Details of the Target
General Information of Target
| Target ID | LDTP03915 | |||||
|---|---|---|---|---|---|---|
| Target Name | Eukaryotic translation initiation factor 2 subunit 3 (EIF2S3) | |||||
| Gene Name | EIF2S3 | |||||
| Gene ID | 1968 | |||||
| Synonyms |
EIF2G; Eukaryotic translation initiation factor 2 subunit 3; EC 3.6.5.3; Eukaryotic translation initiation factor 2 subunit gamma X; eIF2-gamma X; eIF2gX |
|||||
| 3D Structure | ||||||
| Sequence |
MAGGEAGVTLGQPHLSRQDLTTLDVTKLTPLSHEVISRQATINIGTIGHVAHGKSTVVKA
ISGVHTVRFKNELERNITIKLGYANAKIYKLDDPSCPRPECYRSCGSSTPDEFPTDIPGT KGNFKLVRHVSFVDCPGHDILMATMLNGAAVMDAALLLIAGNESCPQPQTSEHLAAIEIM KLKHILILQNKIDLVKESQAKEQYEQILAFVQGTVAEGAPIIPISAQLKYNIEVVCEYIV KKIPVPPRDFTSEPRLIVIRSFDVNKPGCEVDDLKGGVAGGSILKGVLKVGQEIEVRPGI VSKDSEGKLMCKPIFSKIVSLFAEHNDLQYAAPGGLIGVGTKIDPTLCRADRMVGQVLGA VGALPEIFTELEISYFLLRRLLGVRTEGDKKAAKVQKLSKNEVLMVNIGSLSTGGRVSAV KADLGKIVLTNPVCTEVGEKIALSRRVEKHWRLIGWGQIRRGVTIKPTVDDD |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
