V610G | DBIA Probe Info | |||
| SW1783 | SNV: p.R221W | . | |||
| SW620 | SNV: p.E246D | . | |||
| UACC62 | SNV: p.P225L | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K128(10.00) | LDD0277 | [1] | |
|
Probe 1 Probe Info |
 |
Y167(11.77); Y563(7.67) | LDD3495 | [2] | |
|
HPAP Probe Info |
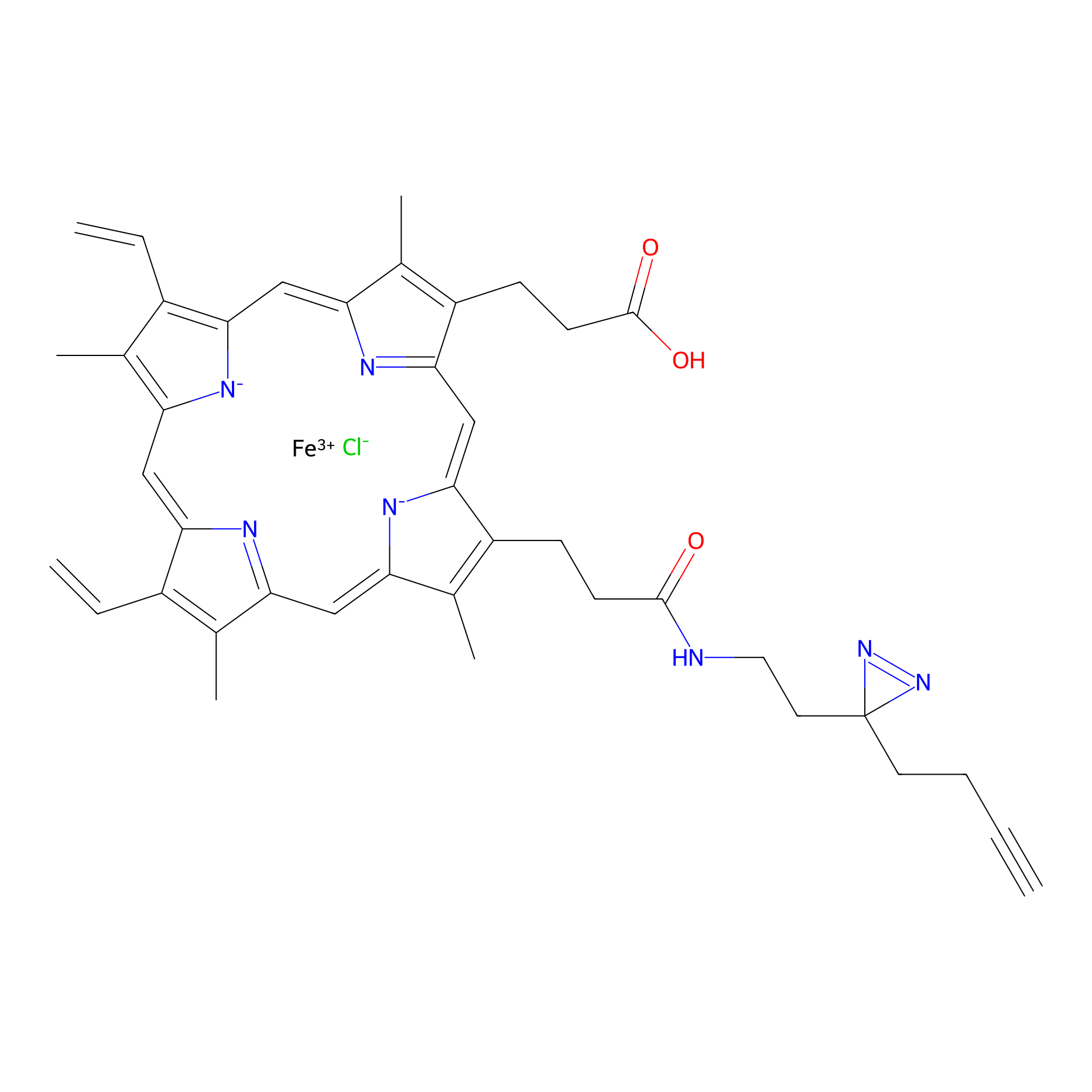 |
3.38 | LDD0062 | [3] | |
|
DBIA Probe Info |
 |
C121(0.90); C125(0.90); C423(1.00) | LDD0531 | [4] | |
|
IA-alkyne Probe Info |
 |
C194(0.00); C317(0.00); C390(0.00) | LDD0162 | [5] | |
|
IPM Probe Info |
 |
C317(0.00); C423(0.00) | LDD0147 | [6] | |
|
TFBX Probe Info |
 |
C317(0.00); C423(0.00) | LDD0148 | [6] | |
|
AOyne Probe Info |
 |
13.40 | LDD0443 | [7] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [8] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C063 Probe Info |
 |
15.03 | LDD1760 | [9] | |
|
C106 Probe Info |
 |
18.25 | LDD1793 | [9] | |
|
C158 Probe Info |
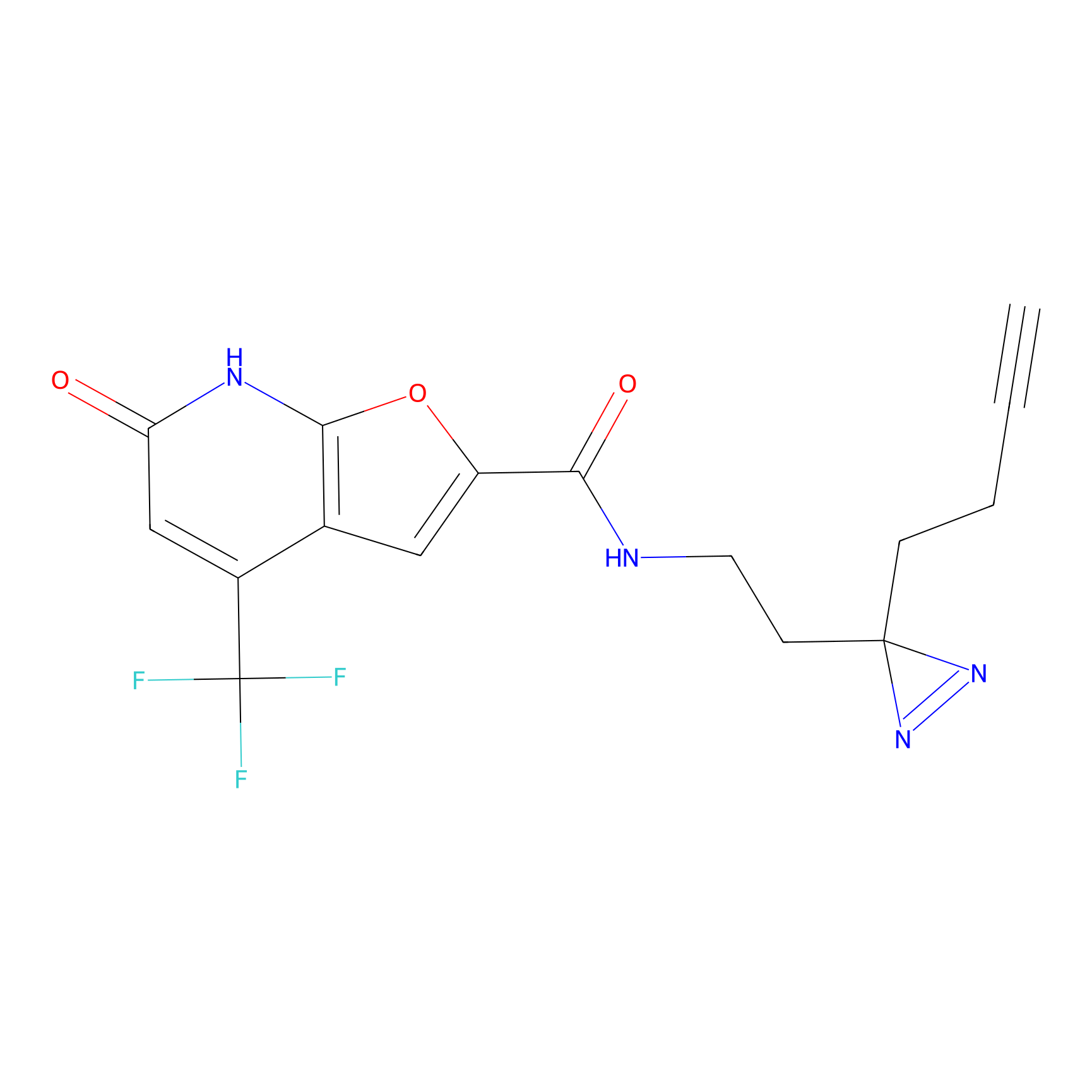 |
9.13 | LDD1838 | [9] | |
|
C173 Probe Info |
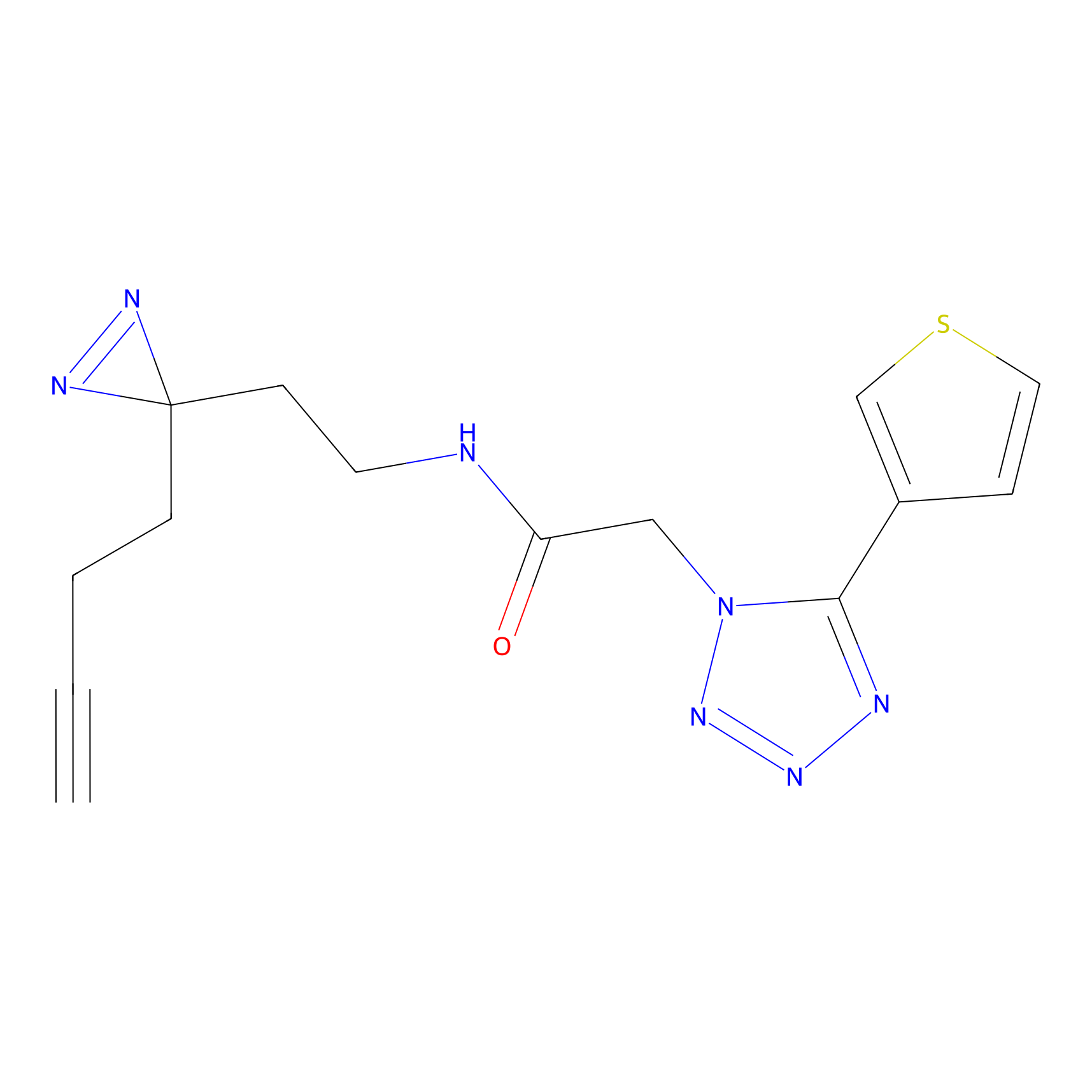 |
5.50 | LDD1853 | [9] | |
|
C201 Probe Info |
 |
97.68 | LDD1877 | [9] | |
|
C205 Probe Info |
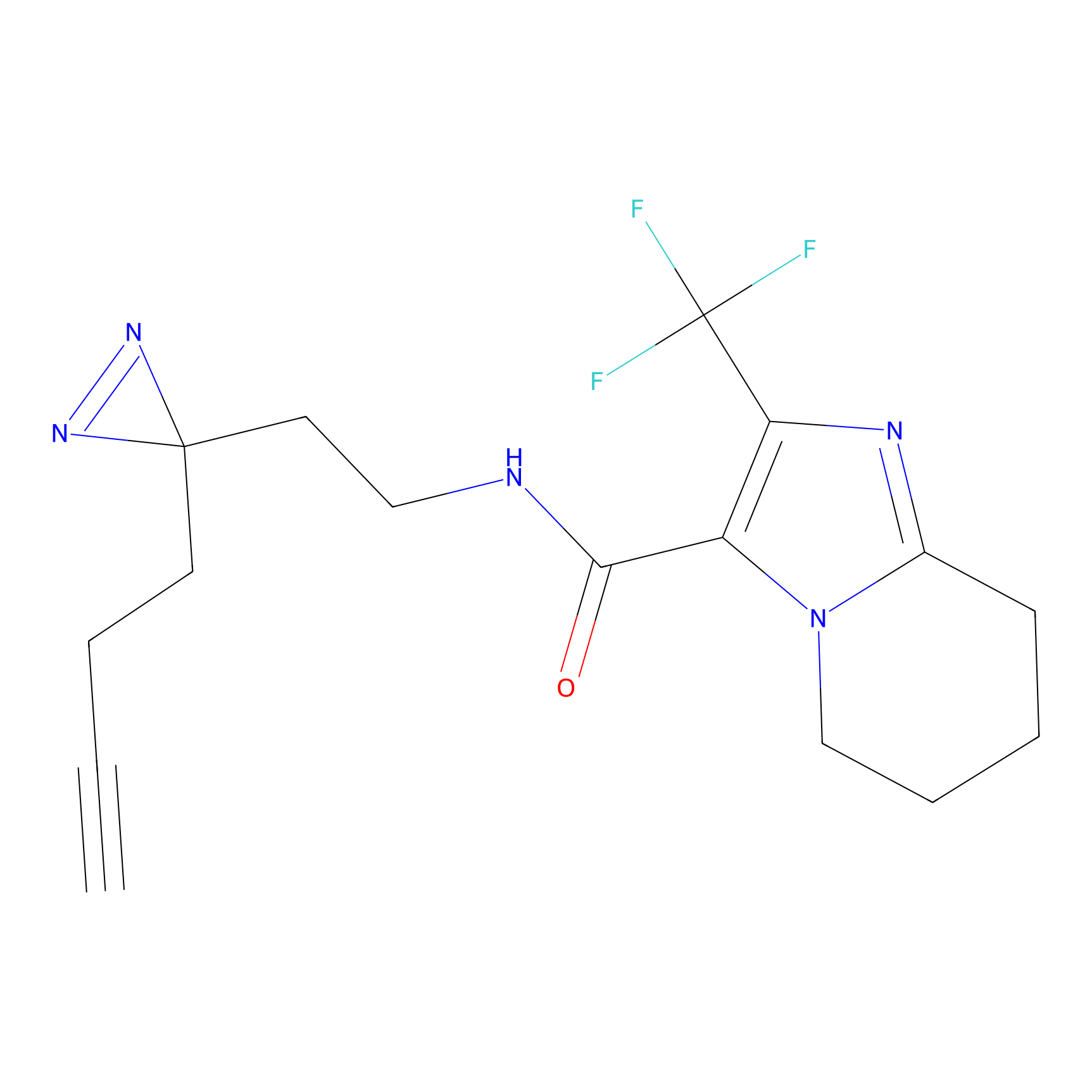 |
7.52 | LDD1880 | [9] | |
|
C206 Probe Info |
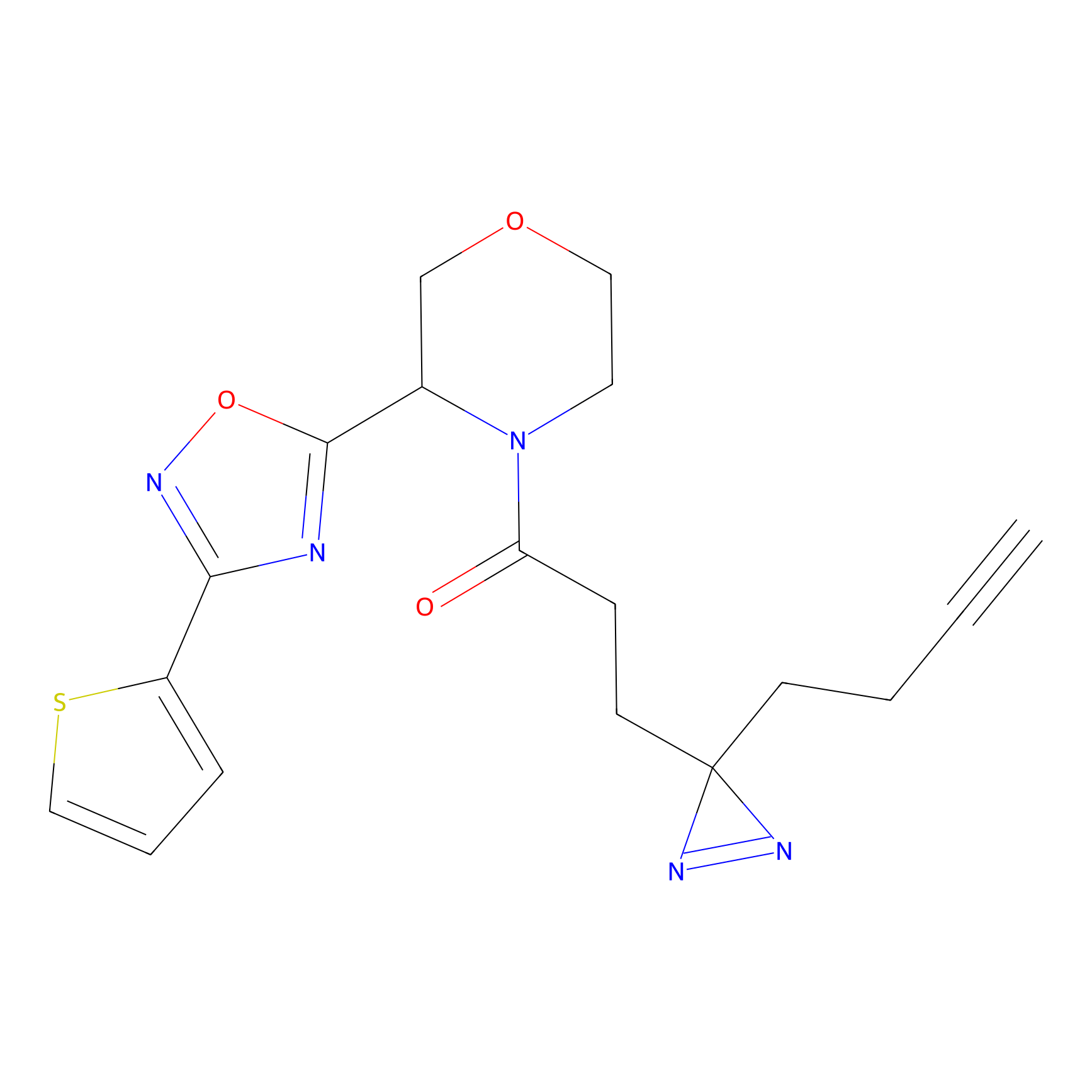 |
21.56 | LDD1881 | [9] | |
|
C207 Probe Info |
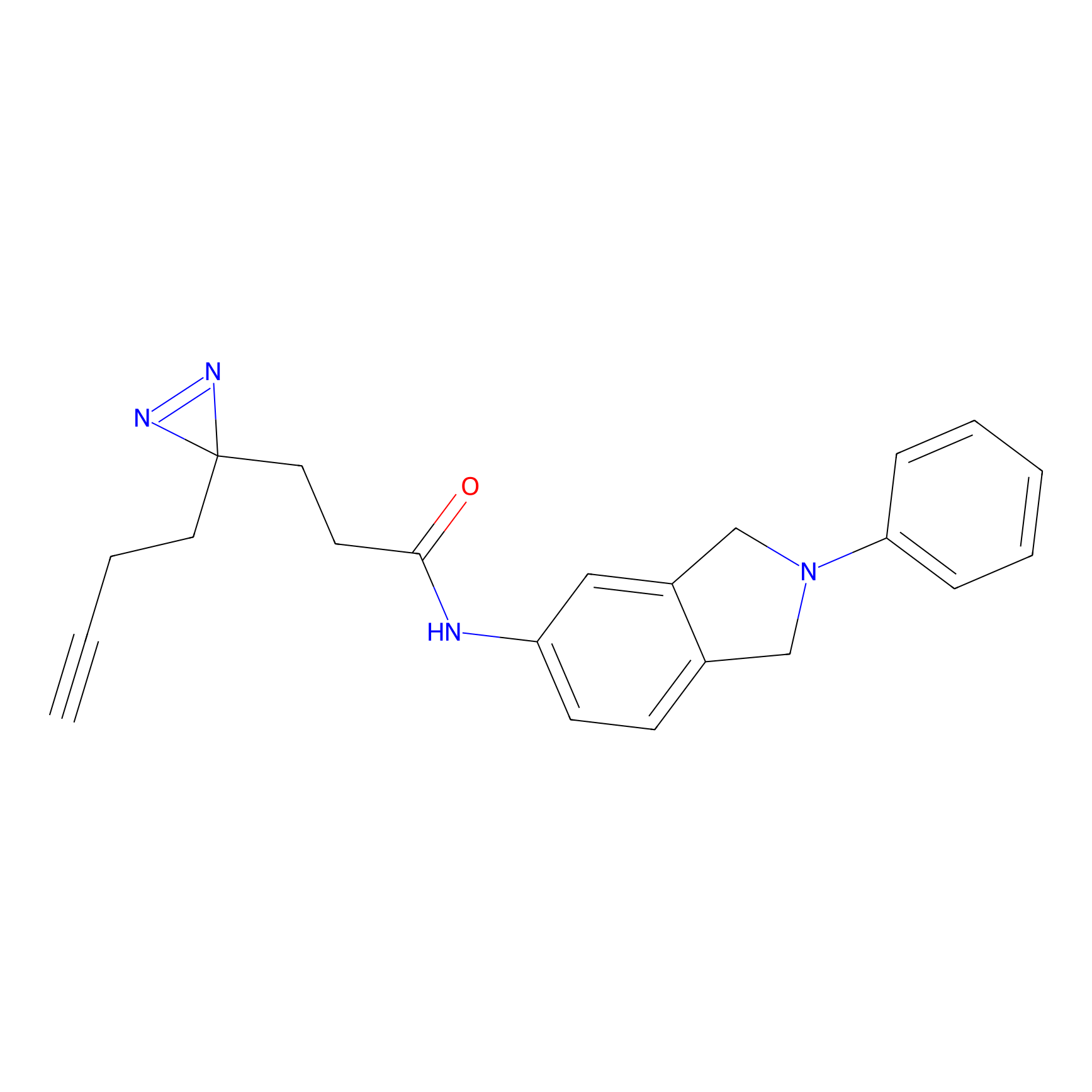 |
21.86 | LDD1882 | [9] | |
|
C313 Probe Info |
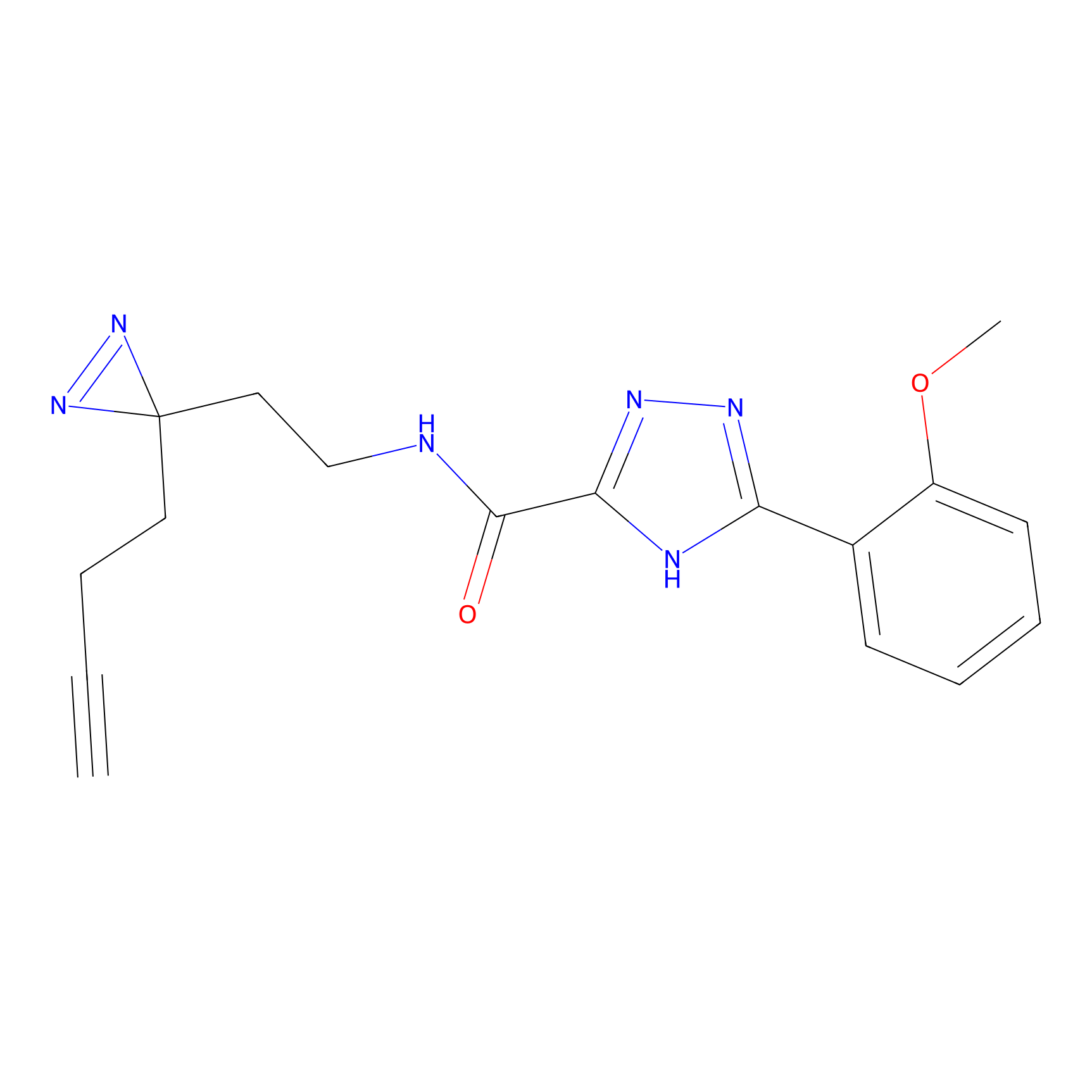 |
28.05 | LDD1980 | [9] | |
|
C356 Probe Info |
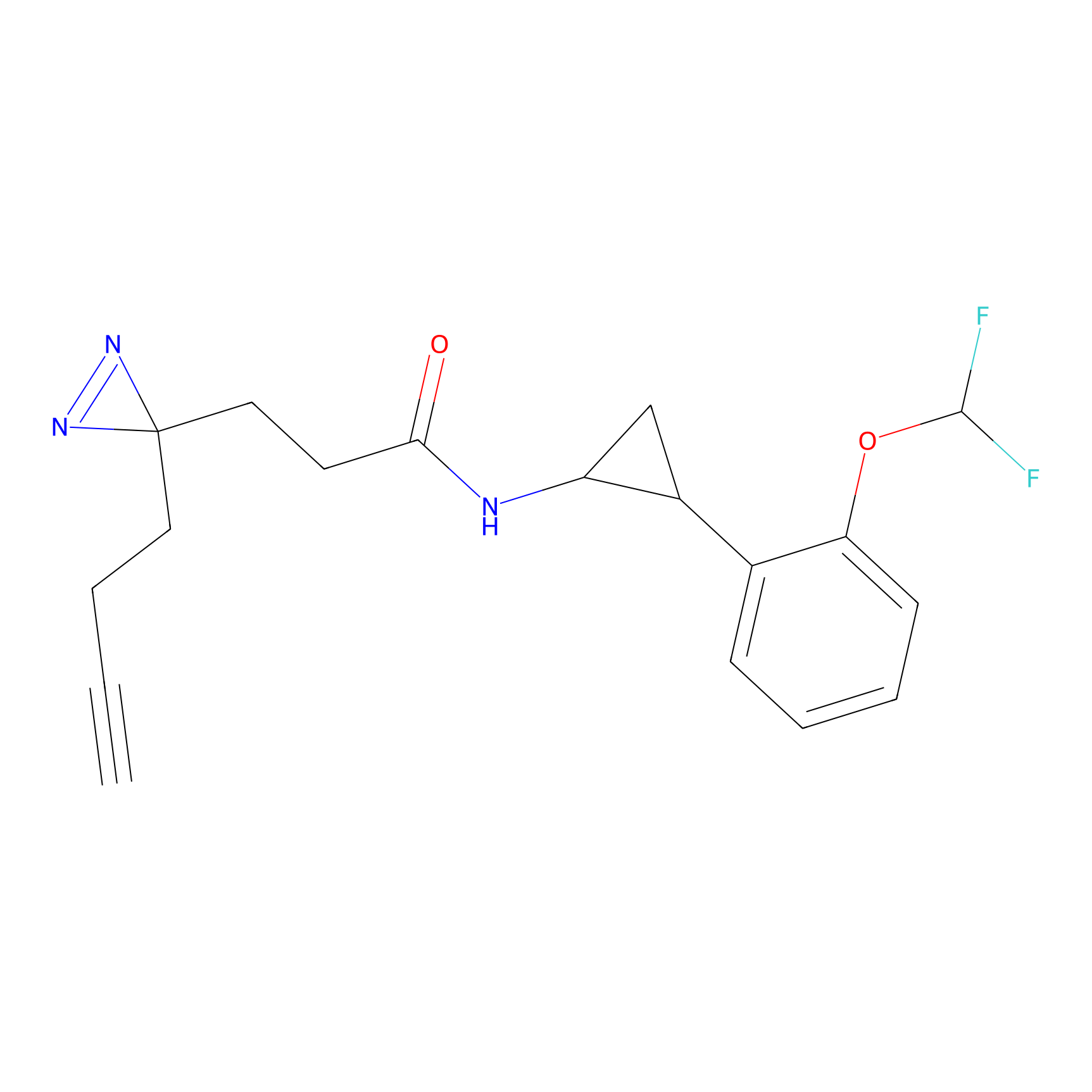 |
14.83 | LDD2017 | [9] | |
|
C361 Probe Info |
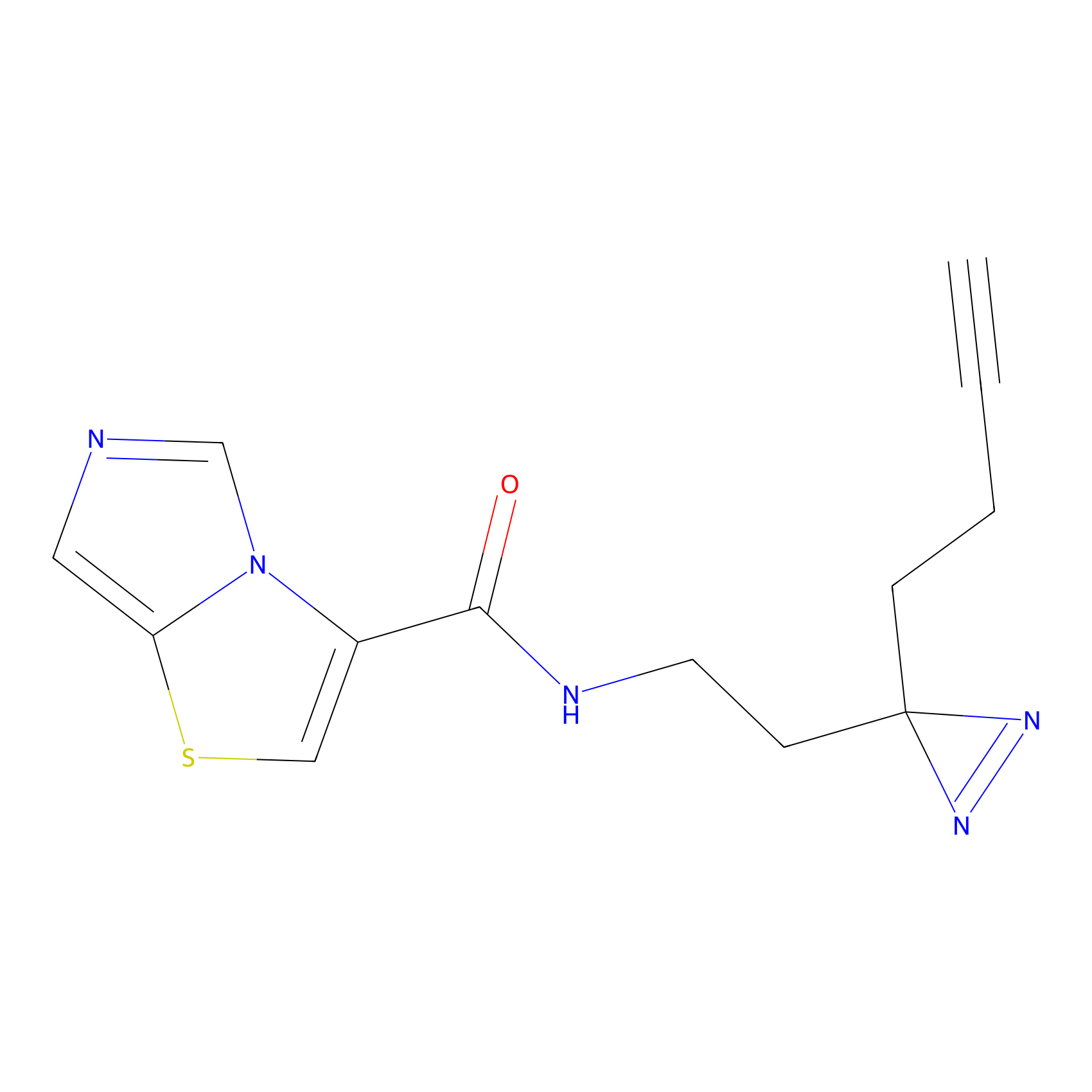 |
17.15 | LDD2022 | [9] | |
|
C362 Probe Info |
 |
31.78 | LDD2023 | [9] | |
|
C363 Probe Info |
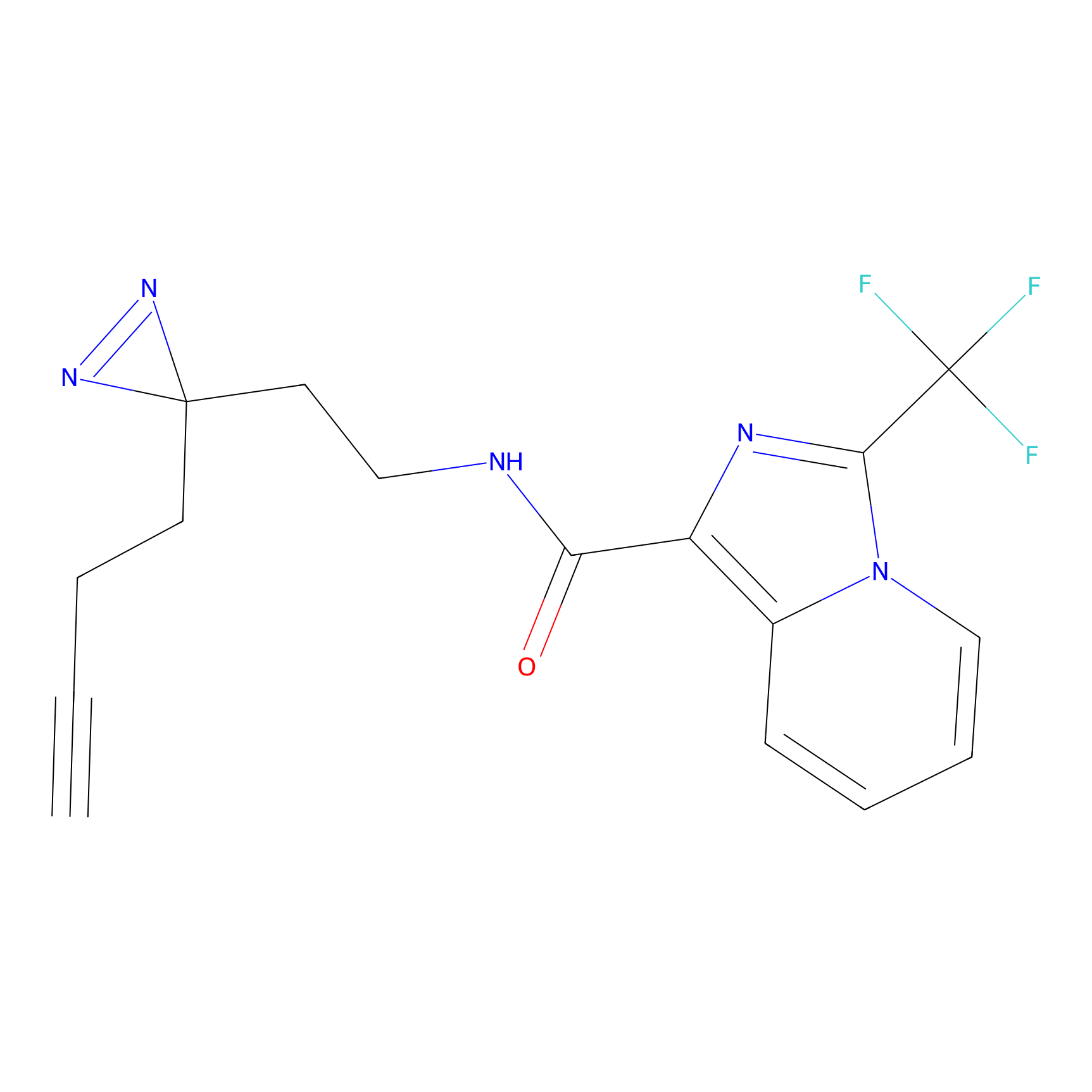 |
27.47 | LDD2024 | [9] | |
|
C364 Probe Info |
 |
20.11 | LDD2025 | [9] | |
|
C388 Probe Info |
 |
37.27 | LDD2047 | [9] | |
|
C403 Probe Info |
 |
19.97 | LDD2061 | [9] | |
|
C413 Probe Info |
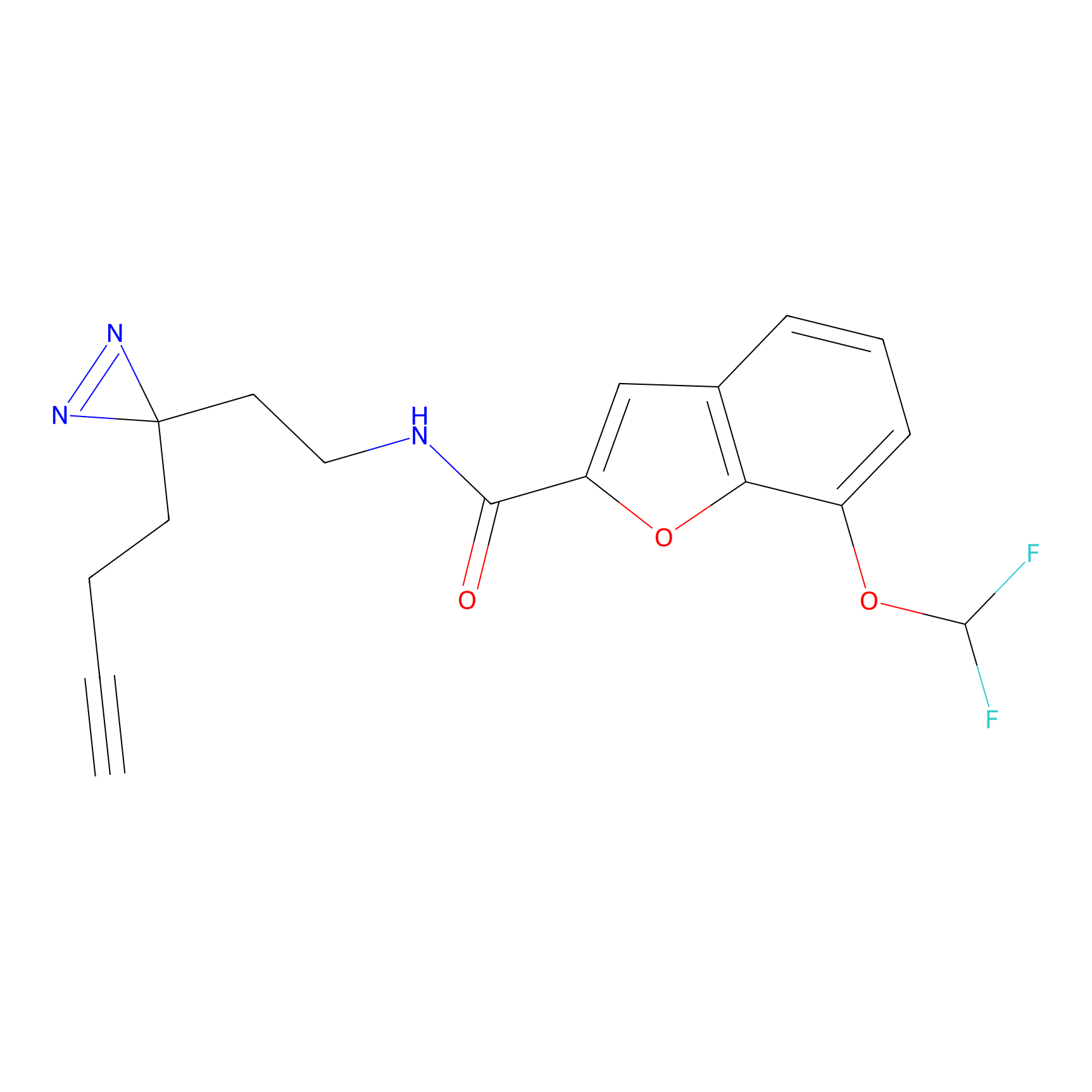 |
23.10 | LDD2069 | [9] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0214 | AC1 | HCT 116 | C121(0.90); C125(0.90); C423(1.00) | LDD0531 | [4] |
| LDCM0215 | AC10 | HCT 116 | C423(1.29) | LDD0532 | [4] |
| LDCM0216 | AC100 | HCT 116 | C121(0.84); C125(0.84) | LDD0533 | [4] |
| LDCM0217 | AC101 | HCT 116 | C121(0.79); C125(0.79) | LDD0534 | [4] |
| LDCM0218 | AC102 | HCT 116 | C121(0.75); C125(0.75) | LDD0535 | [4] |
| LDCM0219 | AC103 | HCT 116 | C121(0.89); C125(0.89) | LDD0536 | [4] |
