Details of the Target
General Information of Target
| Target ID | LDTP06275 | |||||
|---|---|---|---|---|---|---|
| Target Name | Lysosomal-associated transmembrane protein 4A (LAPTM4A) | |||||
| Gene Name | LAPTM4A | |||||
| Gene ID | 9741 | |||||
| Synonyms |
KIAA0108; LAPTM4; MBNT; MTRP; Lysosomal-associated transmembrane protein 4A; Golgi 4-transmembrane-spanning transporter MTP |
|||||
| 3D Structure | ||||||
| Sequence |
MVSMSFKRNRSDRFYSTRCCGCCHVRTGTIILGTWYMVVNLLMAILLTVEVTHPNSMPAV
NIQYEVIGNYYSSERMADNACVLFAVSVLMFIISSMLVYGAISYQVGWLIPFFCYRLFDF VLSCLVAISSLTYLPRIKEYLDQLPDFPYKDDLLALDSSCLLFIVLVFFALFIIFKAYLI NCVWNCYKYINNRNVPEIAVYPAFEAPPQYVLPTYEMAVKMPEKEPPPPYLPA |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
LAPTM4/LAPTM5 transporter family
|
|||||
| Subcellular location |
Endomembrane system
|
|||||
| Function | May function in the transport of nucleosides and/or nucleoside derivatives between the cytosol and the lumen of an intracellular membrane-bound compartment. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FBPP2 Probe Info |
 |
5.05 | LDD0318 | [1] | |
|
TG42 Probe Info |
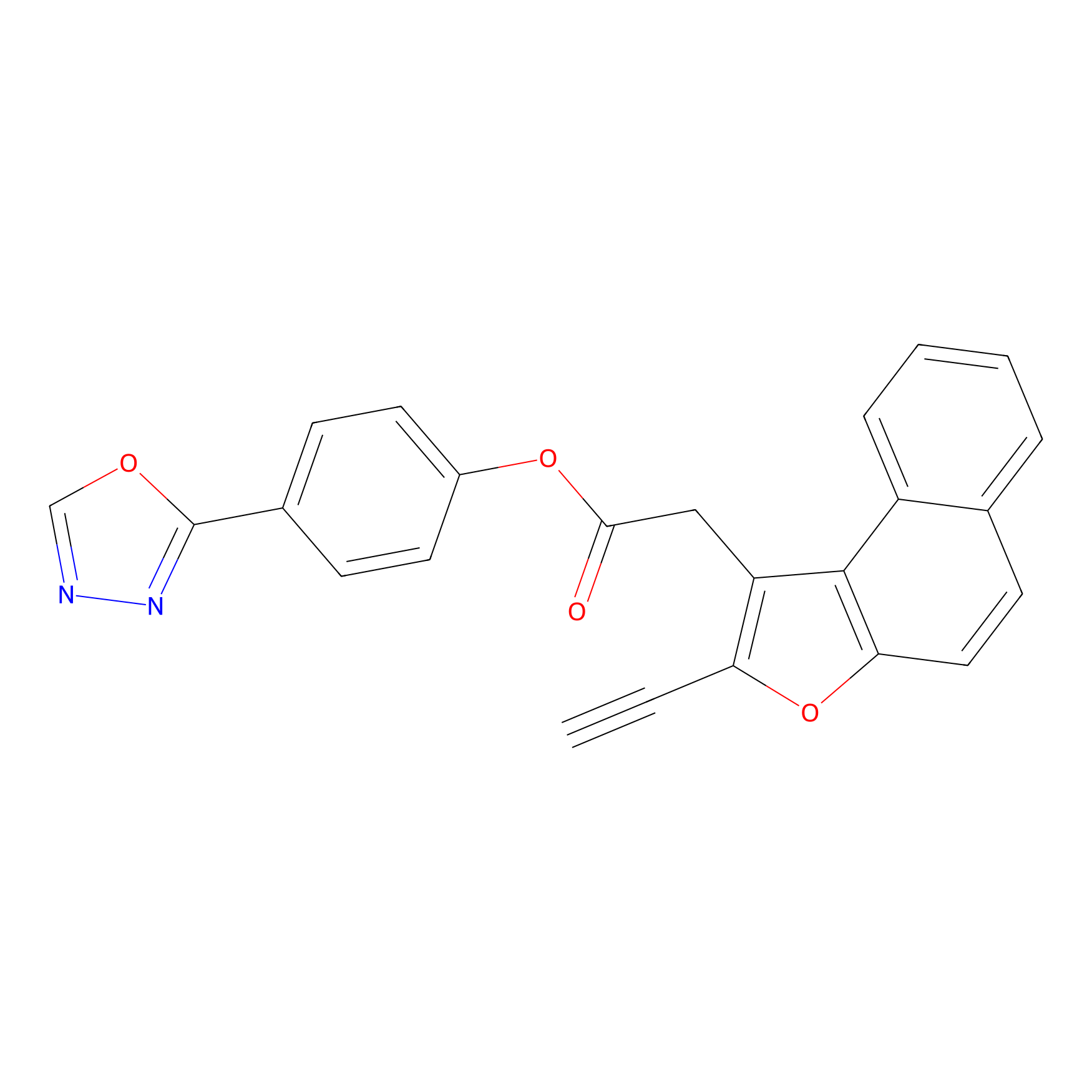 |
7.37 | LDD0326 | [2] | |
|
STPyne Probe Info |
 |
K7(6.17) | LDD0277 | [3] | |
|
Curcusone 37 Probe Info |
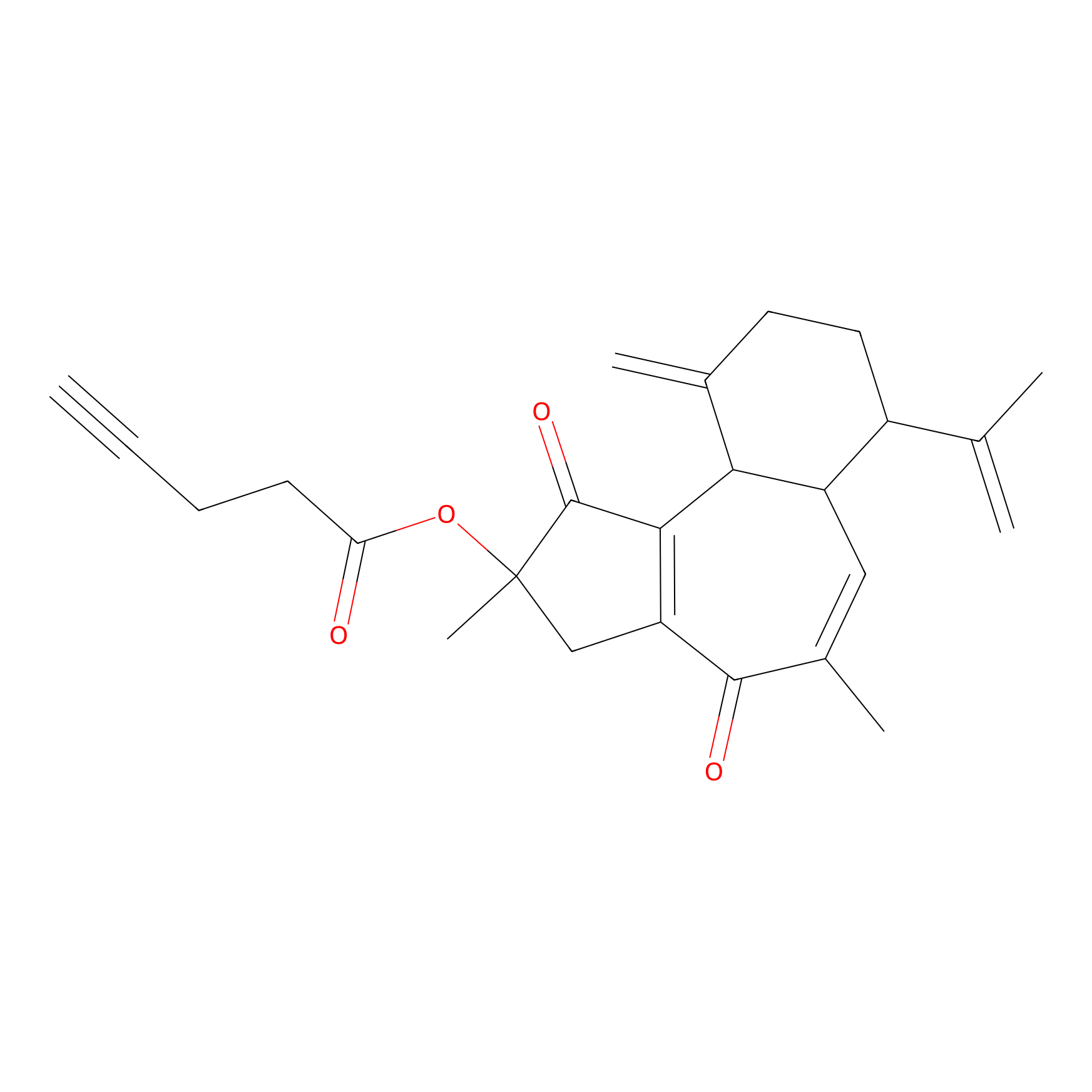 |
5.48 | LDD0188 | [4] | |
|
Jackson_14 Probe Info |
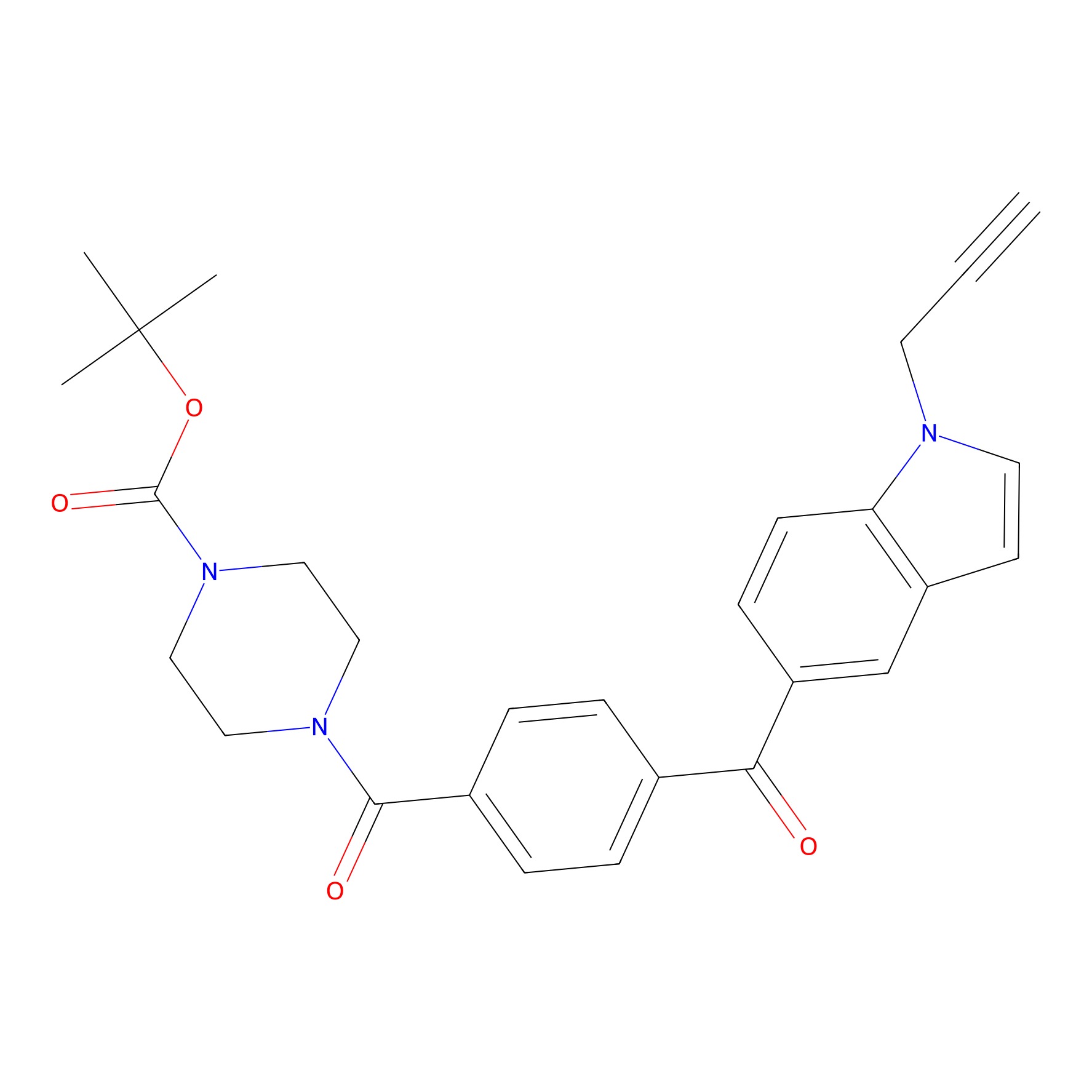 |
2.08 | LDD0123 | [5] | |
|
DBIA Probe Info |
 |