TRAFAC class translation factor GTPase superfamily, Classic translation factor GTPase family, EIF2G subfamily
|
|||||
| Function |
Member of the eIF2 complex that functions in the early steps of protein synthesis by forming a ternary complex with GTP and initiator tRNA. This complex binds to a 40S ribosomal subunit, followed by mRNA binding to form the 43S pre-initiation complex (43S PIC). Junction of the 60S ribosomal subunit to form the 80S initiation complex is preceded by hydrolysis of the GTP bound to eIF2 and release of an eIF2-GDP binary complex. In order for eIF2 to recycle and catalyze another round of initiation, the GDP bound to eIF2 must exchange with GTP by way of a reaction catalyzed by eIF-2B.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
P2 Probe Info |
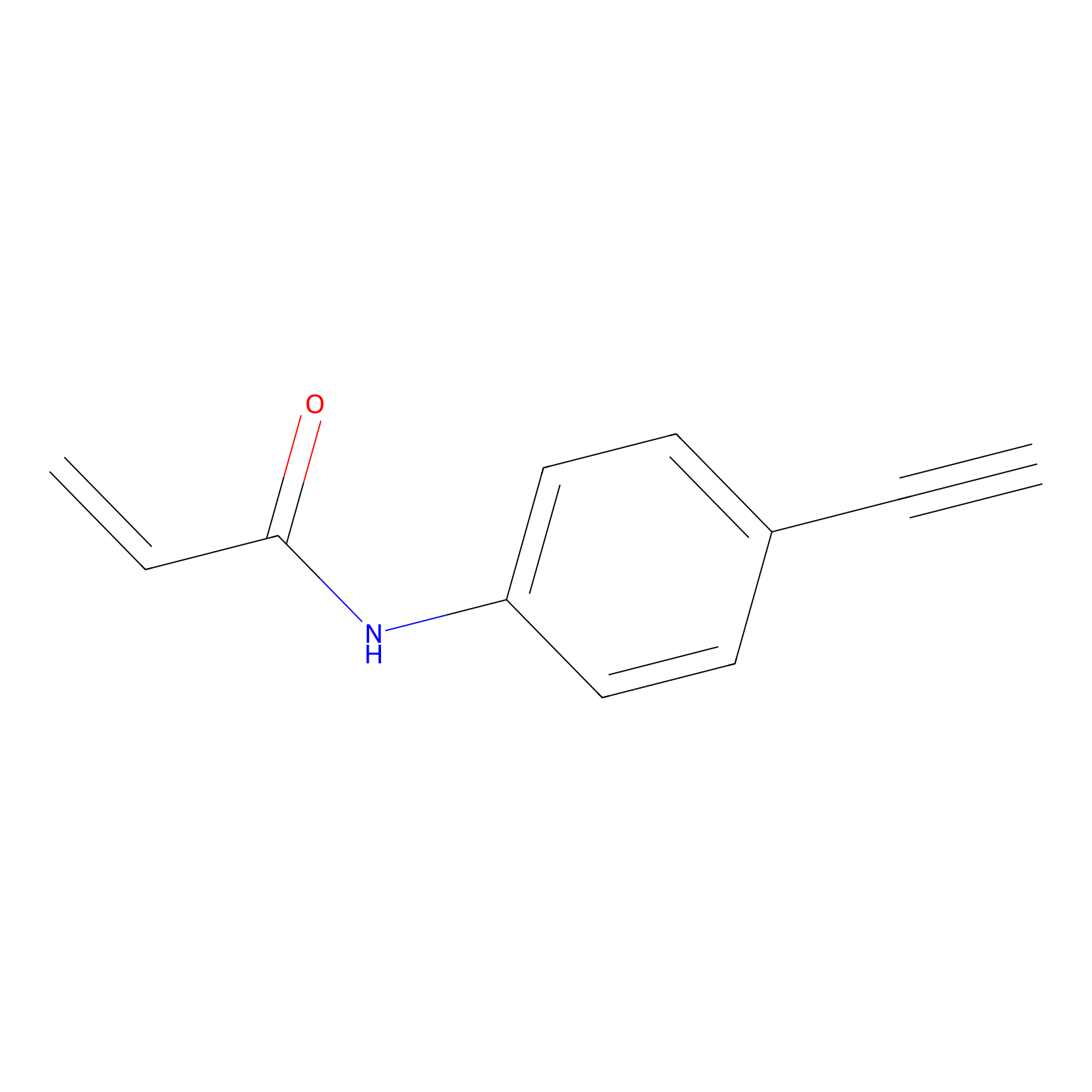 |
3.23 | LDD0449 | [1] | |
|
P3 Probe Info |
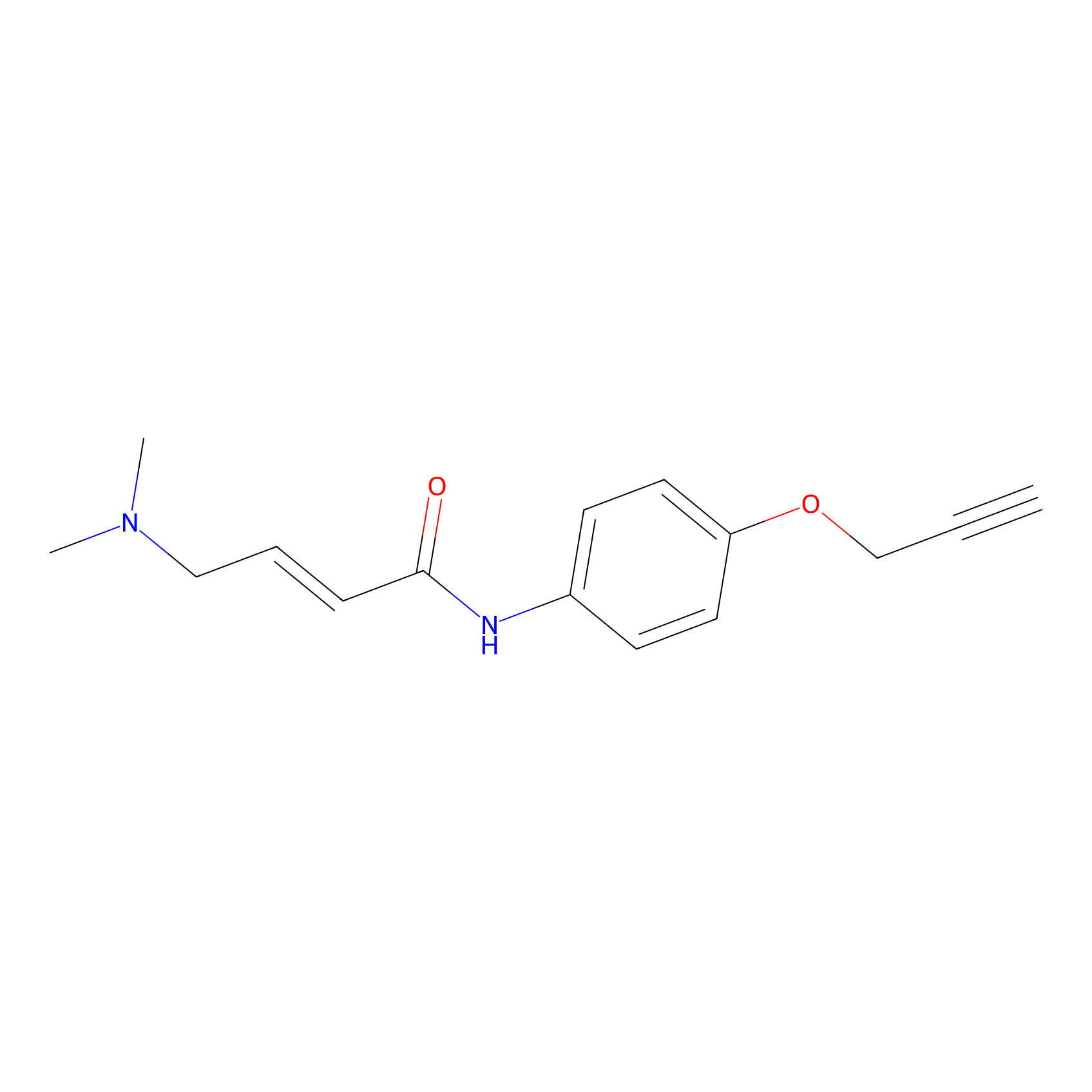 |