| LDCM0220 | AC104 | HCT 116 | C121(0.84); C125(0.84) | LDD0537 | [4] |
| LDCM0221 | AC105 | HCT 116 | C121(0.87); C125(0.87) | LDD0538 | [4] |
| LDCM0222 | AC106 | HCT 116 | C121(0.89); C125(0.89) | LDD0539 | [4] |
| LDCM0223 | AC107 | HCT 116 | C121(1.12); C125(1.12) | LDD0540 | [4] |
| LDCM0224 | AC108 | HCT 116 | C121(0.60); C125(0.60) | LDD0541 | [4] |
| LDCM0225 | AC109 | HCT 116 | C121(0.71); C125(0.71) | LDD0542 | [4] |
| LDCM0226 | AC11 | HCT 116 | C423(1.11) | LDD0543 | [4] |
| LDCM0227 | AC110 | HCT 116 | C121(0.76); C125(0.76) | LDD0544 | [4] |
| LDCM0228 | AC111 | HCT 116 | C121(0.76); C125(0.76) | LDD0545 | [4] |
| LDCM0229 | AC112 | HCT 116 | C121(0.93); C125(0.93) | LDD0546 | [4] |
| LDCM0230 | AC113 | HCT 116 | C423(1.10) | LDD0547 | [4] |
| LDCM0231 | AC114 | HCT 116 | C423(1.07) | LDD0548 | [4] |
| LDCM0232 | AC115 | HCT 116 | C423(1.13) | LDD0549 | [4] |
| LDCM0233 | AC116 | HCT 116 | C423(0.99) | LDD0550 | [4] |
| LDCM0234 | AC117 | HCT 116 | C423(1.22) | LDD0551 | [4] |
| LDCM0235 | AC118 | HCT 116 | C423(1.19) | LDD0552 | [4] |
| LDCM0236 | AC119 | HCT 116 | C423(1.11) | LDD0553 | [4] |
| LDCM0237 | AC12 | HCT 116 | C423(0.72) | LDD0554 | [4] |
| LDCM0238 | AC120 | HCT 116 | C423(1.09) | LDD0555 | [4] |
| LDCM0239 | AC121 | HCT 116 | C423(1.04) | LDD0556 | [4] |
| LDCM0240 | AC122 | HCT 116 | C423(1.01) | LDD0557 | [4] |
| LDCM0241 | AC123 | HCT 116 | C423(1.21) | LDD0558 | [4] |
| LDCM0242 | AC124 | HCT 116 | C423(1.04) | LDD0559 | [4] |
| LDCM0243 | AC125 | HCT 116 | C423(0.96) | LDD0560 | [4] |
| LDCM0244 | AC126 | HCT 116 | C423(1.08) | LDD0561 | [4] |
| LDCM0245 | AC127 | HCT 116 | C423(1.16) | LDD0562 | [4] |
| LDCM0246 | AC128 | HCT 116 | C423(1.71) | LDD0563 | [4] |
| LDCM0247 | AC129 | HCT 116 | C423(1.28) | LDD0564 | [4] |
| LDCM0249 | AC130 | HCT 116 | C423(2.14) | LDD0566 | [4] |
| LDCM0250 | AC131 | HCT 116 | C423(1.14) | LDD0567 | [4] |
| LDCM0251 | AC132 | HCT 116 | C423(1.27) | LDD0568 | [4] |
| LDCM0252 | AC133 | HCT 116 | C423(1.49) | LDD0569 | [4] |
| LDCM0253 | AC134 | HCT 116 | C423(1.62) | LDD0570 | [4] |
| LDCM0254 | AC135 | HCT 116 | C423(1.82) | LDD0571 | [4] |
| LDCM0255 | AC136 | HCT 116 | C423(1.02) | LDD0572 | [4] |
| LDCM0256 | AC137 | HCT 116 | C423(0.92) | LDD0573 | [4] |
| LDCM0257 | AC138 | HCT 116 | C423(1.99) | LDD0574 | [4] |
| LDCM0258 | AC139 | HCT 116 | C423(1.87) | LDD0575 | [4] |
| LDCM0259 | AC14 | HCT 116 | C423(0.74) | LDD0576 | [4] |
| LDCM0260 | AC140 | HCT 116 | C423(1.63) | LDD0577 | [4] |