C19(0.84) | LDD1512 | [6] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C049 Probe Info |
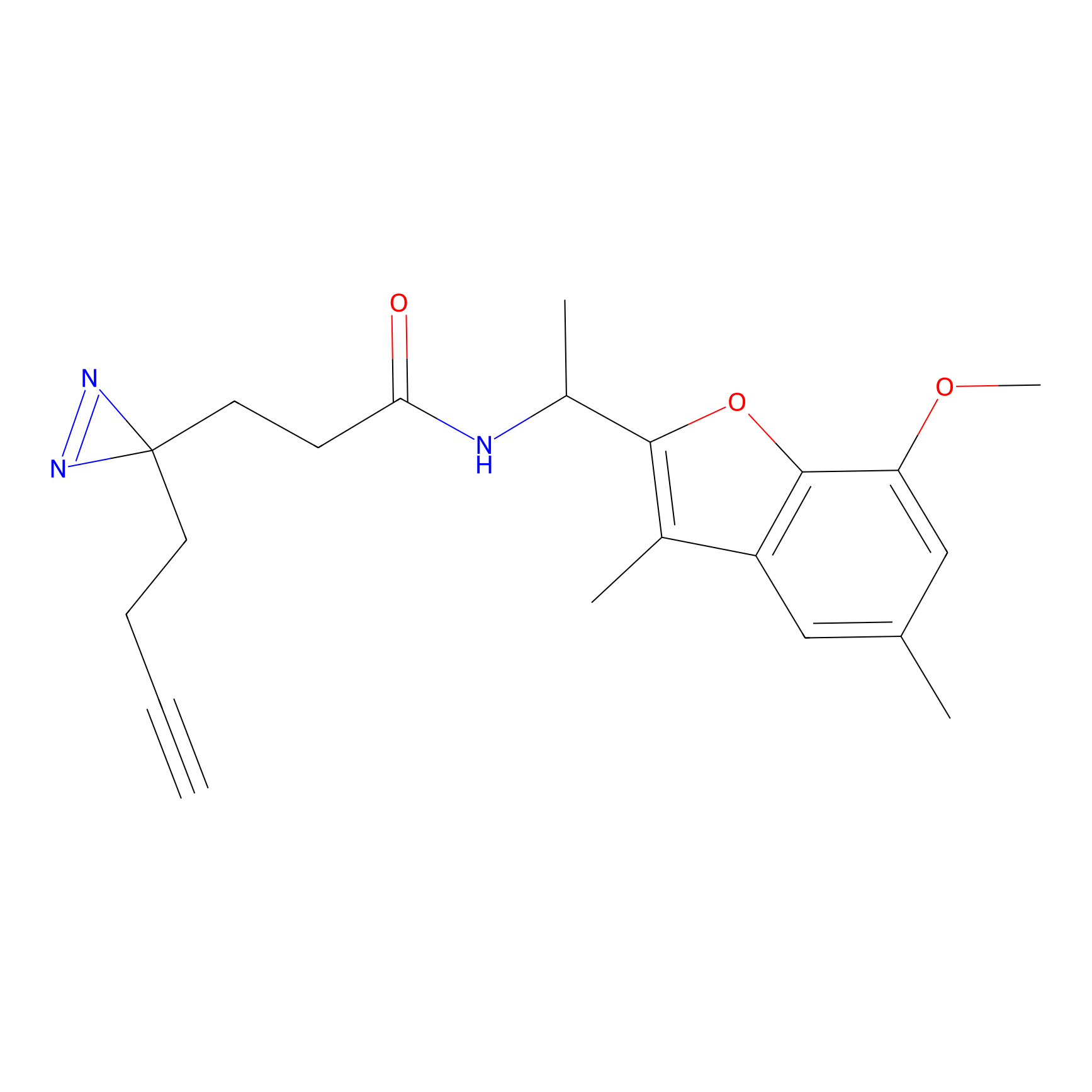 |
10.20 | LDD1747 | [7] | |
|
C055 Probe Info |
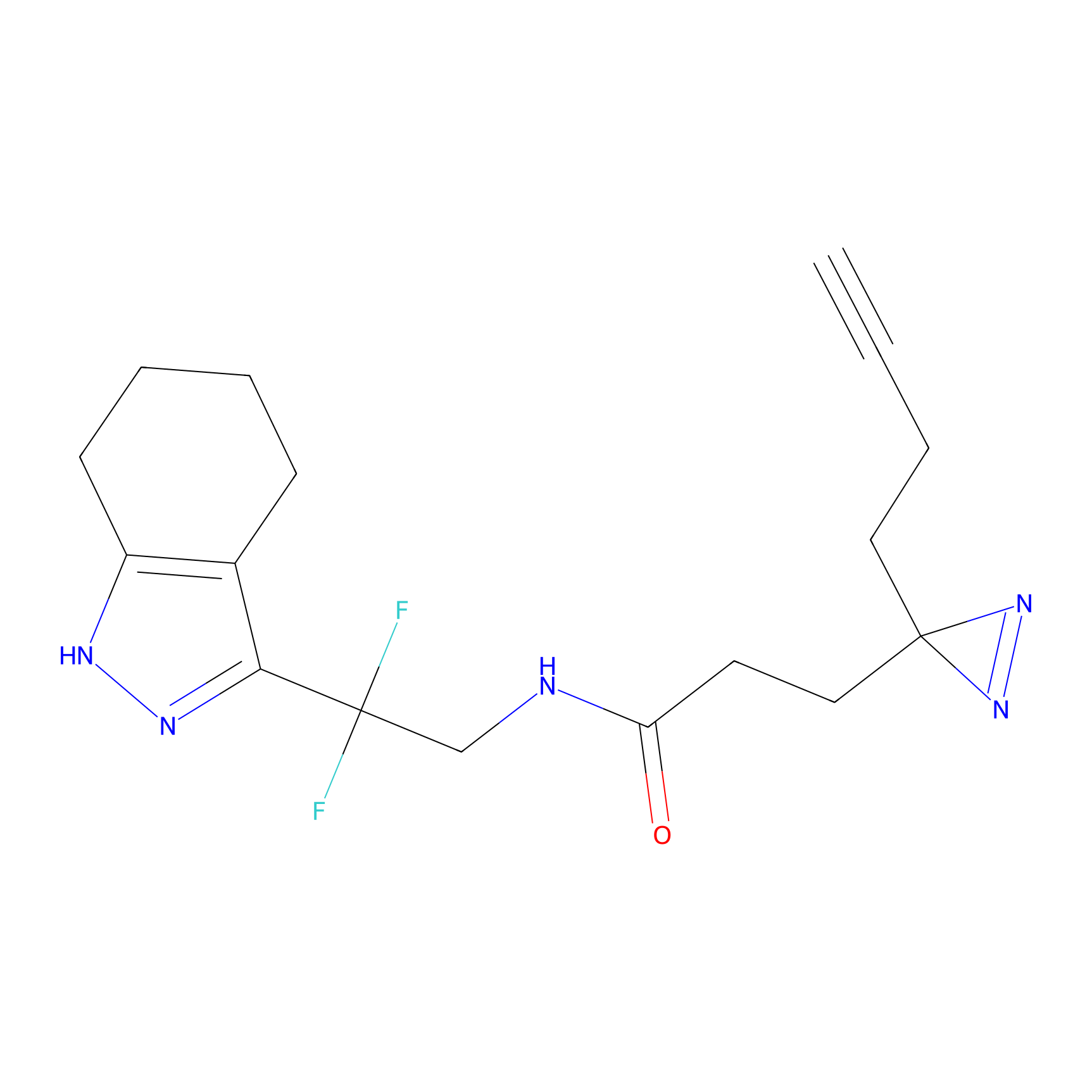 |
15.89 | LDD1752 | [7] | |
|
C056 Probe Info |
 |
25.28 | LDD1753 | [7] | |
|
C285 Probe Info |
 |
34.06 | LDD1955 | [7] | |
|
C286 Probe Info |
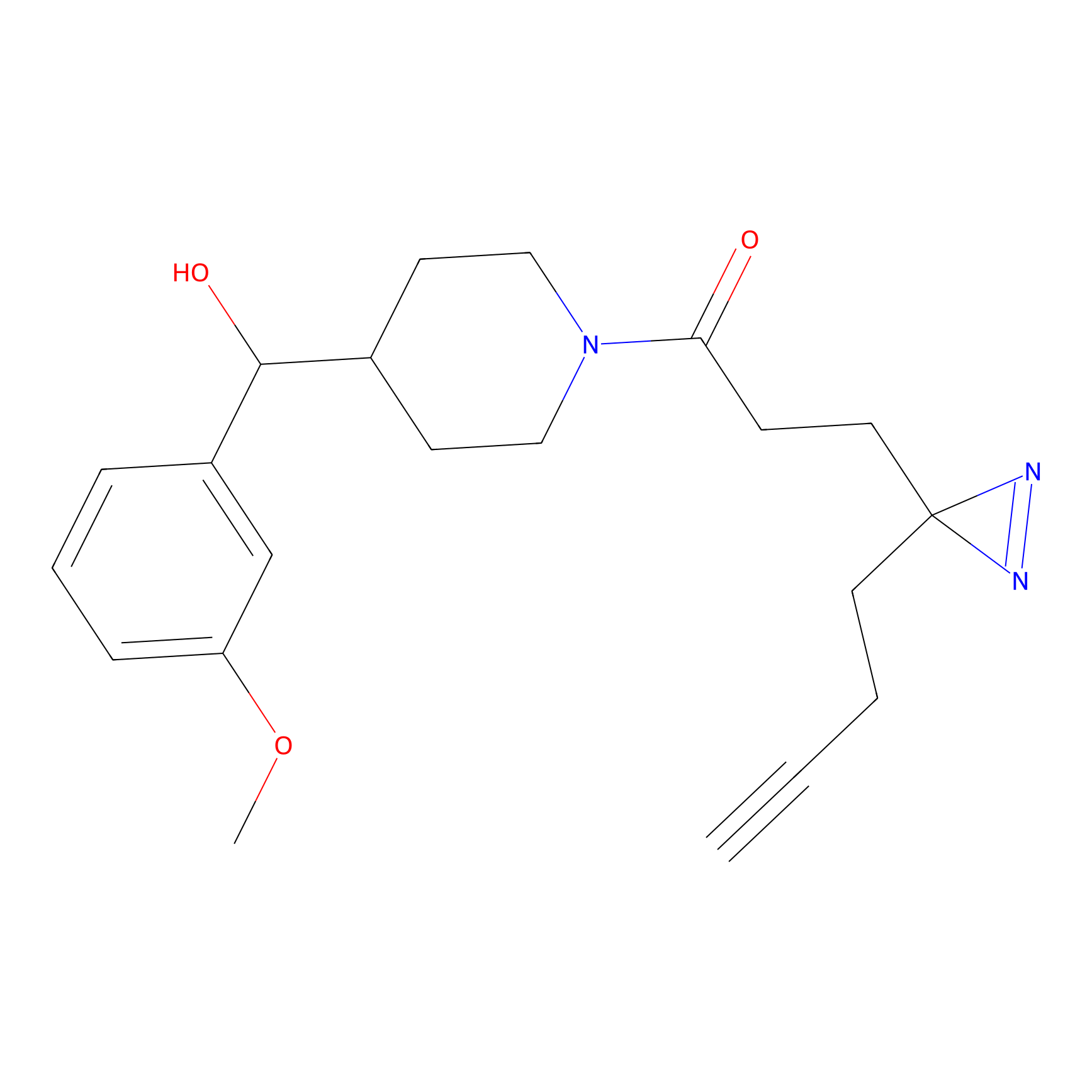 |
5.86 | LDD1956 | [7] | |
|
C287 Probe Info |
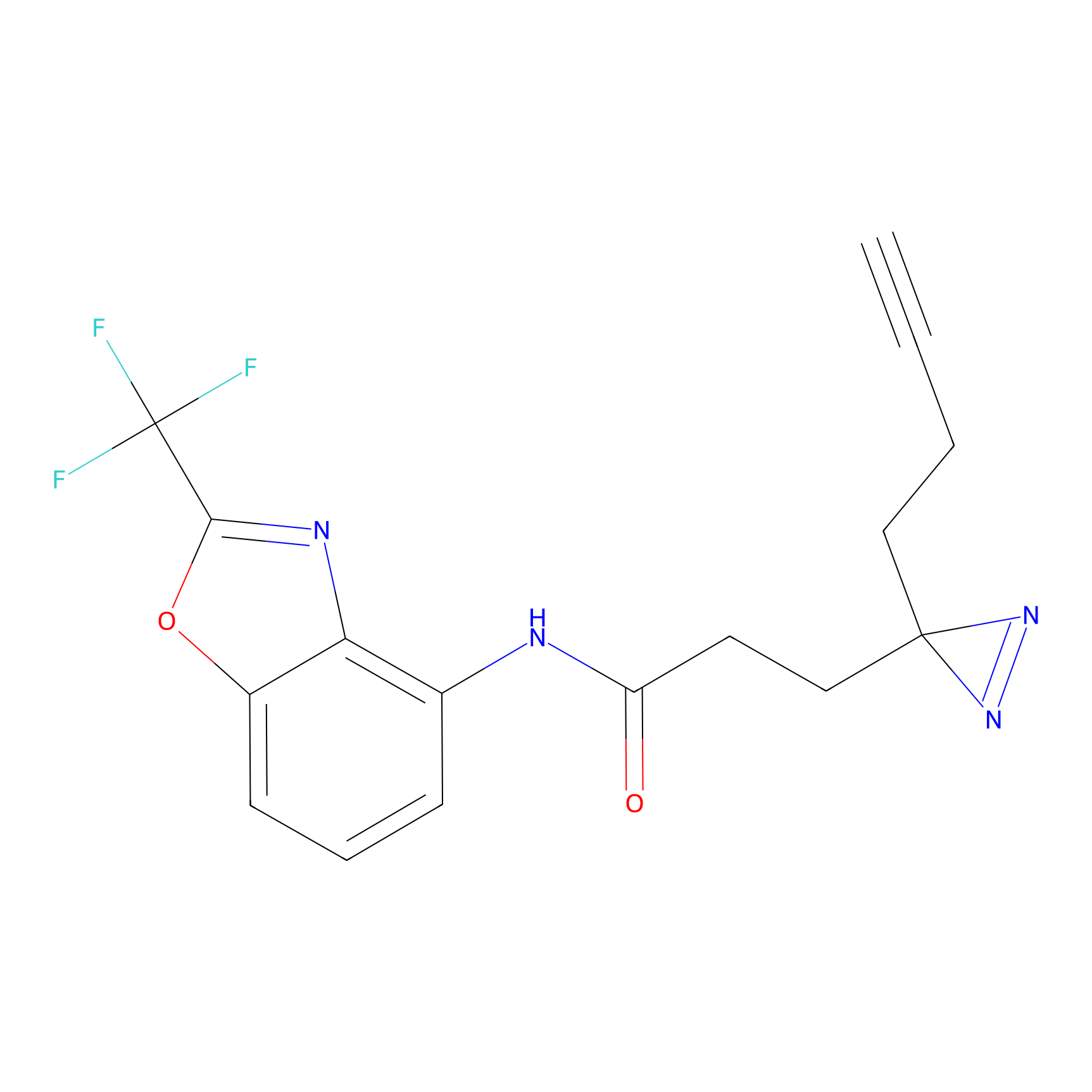 |
31.56 | LDD1957 | [7] | |
|
C288 Probe Info |
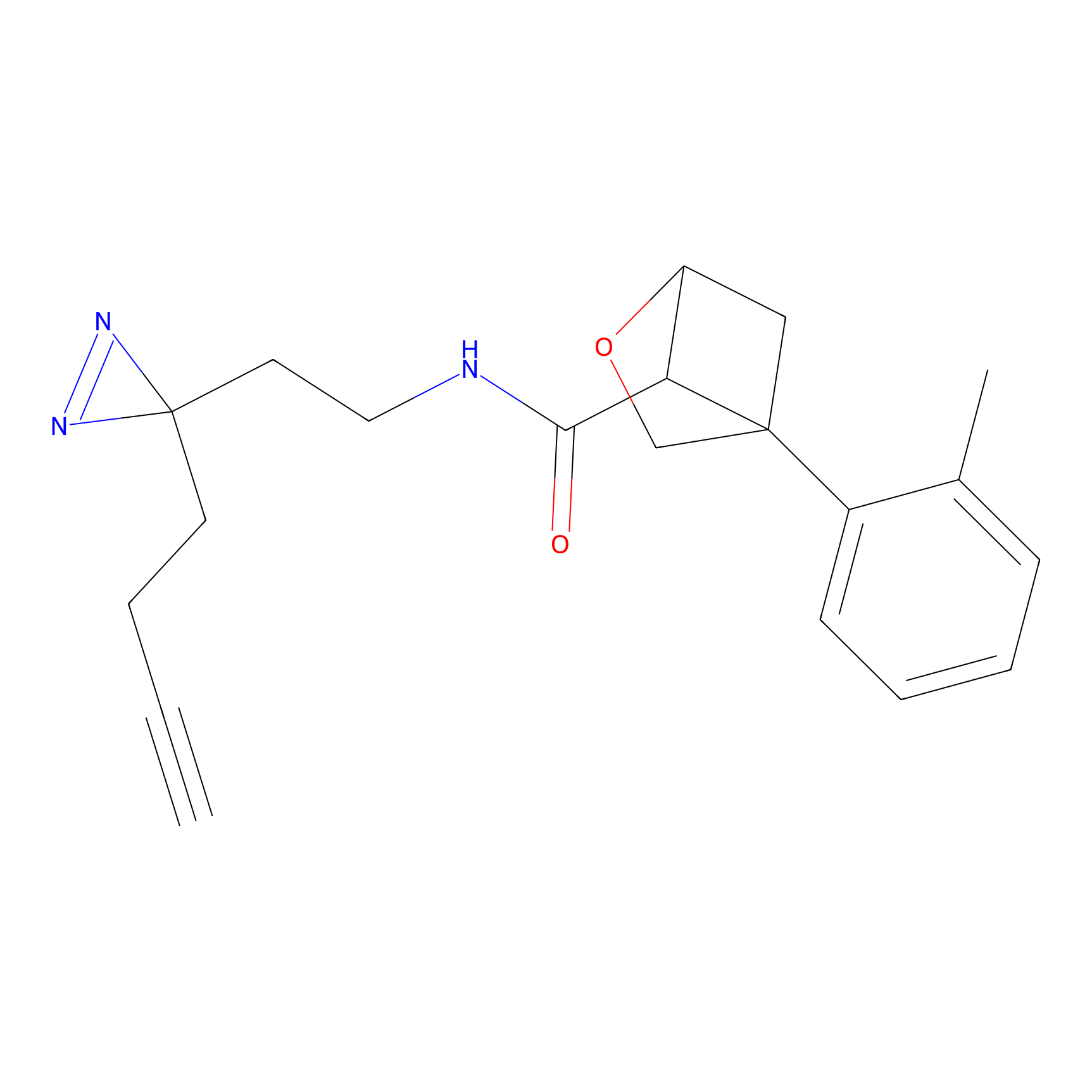 |
18.64 | LDD1958 | [7] | |
|
C289 Probe Info |
 |
80.45 | LDD1959 | [7] | |
|
C290 Probe Info |
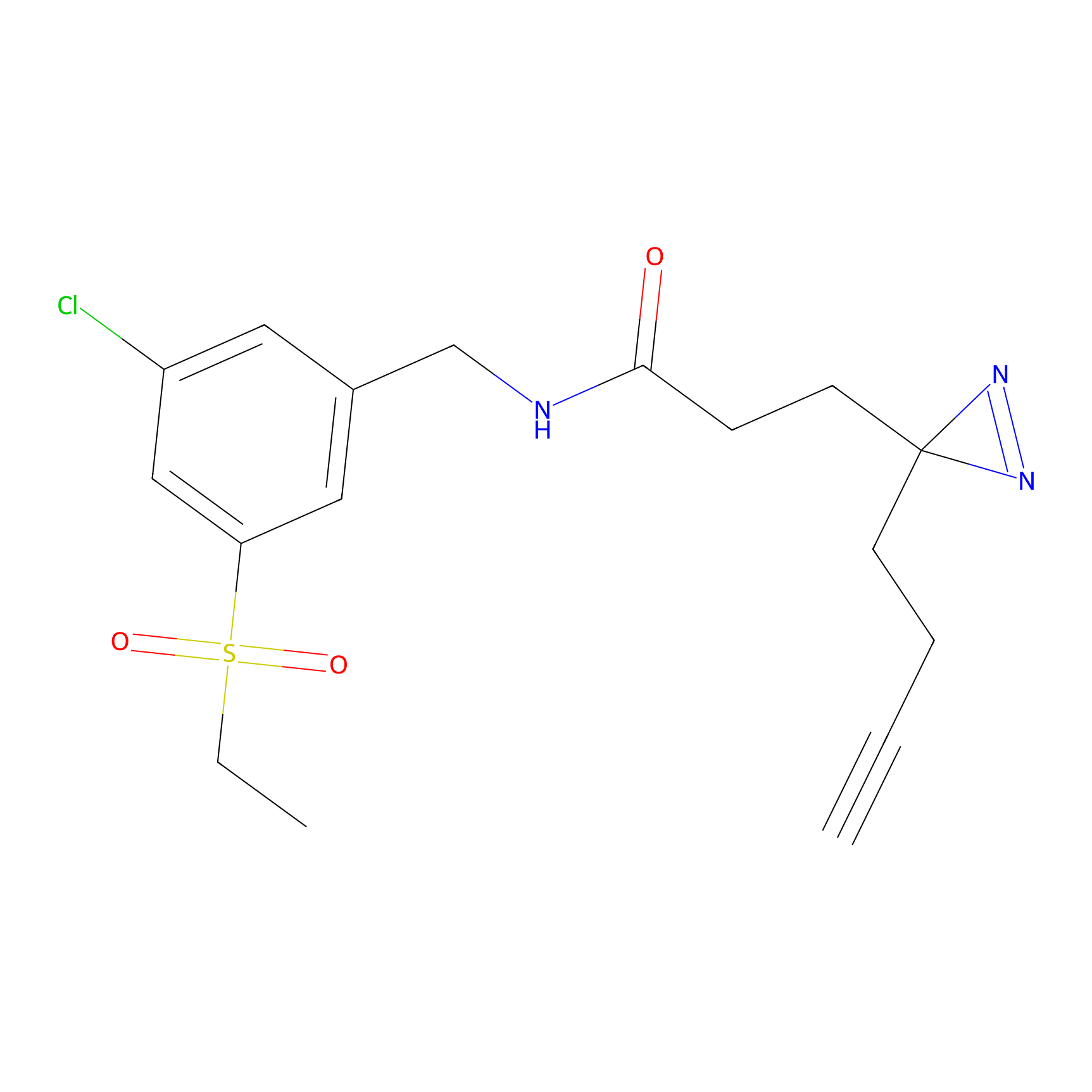 |
7.57 | LDD1960 | [7] | |
|
C299 Probe Info |