3.42 | LDD0450 | [1] | |
|
P8 Probe Info |
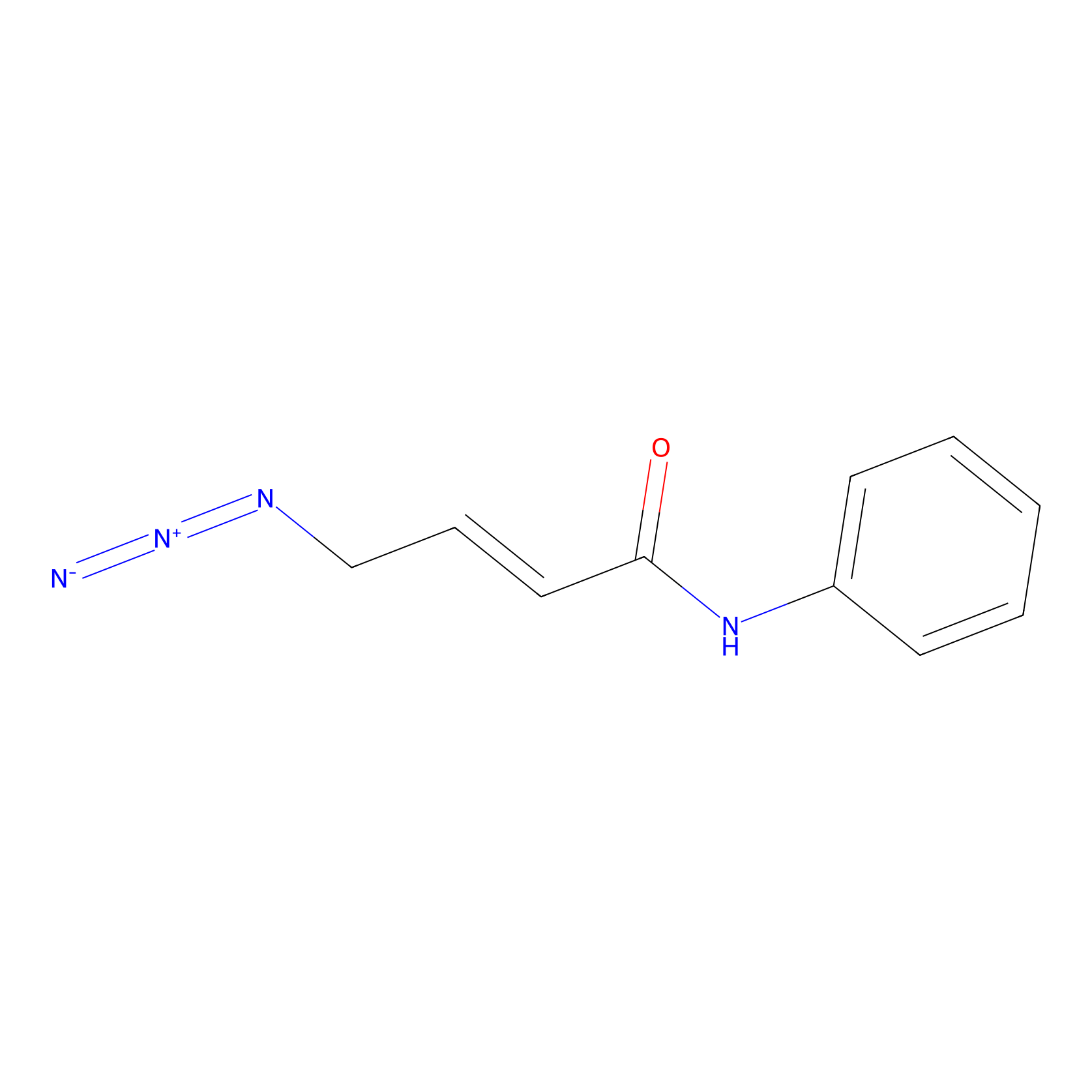 |
3.23 | LDD0455 | [1] | |
|
A-EBA Probe Info |
 |
3.64 | LDD0215 | [2] | |
|
CY4 Probe Info |
 |
30.69 | LDD0244 | [3] | |
|
N1 Probe Info |
 |
27.37 | LDD0242 | [3] | |
|
TH211 Probe Info |
 |
Y330(5.65) | LDD0257 | [4] | |
|
TH214 Probe Info |
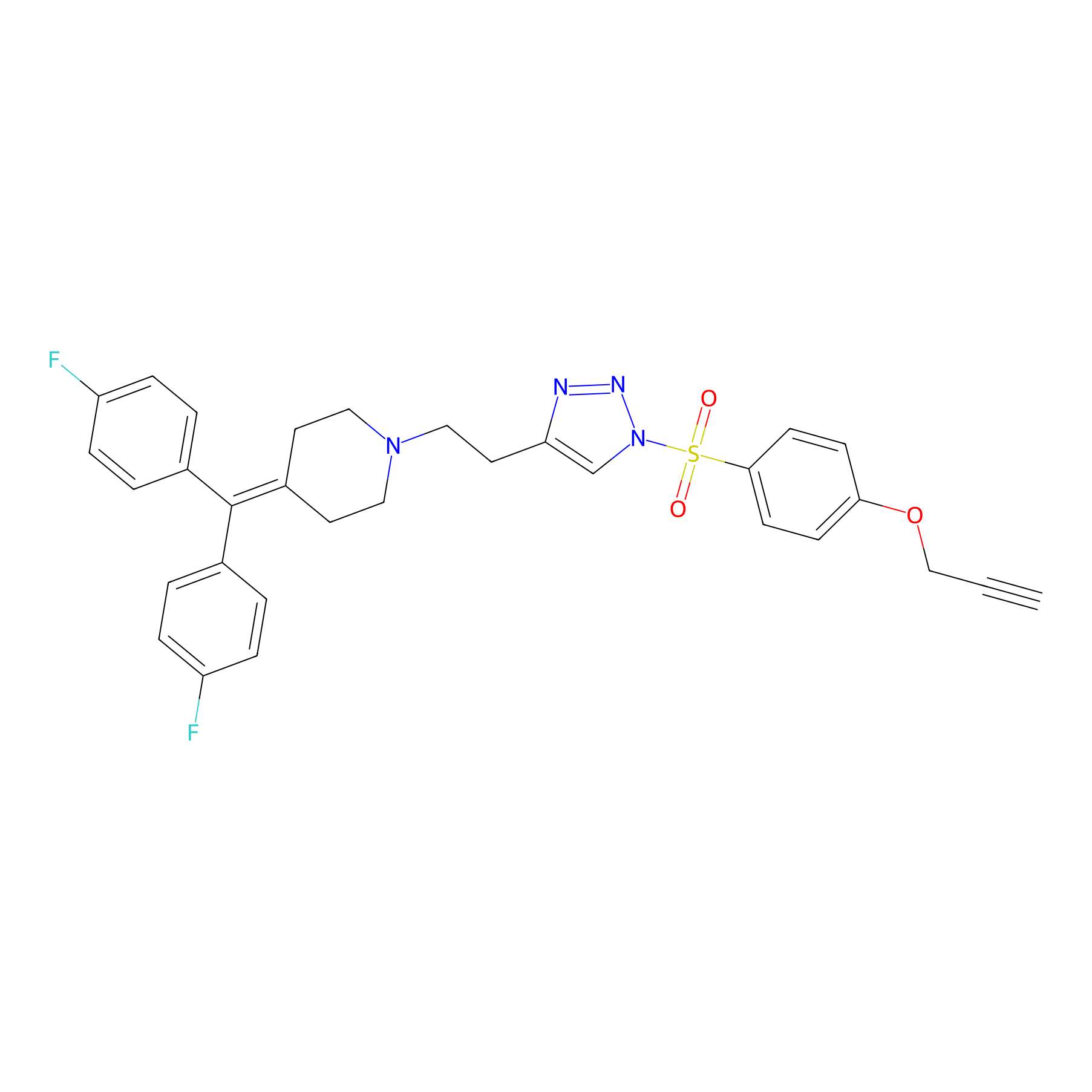 |
Y330(7.87) | LDD0258 | [4] | |
|
TH216 Probe Info |
 |
Y330(11.50) | LDD0259 | [4] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [5] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [5] | |
|
ONAyne Probe Info |
 |
K125(5.00); K400(10.00); K421(0.85) | LDD0274 | [6] | |
|
STPyne Probe Info |
 |
K125(2.36); K183(10.00); K303(1.47); K394(10.00) | LDD0277 | [6] | |
|
AZ-9 Probe Info |
 |
E5(10.00) | LDD2209 | [7] | |
|
OPA-S-S-alkyne Probe Info |
 |
K70(3.79) | LDD3494 | [8] | |
|
JZ128-DTB Probe Info |
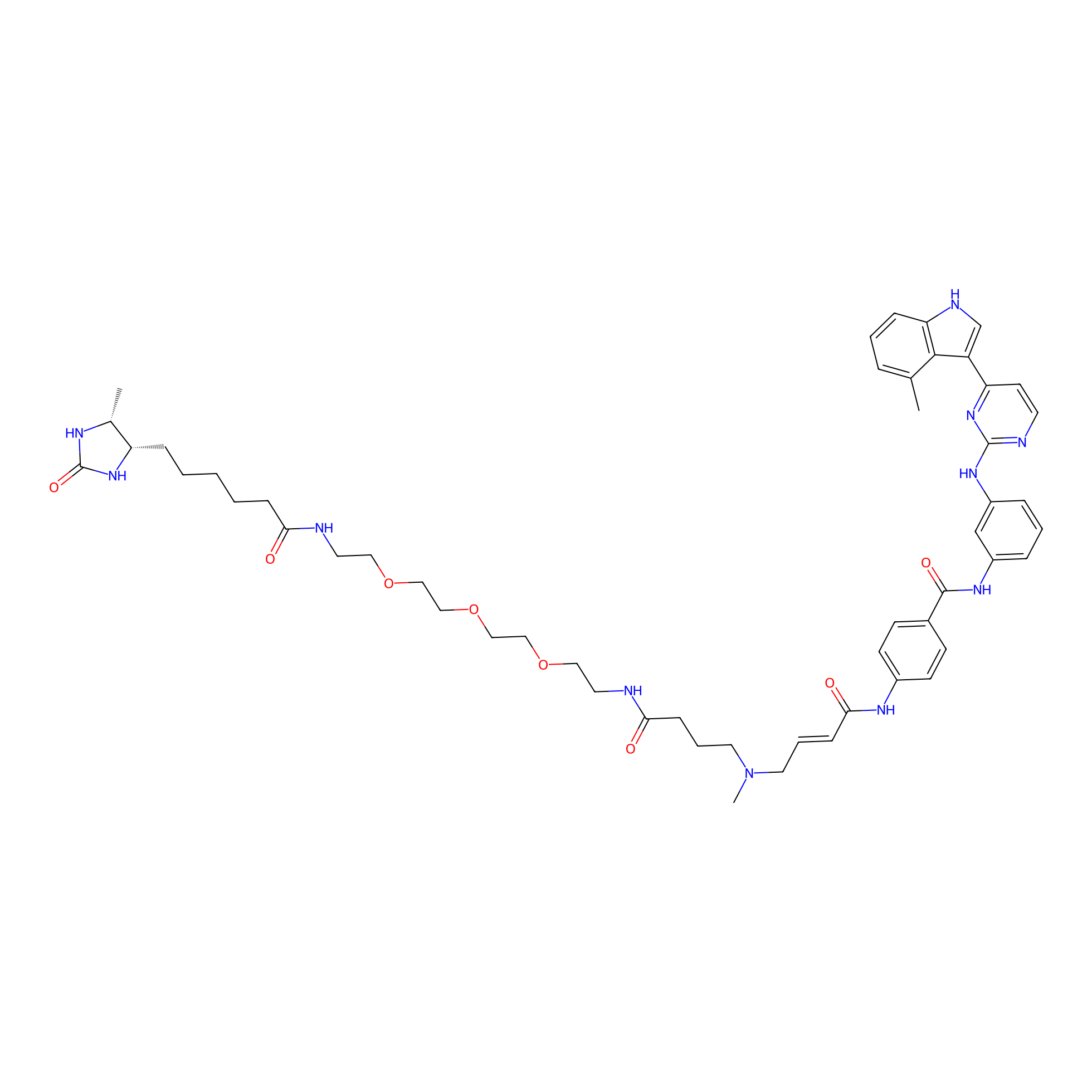 |
N.A. | LDD0462 | [9] | |
|