| LDCM0261 | AC141 | HCT 116 | C423(1.49) | LDD0578 | [4] |
| LDCM0262 | AC142 | HCT 116 | C423(1.14) | LDD0579 | [4] |
| LDCM0263 | AC143 | HCT 116 | C423(1.34) | LDD0580 | [4] |
| LDCM0264 | AC144 | HCT 116 | C423(0.95) | LDD0581 | [4] |
| LDCM0265 | AC145 | HCT 116 | C423(0.98) | LDD0582 | [4] |
| LDCM0266 | AC146 | HCT 116 | C423(1.13) | LDD0583 | [4] |
| LDCM0267 | AC147 | HCT 116 | C423(0.90) | LDD0584 | [4] |
| LDCM0268 | AC148 | HCT 116 | C423(1.03) | LDD0585 | [4] |
| LDCM0269 | AC149 | HCT 116 | C423(1.09) | LDD0586 | [4] |
| LDCM0270 | AC15 | HCT 116 | C423(0.82) | LDD0587 | [4] |
| LDCM0271 | AC150 | HCT 116 | C423(0.85) | LDD0588 | [4] |
| LDCM0272 | AC151 | HCT 116 | C423(1.04) | LDD0589 | [4] |
| LDCM0273 | AC152 | HCT 116 | C423(1.09) | LDD0590 | [4] |
| LDCM0274 | AC153 | HCT 116 | C423(1.60) | LDD0591 | [4] |
| LDCM0621 | AC154 | HCT 116 | C423(1.19) | LDD2158 | [4] |
| LDCM0622 | AC155 | HCT 116 | C423(1.06) | LDD2159 | [4] |
| LDCM0623 | AC156 | HCT 116 | C423(1.19) | LDD2160 | [4] |
| LDCM0624 | AC157 | HCT 116 | C423(1.36) | LDD2161 | [4] |
| LDCM0276 | AC17 | HCT 116 | C423(0.60) | LDD0593 | [4] |
| LDCM0277 | AC18 | HCT 116 | C423(0.97) | LDD0594 | [4] |
| LDCM0278 | AC19 | HCT 116 | C423(0.77) | LDD0595 | [4] |
| LDCM0279 | AC2 | HCT 116 | C121(0.99); C125(0.99); C423(1.15) | LDD0596 | [4] |
| LDCM0280 | AC20 | HCT 116 | C423(0.45) | LDD0597 | [4] |
| LDCM0281 | AC21 | HCT 116 | C423(0.52) | LDD0598 | [4] |
| LDCM0282 | AC22 | HCT 116 | C423(0.55) | LDD0599 | [4] |
| LDCM0283 | AC23 | HCT 116 | C423(0.56) | LDD0600 | [4] |
| LDCM0284 | AC24 | HCT 116 | C423(0.58) | LDD0601 | [4] |
| LDCM0285 | AC25 | HCT 116 | C423(1.67) | LDD0602 | [4] |
| LDCM0286 | AC26 | HCT 116 | C423(1.83) | LDD0603 | [4] |
| LDCM0287 | AC27 | HCT 116 | C423(1.92) | LDD0604 | [4] |
| LDCM0288 | AC28 | HCT 116 | C423(1.46) | LDD0605 | [4] |
| LDCM0289 | AC29 | HCT 116 | C423(1.78) | LDD0606 | [4] |
| LDCM0290 | AC3 | HCT 116 | C423(1.02); C121(1.06); C125(1.06) | LDD0607 | [4] |
| LDCM0291 | AC30 | HCT 116 | C423(1.55) | LDD0608 | [4] |
| LDCM0292 | AC31 | HCT 116 | C423(1.32) | LDD0609 | [4] |
| LDCM0293 | AC32 | HCT 116 | C423(1.51) | LDD0610 | [4] |
| LDCM0294 | AC33 | HCT 116 | C423(1.70) | LDD0611 | [4] |
| LDCM0295 | AC34 | HCT 116 | C423(1.58) | LDD0612 | [4] |
| LDCM0296 | AC35 | HCT 116 | C423(1.13) | LDD0613 | [4] |
| LDCM0297 | AC36 | HCT 116 | C423(0.73) | LDD0614 | [4] |
| LDCM0298 | AC37 | HCT 116 | C423(0.67) | LDD0615 | [4] |
| LDCM0299 | AC38 | HCT 116 | C423(0.71) | LDD0616 | [4] |