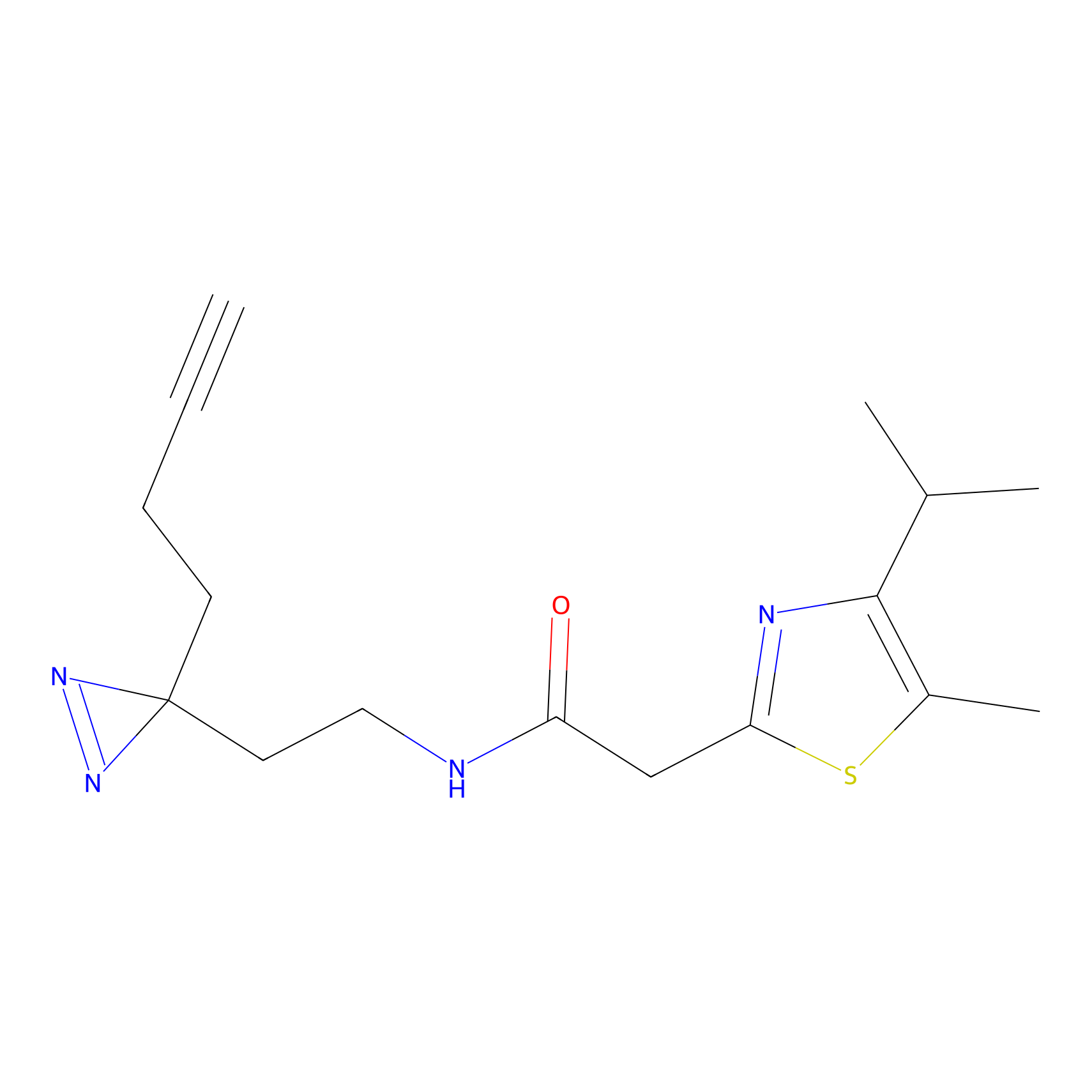 |
29.04 | LDD1968 | [7] | |
|
C302 Probe Info |
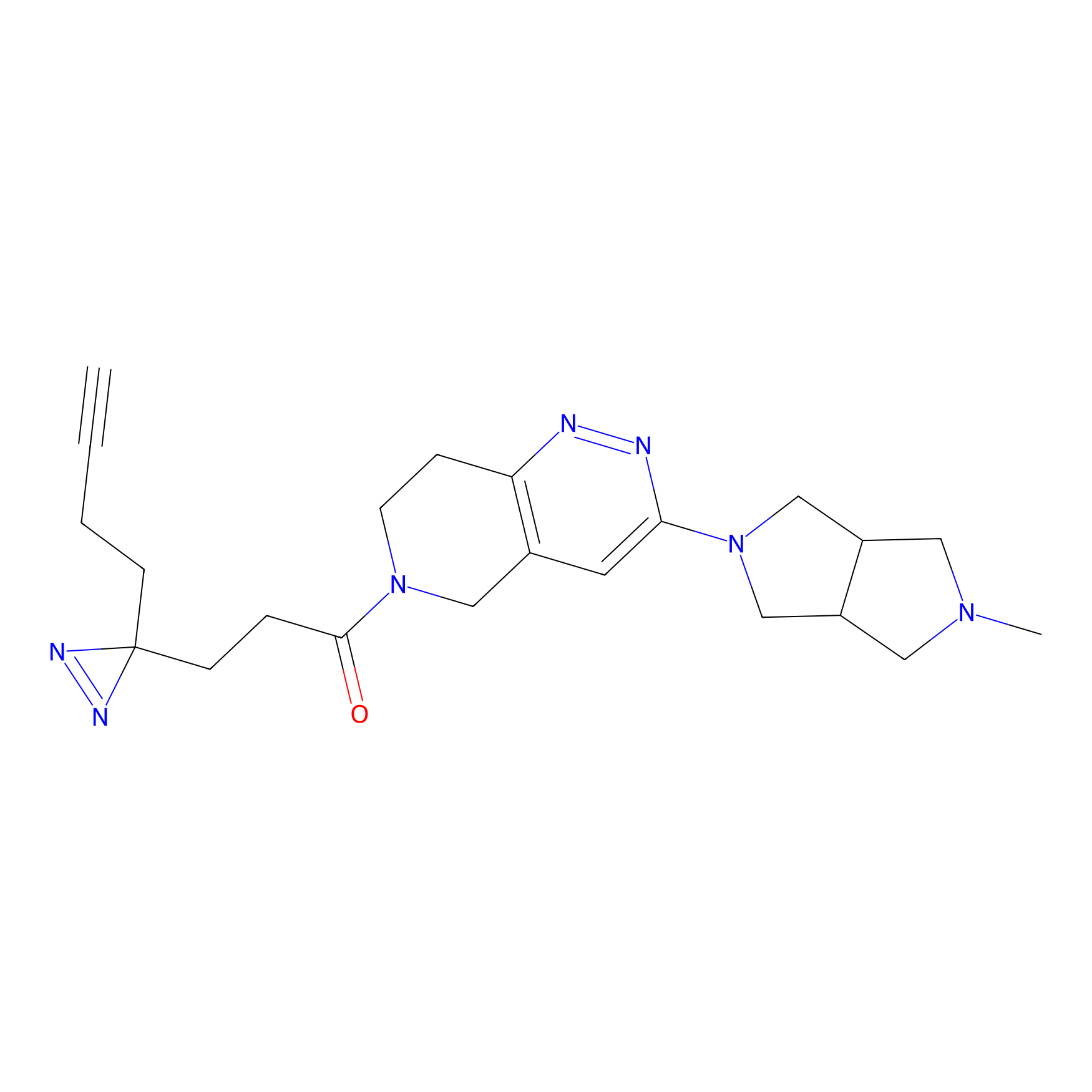 |
5.70 | LDD1971 | [7] | |
|
C303 Probe Info |
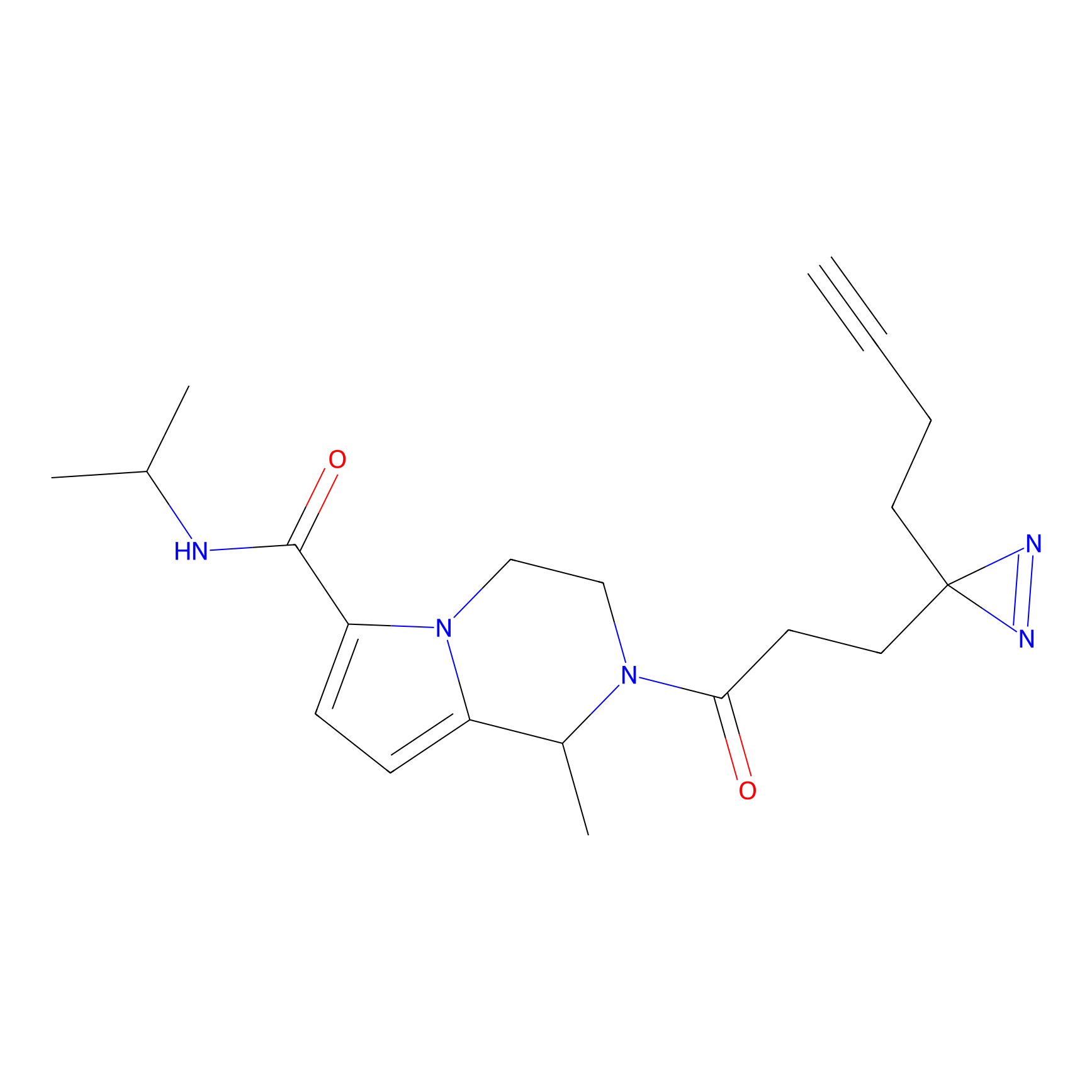 |
5.62 | LDD1972 | [7] | |
|
C304 Probe Info |
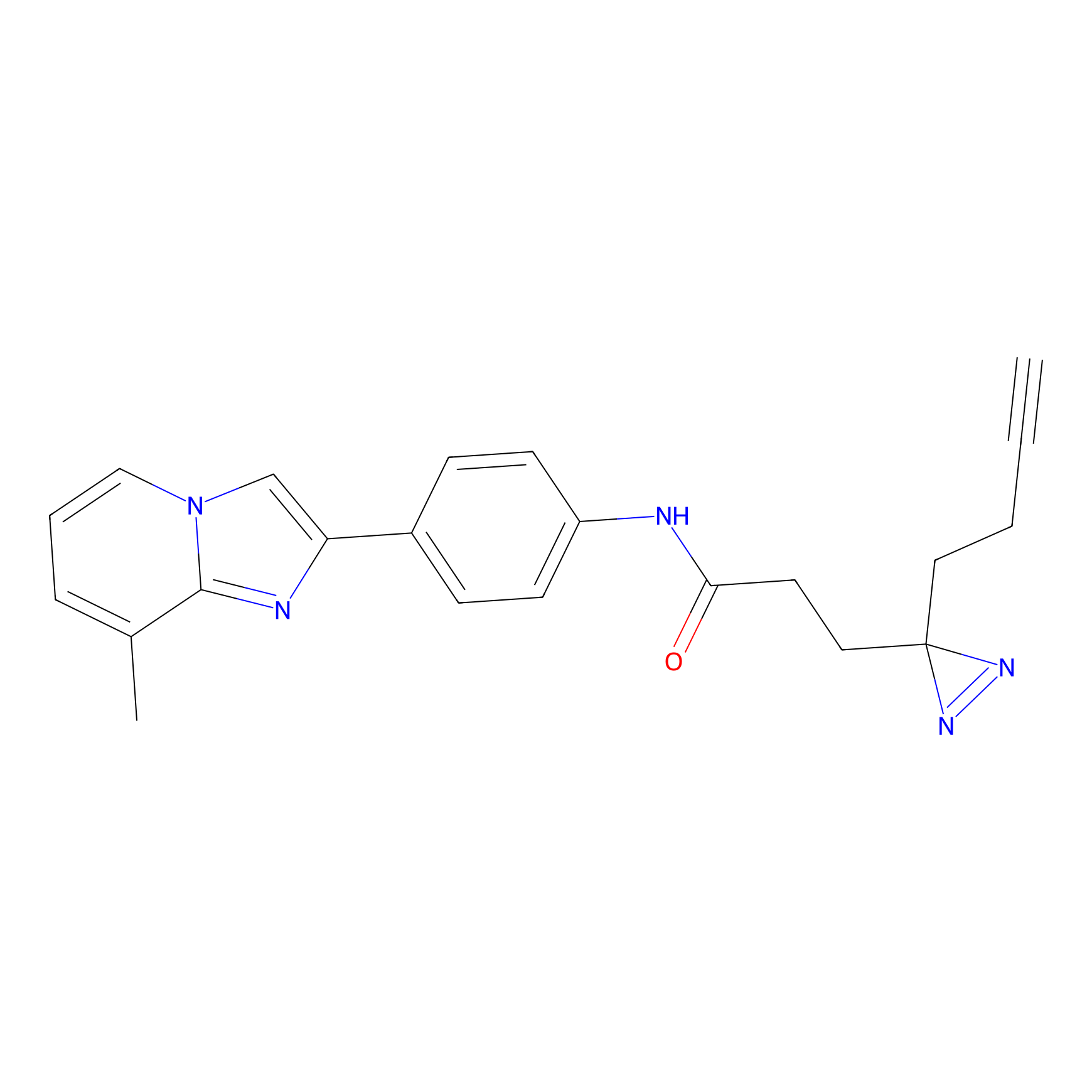 |
5.50 | LDD1973 | [7] | |
|
C305 Probe Info |
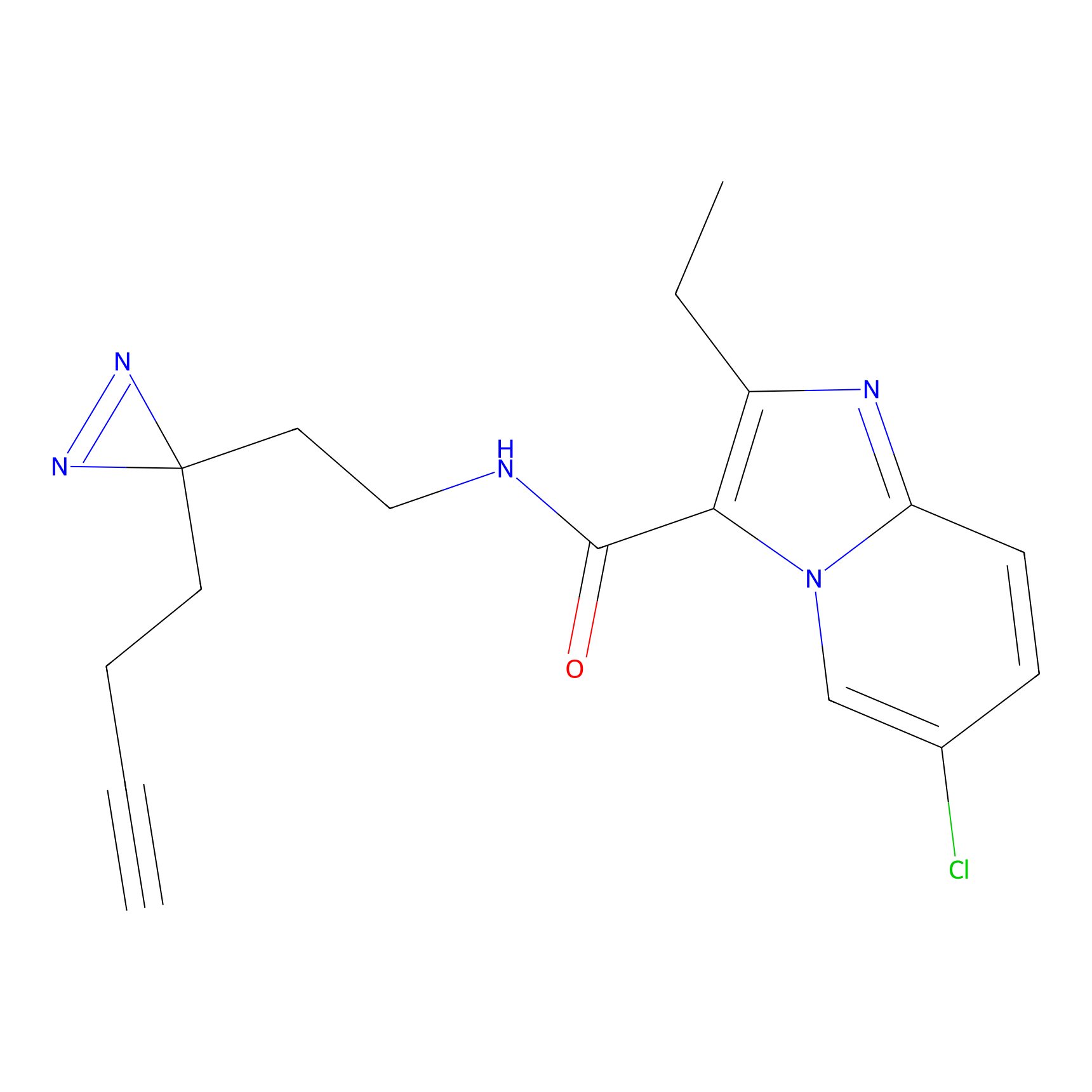 |
25.81 | LDD1974 | [7] | |
|
C307 Probe Info |
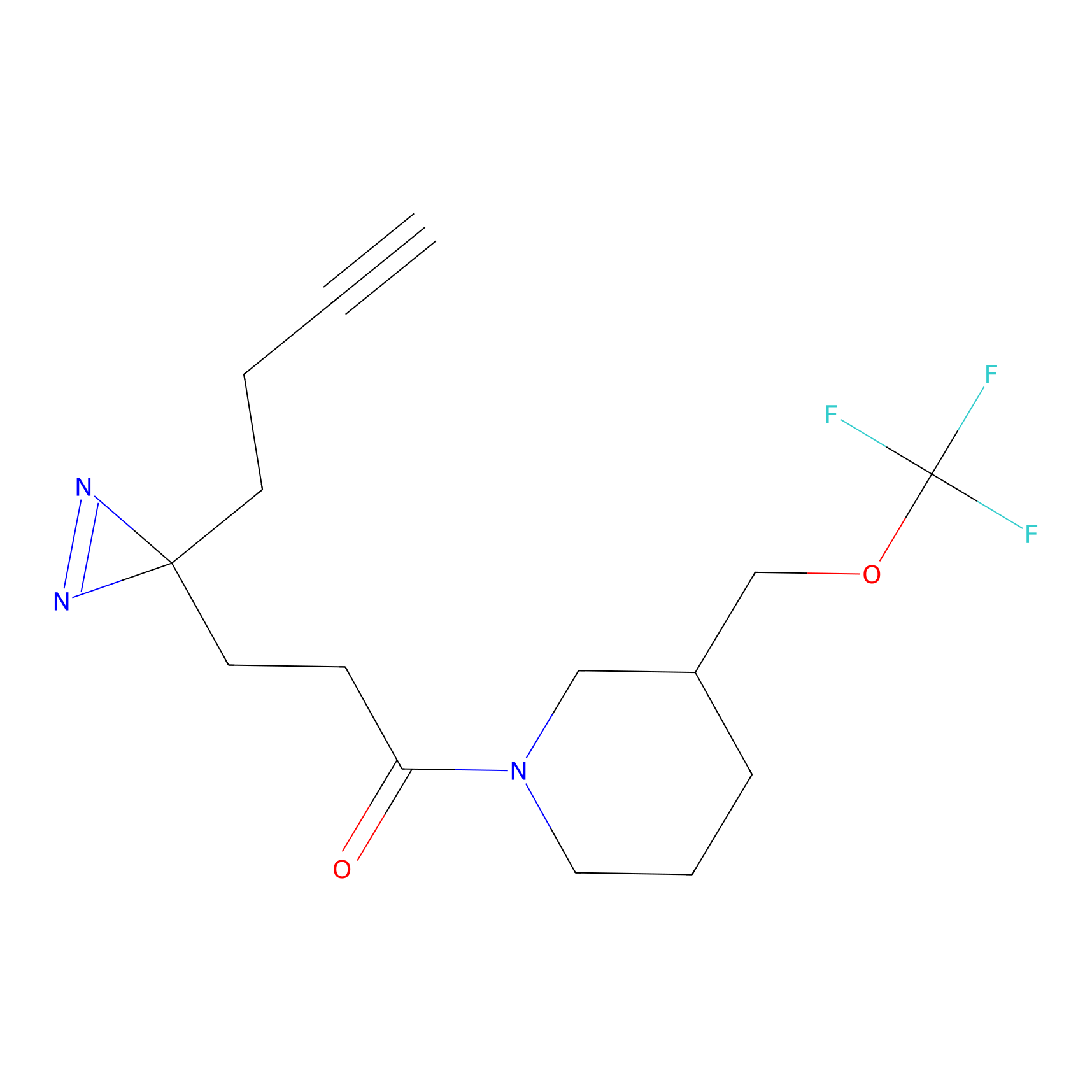 |
9.65 | LDD1975 | [7] | |
|
C310 Probe Info |
 |
30.06 | LDD1977 | [7] | |
|
C362 Probe Info |
 |
99.73 | LDD2023 | [7] | |
|
C363 Probe Info |
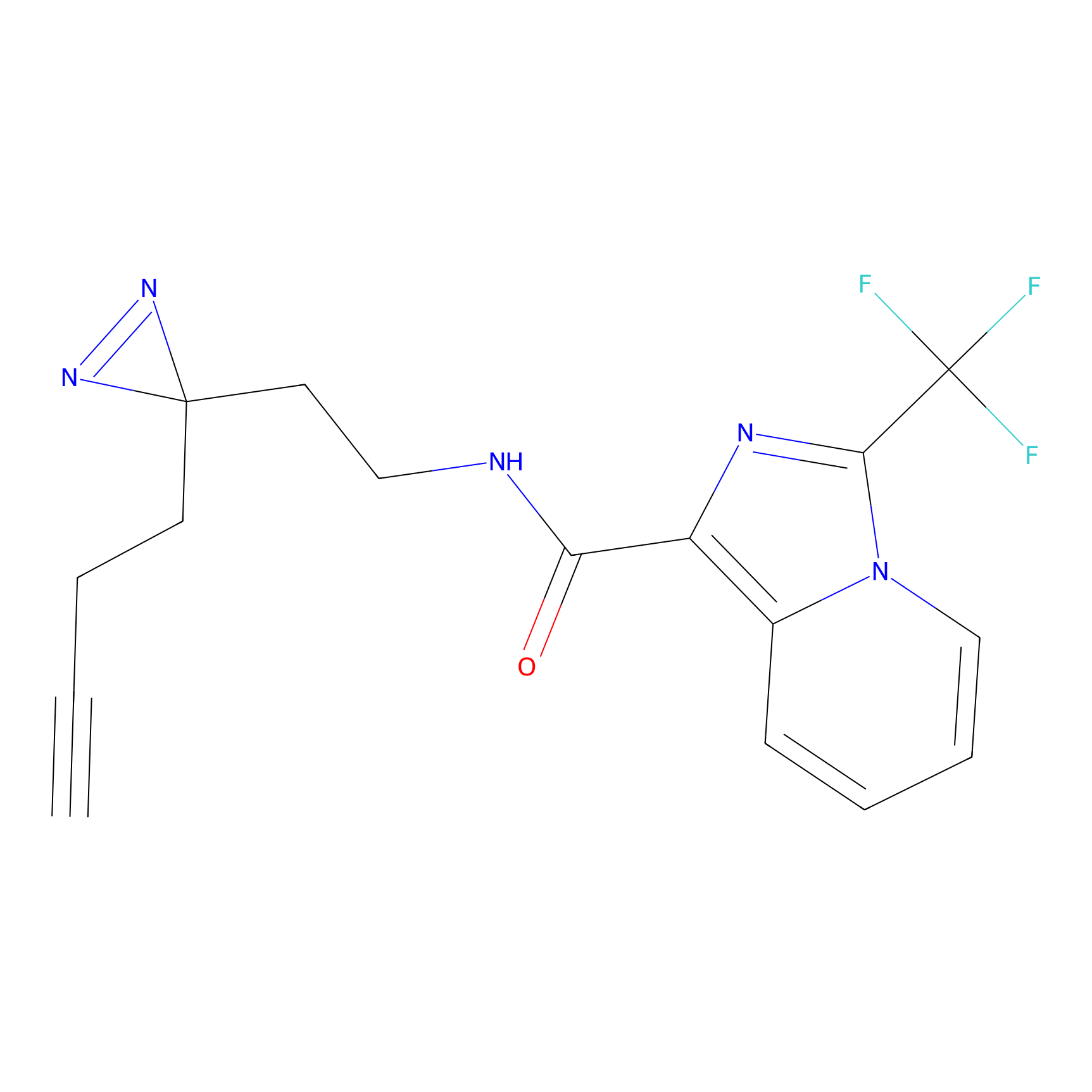 |
99.73 | LDD2024 | [7] | |
|
C364 Probe Info |
 |
99.04 | LDD2025 | [7] | |
|
C366 Probe Info |