THZ1-DTB Probe Info |
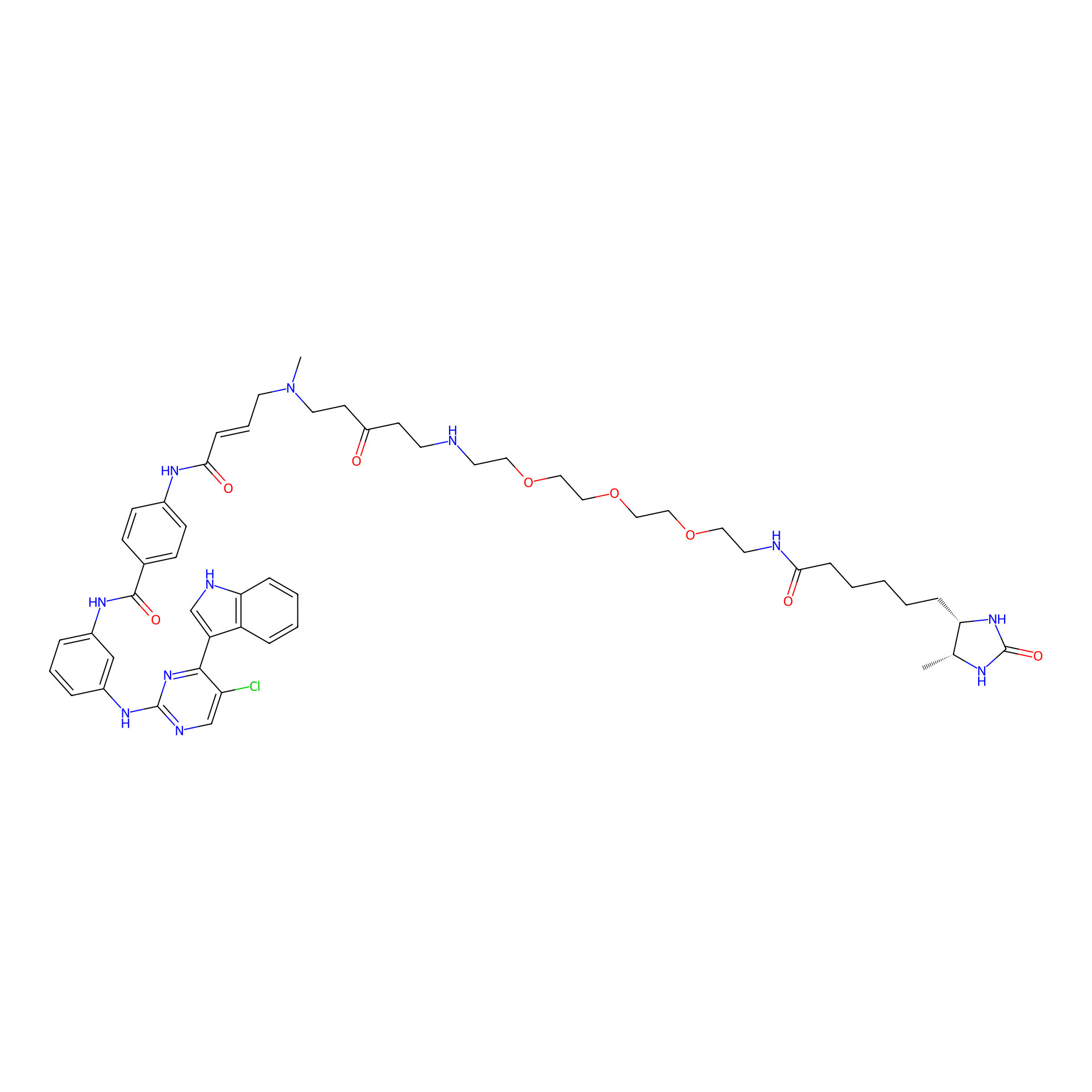 |
C434(1.11); C269(1.09) | LDD0460 | [9] | |
|
AHL-Pu-1 Probe Info |
 |
C96(4.90) | LDD0168 | [10] | |
|
DBIA Probe Info |
 |
C269(1.14); C105(1.07); C348(1.02); C101(1.00) | LDD0078 | [11] | |
|
5E-2FA Probe Info |
 |
H14(0.00); H325(0.00); H33(0.00) | LDD2235 | [12] | |
|
ATP probe Probe Info |
 |
K266(0.00); K275(0.00); K196(0.00); K183(0.00) | LDD0199 | [13] | |
|
m-APA Probe Info |
 |
H14(0.00); H325(0.00); H33(0.00); H65(0.00) | LDD2231 | [12] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C348(0.00); C434(0.00); C311(0.00); C105(0.00) | LDD0038 | [14] | |
|
IA-alkyne Probe Info |
 |
C269(0.00); C105(0.00); C434(0.00); C311(0.00) | LDD0032 | [15] | |
|
IPIAA_H Probe Info |
 |
C434(0.00); C105(0.00) | LDD0030 | [16] | |
|
IPIAA_L Probe Info |
 |
C269(0.00); C434(0.00); C105(0.00) | LDD0031 | [16] | |
|
Lodoacetamide azide Probe Info |
 |
C348(0.00); C434(0.00); C311(0.00); C105(0.00) | LDD0037 | [14] | |