| LDCM0300 | AC39 | HCT 116 | C423(0.82) | LDD0617 | [4] |
| LDCM0301 | AC4 | HCT 116 | C121(1.28); C125(1.28); C423(1.71) | LDD0618 | [4] |
| LDCM0302 | AC40 | HCT 116 | C423(0.75) | LDD0619 | [4] |
| LDCM0303 | AC41 | HCT 116 | C423(0.79) | LDD0620 | [4] |
| LDCM0304 | AC42 | HCT 116 | C423(0.85) | LDD0621 | [4] |
| LDCM0305 | AC43 | HCT 116 | C423(0.77) | LDD0622 | [4] |
| LDCM0306 | AC44 | HCT 116 | C423(0.59) | LDD0623 | [4] |
| LDCM0307 | AC45 | HCT 116 | C423(0.70) | LDD0624 | [4] |
| LDCM0308 | AC46 | HCT 116 | C423(1.17) | LDD0625 | [4] |
| LDCM0309 | AC47 | HCT 116 | C423(0.75) | LDD0626 | [4] |
| LDCM0310 | AC48 | HCT 116 | C423(1.23) | LDD0627 | [4] |
| LDCM0311 | AC49 | HCT 116 | C423(1.26) | LDD0628 | [4] |
| LDCM0312 | AC5 | HCT 116 | C121(1.26); C125(1.26); C423(1.44) | LDD0629 | [4] |
| LDCM0313 | AC50 | HCT 116 | C423(1.31) | LDD0630 | [4] |
| LDCM0314 | AC51 | HCT 116 | C423(0.94) | LDD0631 | [4] |
| LDCM0315 | AC52 | HCT 116 | C423(0.98) | LDD0632 | [4] |
| LDCM0316 | AC53 | HCT 116 | C423(0.85) | LDD0633 | [4] |
| LDCM0317 | AC54 | HCT 116 | C423(1.64) | LDD0634 | [4] |
| LDCM0318 | AC55 | HCT 116 | C423(0.95) | LDD0635 | [4] |
| LDCM0319 | AC56 | HCT 116 | C423(1.16) | LDD0636 | [4] |
| LDCM0320 | AC57 | HCT 116 | C423(1.11) | LDD0637 | [4] |
| LDCM0321 | AC58 | HCT 116 | C423(0.88) | LDD0638 | [4] |
| LDCM0322 | AC59 | HCT 116 | C423(1.02) | LDD0639 | [4] |
| LDCM0323 | AC6 | HCT 116 | C423(0.80) | LDD0640 | [4] |
| LDCM0324 | AC60 | HCT 116 | C423(1.21) | LDD0641 | [4] |
| LDCM0325 | AC61 | HCT 116 | C423(0.95) | LDD0642 | [4] |
| LDCM0326 | AC62 | HCT 116 | C423(1.11) | LDD0643 | [4] |
| LDCM0327 | AC63 | HCT 116 | C423(0.93) | LDD0644 | [4] |
| LDCM0328 | AC64 | HCT 116 | C423(1.23) | LDD0645 | [4] |
| LDCM0329 | AC65 | HCT 116 | C423(1.08) | LDD0646 | [4] |
| LDCM0330 | AC66 | HCT 116 | C423(0.90) | LDD0647 | [4] |
| LDCM0331 | AC67 | HCT 116 | C423(1.01) | LDD0648 | [4] |
| LDCM0334 | AC7 | HCT 116 | C423(0.59) | LDD0651 | [4] |
| LDCM0345 | AC8 | HCT 116 | C423(0.98) | LDD0662 | [4] |
| LDCM0349 | AC83 | HCT 116 | C121(1.15); C125(1.15) | LDD0666 | [4] |
| LDCM0350 | AC84 | HCT 116 | C121(1.47); C125(1.47) | LDD0667 | [4] |
| LDCM0351 | AC85 | HCT 116 | C121(1.45); C125(1.45) | LDD0668 | [4] |
| LDCM0352 | AC86 | HCT 116 | C121(1.27); C125(1.27) | LDD0669 | [4] |
| LDCM0353 | AC87 | HCT 116 | C121(0.98); C125(0.98) | LDD0670 | [4] |
| LDCM0354 | AC88 | HCT 116 | C121(0.93); C125(0.93) | LDD0671 | [4] |
| LDCM0355 | AC89 | HCT 116 | C121(1.84); C125(1.84) | LDD0672 | [4] |