 |
20.68 | LDD2027 | [7] | |
|
C367 Probe Info |
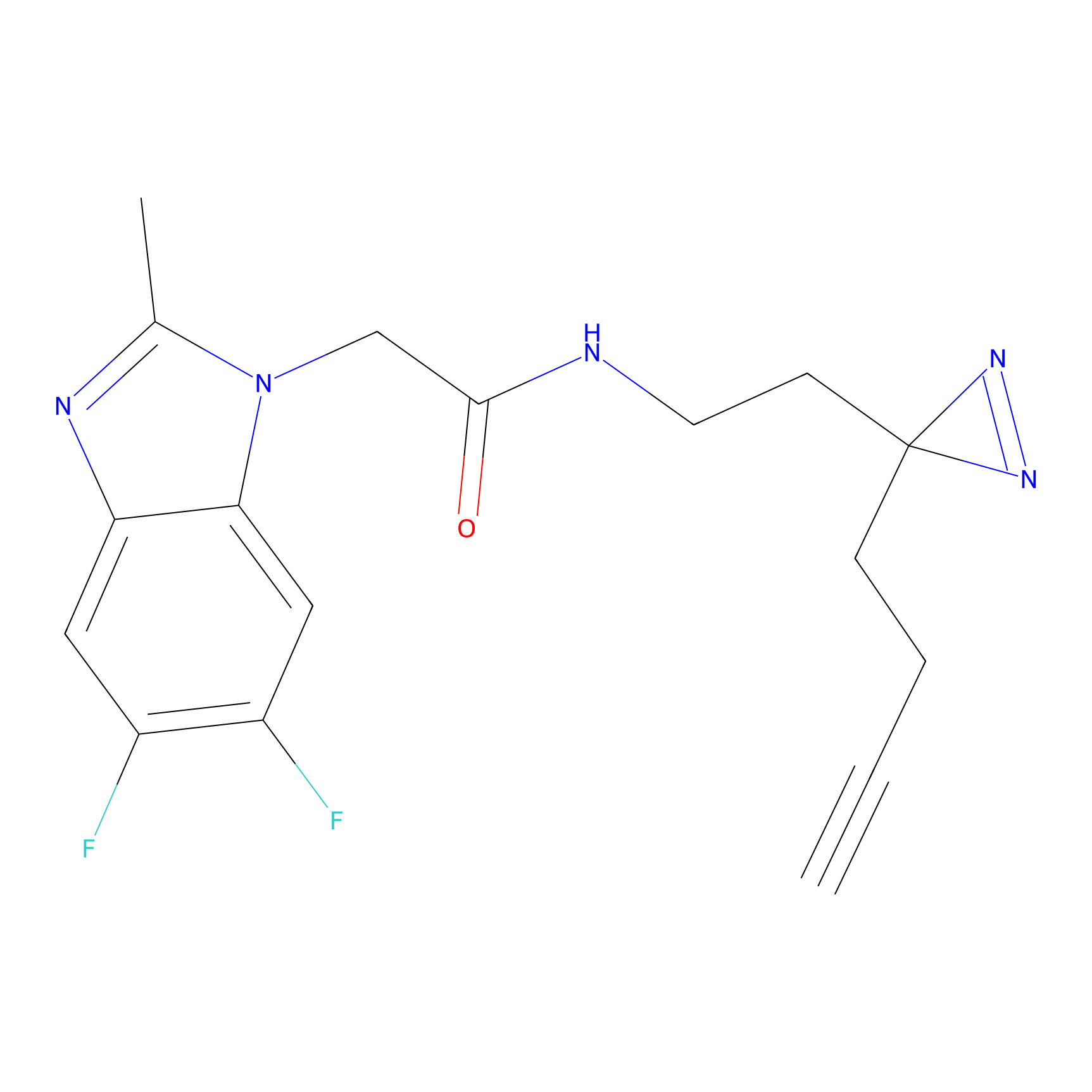 |
13.27 | LDD2028 | [7] | |
|
A-DA Probe Info |
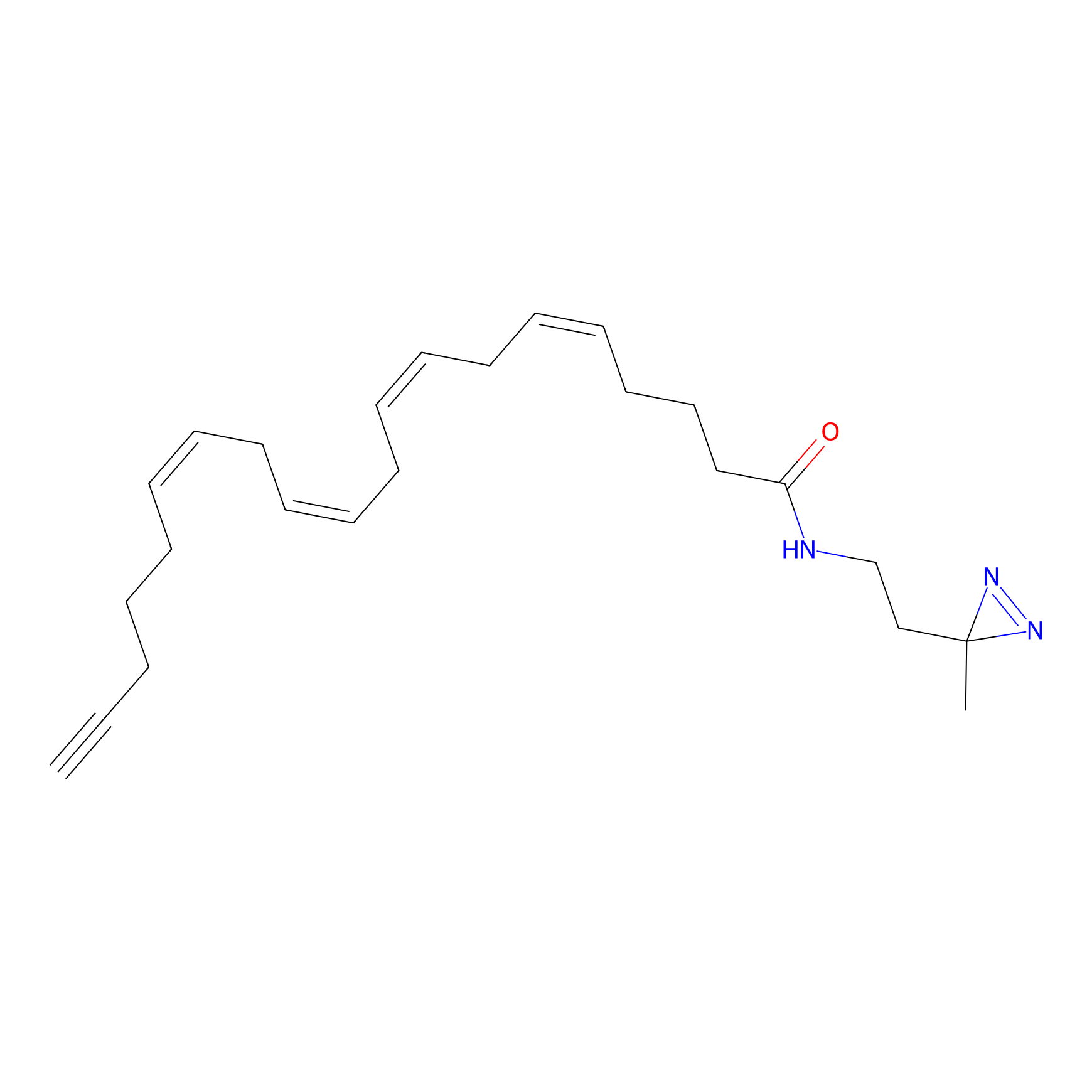 |
3.41 | LDD0332 | [8] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0259 | AC14 | HEK-293T | C19(0.84) | LDD1512 | [6] |
| LDCM0282 | AC22 | HEK-293T | C19(0.95) | LDD1521 | [6] |
| LDCM0284 | AC24 | HEK-293T | C19(0.92) | LDD1523 | [6] |
| LDCM0291 | AC30 | HEK-293T | C19(0.98) | LDD1530 | [6] |
| LDCM0293 | AC32 | HEK-293T | C19(0.97) | LDD1532 | [6] |
| LDCM0299 | AC38 | HEK-293T | C19(1.04) | LDD1538 | [6] |
| LDCM0302 | AC40 | HEK-293T | C19(1.06) | LDD1541 | [6] |
| LDCM0308 | AC46 | HEK-293T | C19(1.16) | LDD1547 | [6] |