|
BTD Probe Info |
 |
N.A. | LDD0004 | [17] | |
|
IPM Probe Info |
 |
C434(0.00); C269(0.00); C105(0.00) | LDD0025 | [18] | |
|
JW-RF-010 Probe Info |
 |
C105(0.00); C269(0.00); C434(0.00) | LDD0026 | [18] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [19] | |
|
TFBX Probe Info |
 |
C434(0.00); C105(0.00) | LDD0027 | [18] | |
|
WYneN Probe Info |
 |
C96(0.00); C348(0.00) | LDD0021 | [17] | |
|
WYneO Probe Info |
 |
C269(0.00); C105(0.00) | LDD0022 | [17] | |
|
Compound 10 Probe Info |
 |
C105(0.00); C434(0.00) | LDD2216 | [20] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [20] | |
|
ENE Probe Info |
 |
C269(0.00); C348(0.00) | LDD0006 | [17] | |
|
NHS Probe Info |
 |
K303(0.00); K196(0.00) | LDD0010 | [17] | |
|
PF-06672131 Probe Info |
 |
N.A. | LDD0017 | [21] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [22] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [17] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [23] | |
|
1c-yne Probe Info |
 |
K266(0.00); K275(0.00); K27(0.00); K342(0.00) | LDD0228 | [24] | |
|
Acrolein Probe Info |
 |
C105(0.00); C311(0.00) | LDD0217 | [25] | |
|
Methacrolein Probe Info |
 |
C311(0.00); C105(0.00) | LDD0218 | [25] | |
|
W1 Probe Info |
 |
C105(0.00); C96(0.00); C348(0.00) | LDD0236 | [26] | |
|
AOyne Probe Info |
 |
5.50 | LDD0443 | [27] | |
|
NAIA_5 Probe Info |
 |
C434(0.00); C311(0.00); C348(0.00); C96(0.00) | LDD2223 | [19] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FFF probe13 Probe Info |
 |
6.58 | LDD0475 | [28] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [29] | |
|
DA-2 Probe Info |
 |
N.A. | LDD0070 | [30] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0502 | 1-(Cyanoacetyl)piperidine | MDA-MB-231 | C348(1.40) | LDD2095 | [31] |
| LDCM0537 | 2-Cyano-N,N-dimethylacetamide | MDA-MB-231 | C269(0.76) | LDD2130 | [31] |