| LDCM0357 | AC90 | HCT 116 | C121(1.22); C125(1.22) | LDD0674 | [4] |
| LDCM0358 | AC91 | HCT 116 | C121(1.37); C125(1.37) | LDD0675 | [4] |
| LDCM0359 | AC92 | HCT 116 | C121(1.81); C125(1.81) | LDD0676 | [4] |
| LDCM0360 | AC93 | HCT 116 | C121(1.18); C125(1.18) | LDD0677 | [4] |
| LDCM0361 | AC94 | HCT 116 | C121(1.06); C125(1.06) | LDD0678 | [4] |
| LDCM0362 | AC95 | HCT 116 | C121(1.39); C125(1.39) | LDD0679 | [4] |
| LDCM0363 | AC96 | HCT 116 | C121(1.28); C125(1.28) | LDD0680 | [4] |
| LDCM0364 | AC97 | HCT 116 | C121(1.33); C125(1.33) | LDD0681 | [4] |
| LDCM0365 | AC98 | HCT 116 | C121(0.87); C125(0.87) | LDD0682 | [4] |
| LDCM0366 | AC99 | HCT 116 | C121(0.79); C125(0.79) | LDD0683 | [4] |
| LDCM0248 | AKOS034007472 | HCT 116 | C423(1.06) | LDD0565 | [4] |
| LDCM0356 | AKOS034007680 | HCT 116 | C423(1.76) | LDD0673 | [4] |
| LDCM0275 | AKOS034007705 | HCT 116 | C423(1.09) | LDD0592 | [4] |
| LDCM0020 | ARS-1620 | HCC44 | C317(1.04) | LDD2171 | [4] |
| LDCM0632 | CL-Sc | Hep-G2 | C317(0.99) | LDD2227 | [8] |
| LDCM0367 | CL1 | HCT 116 | C423(1.36) | LDD0684 | [4] |
| LDCM0368 | CL10 | HCT 116 | C423(1.49) | LDD0685 | [4] |
| LDCM0369 | CL100 | HCT 116 | C423(1.02); C121(1.08); C125(1.08) | LDD0686 | [4] |
| LDCM0370 | CL101 | HCT 116 | C423(1.04) | LDD0687 | [4] |
| LDCM0371 | CL102 | HCT 116 | C423(0.65) | LDD0688 | [4] |
| LDCM0372 | CL103 | HCT 116 | C423(0.90) | LDD0689 | [4] |
| LDCM0373 | CL104 | HCT 116 | C423(0.84) | LDD0690 | [4] |
| LDCM0374 | CL105 | HCT 116 | C423(0.46) | LDD0691 | [4] |
| LDCM0375 | CL106 | HCT 116 | C423(0.78) | LDD0692 | [4] |
| LDCM0376 | CL107 | HCT 116 | C423(0.88) | LDD0693 | [4] |
| LDCM0377 | CL108 | HCT 116 | C423(0.61) | LDD0694 | [4] |
| LDCM0378 | CL109 | HCT 116 | C423(0.82) | LDD0695 | [4] |
| LDCM0379 | CL11 | HCT 116 | C423(1.08) | LDD0696 | [4] |
| LDCM0380 | CL110 | HCT 116 | C423(0.70) | LDD0697 | [4] |
| LDCM0381 | CL111 | HCT 116 | C423(0.84) | LDD0698 | [4] |
| LDCM0382 | CL112 | HCT 116 | C423(0.94) | LDD0699 | [4] |
| LDCM0383 | CL113 | HCT 116 | C423(1.71) | LDD0700 | [4] |
| LDCM0384 | CL114 | HCT 116 | C423(1.48) | LDD0701 | [4] |
| LDCM0385 | CL115 | HCT 116 | C423(1.36) | LDD0702 | [4] |
| LDCM0386 | CL116 | HCT 116 | C423(2.04) | LDD0703 | [4] |
| LDCM0387 | CL117 | HCT 116 | C423(0.62) | LDD0704 | [4] |
| LDCM0388 | CL118 | HCT 116 | C423(0.86) | LDD0705 | [4] |
| LDCM0389 | CL119 | HCT 116 | C423(0.77) | LDD0706 | [4] |
| LDCM0390 | CL12 | HCT 116 | C423(1.47) | LDD0707 | [4] |