| LDCM0310 | AC48 | HEK-293T | C19(0.90) | LDD1549 | [6] |
| LDCM0317 | AC54 | HEK-293T | C19(0.62) | LDD1556 | [6] |
| LDCM0319 | AC56 | HEK-293T | C19(1.04) | LDD1558 | [6] |
| LDCM0323 | AC6 | HEK-293T | C19(0.73) | LDD1562 | [6] |
| LDCM0326 | AC62 | HEK-293T | C19(1.13) | LDD1565 | [6] |
| LDCM0328 | AC64 | HEK-293T | C19(0.99) | LDD1567 | [6] |
| LDCM0345 | AC8 | HEK-293T | C19(0.89) | LDD1569 | [6] |
| LDCM0275 | AKOS034007705 | HEK-293T | C19(0.93) | LDD1514 | [6] |
| LDCM0368 | CL10 | HEK-293T | C19(1.52) | LDD1572 | [6] |
| LDCM0390 | CL12 | HEK-293T | C19(0.91) | LDD1594 | [6] |
| LDCM0410 | CL22 | HEK-293T | C19(0.88) | LDD1614 | [6] |
| LDCM0412 | CL24 | HEK-293T | C19(0.85) | LDD1616 | [6] |
| LDCM0423 | CL34 | HEK-293T | C19(1.13) | LDD1627 | [6] |