| LDCM0510 | 3-(4-(Hydroxydiphenylmethyl)piperidin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C348(1.18) | LDD2103 | [31] |
| LDCM0538 | 4-(Cyanoacetyl)morpholine | MDA-MB-231 | C105(0.54) | LDD2131 | [31] |
| LDCM0025 | 4SU-RNA | HEK-293T | C96(4.90) | LDD0168 | [10] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C311(2.17); C269(2.03); C96(3.90) | LDD0169 | [10] |
| LDCM0214 | AC1 | HCT 116 | C101(1.04); C105(1.23); C236(0.99); C269(1.22) | LDD0531 | [11] |
| LDCM0215 | AC10 | HCT 116 | C101(1.28); C105(0.97); C348(1.12); C434(0.74) | LDD0532 | [11] |
| LDCM0216 | AC100 | HCT 116 | C101(0.89); C105(1.01); C269(0.68); C348(0.81) | LDD0533 | [11] |
| LDCM0217 | AC101 | HCT 116 | C101(0.94); C105(1.02); C269(0.75); C348(0.89) | LDD0534 | [11] |
| LDCM0218 | AC102 | HCT 116 | C101(0.96); C105(1.14); C269(0.73); C348(0.92) | LDD0535 | [11] |
| LDCM0219 | AC103 | HCT 116 | C101(0.80); C105(1.17); C269(0.78); C348(0.79) | LDD0536 | [11] |
| LDCM0220 | AC104 | HCT 116 | C101(0.95); C105(1.12); C269(0.79); C348(1.13) | LDD0537 | [11] |