| LDCM0391 | CL120 | HCT 116 | C423(0.76) | LDD0708 | [4] |
| LDCM0392 | CL121 | HCT 116 | C423(0.76) | LDD0709 | [4] |
| LDCM0393 | CL122 | HCT 116 | C423(0.68) | LDD0710 | [4] |
| LDCM0394 | CL123 | HCT 116 | C423(0.66) | LDD0711 | [4] |
| LDCM0395 | CL124 | HCT 116 | C423(1.01) | LDD0712 | [4] |
| LDCM0396 | CL125 | HCT 116 | C423(0.95) | LDD0713 | [4] |
| LDCM0397 | CL126 | HCT 116 | C423(0.66) | LDD0714 | [4] |
| LDCM0398 | CL127 | HCT 116 | C423(0.90) | LDD0715 | [4] |
| LDCM0399 | CL128 | HCT 116 | C423(0.87) | LDD0716 | [4] |
| LDCM0400 | CL13 | HCT 116 | C423(1.25) | LDD0717 | [4] |
| LDCM0401 | CL14 | HCT 116 | C423(1.31) | LDD0718 | [4] |
| LDCM0402 | CL15 | HCT 116 | C423(1.15) | LDD0719 | [4] |
| LDCM0403 | CL16 | HCT 116 | C423(1.54) | LDD0720 | [4] |
| LDCM0404 | CL17 | HCT 116 | C423(0.81) | LDD0721 | [4] |
| LDCM0405 | CL18 | HCT 116 | C423(0.80) | LDD0722 | [4] |
| LDCM0406 | CL19 | HCT 116 | C423(0.89) | LDD0723 | [4] |
| LDCM0407 | CL2 | HCT 116 | C423(1.11) | LDD0724 | [4] |
| LDCM0408 | CL20 | HCT 116 | C423(1.08) | LDD0725 | [4] |
| LDCM0409 | CL21 | HCT 116 | C423(0.84) | LDD0726 | [4] |
| LDCM0410 | CL22 | HCT 116 | C423(0.83) | LDD0727 | [4] |
| LDCM0411 | CL23 | HCT 116 | C423(1.06) | LDD0728 | [4] |
| LDCM0412 | CL24 | HCT 116 | C423(1.03) | LDD0729 | [4] |
| LDCM0413 | CL25 | HCT 116 | C423(0.87) | LDD0730 | [4] |
| LDCM0414 | CL26 | HCT 116 | C423(1.32) | LDD0731 | [4] |
| LDCM0415 | CL27 | HCT 116 | C423(1.01) | LDD0732 | [4] |
| LDCM0416 | CL28 | HCT 116 | C423(1.00) | LDD0733 | [4] |
| LDCM0417 | CL29 | HCT 116 | C423(0.94) | LDD0734 | [4] |
| LDCM0418 | CL3 | HCT 116 | C423(1.06) | LDD0735 | [4] |
| LDCM0419 | CL30 | HCT 116 | C423(1.15) | LDD0736 | [4] |
| LDCM0420 | CL31 | HCT 116 | C121(1.19); C125(1.19) | LDD0737 | [4] |
| LDCM0421 | CL32 | HCT 116 | C121(0.98); C125(0.98) | LDD0738 | [4] |
| LDCM0422 | CL33 | HCT 116 | C121(1.03); C125(1.03) | LDD0739 | [4] |
| LDCM0423 | CL34 | HCT 116 | C121(1.06); C125(1.06) | LDD0740 | [4] |
| LDCM0424 | CL35 | HCT 116 | C121(1.06); C125(1.06) | LDD0741 | [4] |
| LDCM0425 | CL36 | HCT 116 | C121(1.30); C125(1.30) | LDD0742 | [4] |
| LDCM0426 | CL37 | HCT 116 | C121(1.24); C125(1.24) | LDD0743 | [4] |
| LDCM0428 | CL39 | HCT 116 | C121(0.95); C125(0.95) | LDD0745 | [4] |
| LDCM0429 | CL4 | HCT 116 | C423(1.13) | LDD0746 | [4] |
| LDCM0430 | CL40 | HCT 116 | C121(0.78); C125(0.78) | LDD0747 | [4] |
| LDCM0431 | CL41 | HCT 116 | C121(1.19); C125(1.19) | LDD0748 | [4] |
| LDCM0432 | CL42 | HCT 116 | C121(1.23); C125(1.23) | LDD0749 | [4] |