| LDCM0425 | CL36 | HEK-293T | C19(1.24) | LDD1629 | [6] |
| LDCM0436 | CL46 | HEK-293T | C19(1.11) | LDD1640 | [6] |
| LDCM0438 | CL48 | HEK-293T | C19(0.92) | LDD1642 | [6] |
| LDCM0449 | CL58 | HEK-293T | C19(1.27) | LDD1652 | [6] |
| LDCM0452 | CL60 | HEK-293T | C19(1.10) | LDD1655 | [6] |
| LDCM0463 | CL70 | HEK-293T | C19(0.97) | LDD1666 | [6] |
| LDCM0465 | CL72 | HEK-293T | C19(0.99) | LDD1668 | [6] |
| LDCM0476 | CL82 | HEK-293T | C19(1.01) | LDD1679 | [6] |
| LDCM0478 | CL84 | HEK-293T | C19(1.12) | LDD1681 | [6] |
| LDCM0489 | CL94 | HEK-293T | C19(1.01) | LDD1692 | [6] |
| LDCM0491 | CL96 | HEK-293T | C19(1.23) | LDD1694 | [6] |
| LDCM0033 | Curcusone1d | MCF-7 | 5.48 | LDD0188 | [4] |
| LDCM0016 | Ranjitkar_cp1 | MDA-MB-231 | 2.08 | LDD0123 | [5] |
| LDCM0137 | SR-4995 | HEK-293T | 3.41 | LDD0332 | [8] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Cytochrome b-c1 complex subunit 7 (UQCRB) | UQCRB/QCR7 family | P14927 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Krueppel-like factor 11 (KLF11) | Sp1 C2H2-type zinc-finger protein family | O14901 | |||
GPCR
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Free fatty acid receptor 2 (FFAR2) | G-protein coupled receptor 1 family | O15552 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Protein BRICK1 (BRK1) | BRK1 family | Q8WUW1 | |||
| Protein TMED8 (TMED8) | . | Q6PL24 | |||
| Sprouty-related, EVH1 domain-containing protein 1 (SPRED1) | . | Q7Z699 | |||
References