| LDCM0433 | CL43 | HCT 116 | C121(1.32); C125(1.32) | LDD0750 | [4] |
| LDCM0434 | CL44 | HCT 116 | C121(1.17); C125(1.17) | LDD0751 | [4] |
| LDCM0435 | CL45 | HCT 116 | C121(1.12); C125(1.12) | LDD0752 | [4] |
| LDCM0436 | CL46 | HEK-293T | C125(1.26) | LDD1034 | [4] |
| LDCM0437 | CL47 | HEK-293T | C125(1.14) | LDD1035 | [4] |
| LDCM0438 | CL48 | HEK-293T | C125(0.93) | LDD1036 | [4] |
| LDCM0439 | CL49 | HEK-293T | C125(1.13) | LDD1037 | [4] |
| LDCM0440 | CL5 | HCT 116 | C423(1.02) | LDD0757 | [4] |
| LDCM0441 | CL50 | HEK-293T | C125(1.37) | LDD1039 | [4] |
| LDCM0442 | CL51 | HEK-293T | C125(1.31) | LDD1040 | [4] |
| LDCM0443 | CL52 | HEK-293T | C125(0.99) | LDD1041 | [4] |
| LDCM0444 | CL53 | HEK-293T | C125(1.15) | LDD1042 | [4] |
| LDCM0445 | CL54 | HEK-293T | C125(1.32) | LDD1043 | [4] |
| LDCM0446 | CL55 | HEK-293T | C125(1.16) | LDD1044 | [4] |
| LDCM0447 | CL56 | HEK-293T | C125(0.93) | LDD1045 | [4] |
| LDCM0448 | CL57 | HEK-293T | C125(1.37) | LDD1046 | [4] |
| LDCM0449 | CL58 | HEK-293T | C125(1.05) | LDD1047 | [4] |
| LDCM0450 | CL59 | HEK-293T | C125(1.22) | LDD1048 | [4] |
| LDCM0451 | CL6 | HCT 116 | C423(1.20) | LDD0768 | [4] |
| LDCM0452 | CL60 | HEK-293T | C125(1.37) | LDD1050 | [4] |
| LDCM0453 | CL61 | HCT 116 | C423(0.81) | LDD0770 | [4] |
| LDCM0454 | CL62 | HCT 116 | C423(0.94) | LDD0771 | [4] |
| LDCM0455 | CL63 | HCT 116 | C423(1.26) | LDD0772 | [4] |
| LDCM0456 | CL64 | HCT 116 | C423(0.71) | LDD0773 | [4] |
| LDCM0457 | CL65 | HCT 116 | C423(1.05) | LDD0774 | [4] |
| LDCM0458 | CL66 | HCT 116 | C423(1.20) | LDD0775 | [4] |
| LDCM0459 | CL67 | HCT 116 | C423(0.89) | LDD0776 | [4] |
| LDCM0460 | CL68 | HCT 116 | C423(0.73) | LDD0777 | [4] |
| LDCM0461 | CL69 | HCT 116 | C423(1.56) | LDD0778 | [4] |
| LDCM0462 | CL7 | HCT 116 | C423(1.16) | LDD0779 | [4] |
| LDCM0463 | CL70 | HCT 116 | C423(1.05) | LDD0780 | [4] |
| LDCM0464 | CL71 | HCT 116 | C423(0.95) | LDD0781 | [4] |
| LDCM0465 | CL72 | HCT 116 | C423(0.92) | LDD0782 | [4] |
| LDCM0466 | CL73 | HCT 116 | C423(0.87) | LDD0783 | [4] |
| LDCM0467 | CL74 | HCT 116 | C423(1.24) | LDD0784 | [4] |
| LDCM0469 | CL76 | HCT 116 | C121(1.09); C125(1.09); C423(1.13) | LDD0786 | [4] |
| LDCM0470 | CL77 | HCT 116 | C121(0.84); C125(0.84); C423(0.79) | LDD0787 | [4] |
| LDCM0471 | CL78 | HCT 116 | C121(1.09); C125(1.09); C423(0.72) | LDD0788 | [4] |
| LDCM0472 | CL79 | HCT 116 | C121(1.21); C125(1.21); C423(1.19) | LDD0789 | [4] |
| LDCM0473 | CL8 | HCT 116 | C423(1.25) | LDD0790 | [4] |
| LDCM0474 | CL80 | HCT 116 | C121(0.84); C125(0.84); C423(0.79) | LDD0791 | [4] |
| LDCM0475 | CL81 | HCT 116 | C121(1.14); C125(1.14); C423(0.91) | LDD0792 | [4] |
| LDCM0476 | CL82 | HCT 116 | C121(1.56); C125(1.56); C423(1.10) | LDD0793 | [4] |
| LDCM0477 | CL83 | HCT 116 | C121(1.26); C125(1.26); C423(0.92) | LDD0794 | [4] |
| LDCM0478 | CL84 | HCT 116 | C121(1.47); C125(1.47); C423(0.99) | LDD0795 | [4] |
| LDCM0479 | CL85 | HCT 116 | C121(1.05); C125(1.05); C423(1.40) | LDD0796 | [4] |
| LDCM0480 | CL86 | HCT 116 | C121(1.12); C125(1.12); C423(1.23) | LDD0797 | [4] |
| LDCM0481 | CL87 | HCT 116 | C121(1.12); C125(1.12); C423(0.91) | LDD0798 | [4] |
| LDCM0482 | CL88 | HCT 116 | C121(1.46); C125(1.46); C423(1.21) | LDD0799 | [4] |
| LDCM0483 | CL89 | HCT 116 | C121(1.97); C125(1.97); C423(1.02) | LDD0800 | [4] |
| LDCM0484 | CL9 | HCT 116 | C423(1.16) | LDD0801 | [4] |
| LDCM0485 | CL90 | HCT 116 | C121(0.92); C125(0.92); C423(1.50) | LDD0802 | [4] |
| LDCM0486 | CL91 | HCT 116 | C121(1.00); C125(1.00); C423(1.15) | LDD0803 | [4] |
| LDCM0487 | CL92 | HCT 116 | C121(0.76); C125(0.76); C423(0.74) | LDD0804 | [4] |
| LDCM0488 | CL93 | HCT 116 | C121(0.77); C125(0.77); C423(0.98) | LDD0805 | [4] |
| LDCM0489 | CL94 | HCT 116 | C121(0.82); C125(0.82); C423(1.48) | LDD0806 | [4] |
| LDCM0490 | CL95 | HCT 116 | C121(0.87); C125(0.87); C423(0.98) | LDD0807 | [4] |
| LDCM0491 | CL96 | HCT 116 | C121(1.08); C125(1.08); C423(0.94) | LDD0808 | [4] |
| LDCM0492 | CL97 | HCT 116 | C121(0.99); C125(0.99); C423(1.04) | LDD0809 | [4] |
| LDCM0493 | CL98 | HCT 116 | C121(0.98); C125(0.98); C423(1.20) | LDD0810 | [4] |
| LDCM0494 | CL99 | HCT 116 | C121(1.06); C125(1.06); C423(1.20) | LDD0811 | [4] |
| LDCM0495 | E2913 | HEK-293T | C423(1.01); C317(1.04); C125(1.01) | LDD1698 | [10] |
| LDCM0468 | Fragment33 | HCT 116 | C423(1.09) | LDD0785 | [4] |
| LDCM0427 | Fragment51 | HCT 116 | C121(1.24); C125(1.24) | LDD0744 | [4] |
| LDCM0022 | KB02 | HEK-293T | C423(0.89); C317(0.98) | LDD1492 | [10] |
| LDCM0023 | KB03 | HEK-293T | C423(0.95); C317(1.05) | LDD1497 | [10] |
| LDCM0024 | KB05 | G361 | C423(2.22) | LDD3311 | [11] |
| LDCM0014 | Panhematin | HEK-293T | 3.38 | LDD0062 | [3] |
| LDCM0021 | THZ1 | HCT 116 | C317(1.04) | LDD2173 | [4] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| AP-1 complex subunit mu-1 (AP1M1) | Adaptor complexes medium subunit family | Q9BXS5 | |||
| Sodium channel modifier 1 (SCNM1) | . | Q9BWG6 | |||
Transcription factor
Other
References