| LDCM0221 | AC105 | HCT 116 | C101(1.05); C105(1.00); C269(0.79); C348(0.89) | LDD0538 | [11] |
| LDCM0222 | AC106 | HCT 116 | C101(0.80); C105(1.12); C269(0.70); C348(0.69) | LDD0539 | [11] |
| LDCM0223 | AC107 | HCT 116 | C101(0.95); C105(1.17); C269(0.75); C348(0.82) | LDD0540 | [11] |
| LDCM0224 | AC108 | HCT 116 | C101(1.11); C105(0.97); C269(0.86); C348(0.90) | LDD0541 | [11] |
| LDCM0225 | AC109 | HCT 116 | C101(1.03); C105(0.92); C269(0.79); C348(1.03) | LDD0542 | [11] |
| LDCM0226 | AC11 | HCT 116 | C101(1.05); C105(1.07); C348(1.08); C434(0.68) | LDD0543 | [11] |
| LDCM0227 | AC110 | HCT 116 | C101(0.89); C105(0.84); C269(0.74); C348(0.74) | LDD0544 | [11] |
| LDCM0228 | AC111 | HCT 116 | C101(0.89); C105(0.91); C269(0.73); C348(0.73) | LDD0545 | [11] |
| LDCM0229 | AC112 | HCT 116 | C101(0.79); C105(1.07); C269(0.68); C348(0.64) | LDD0546 | [11] |
| LDCM0230 | AC113 | HCT 116 | C101(1.59); C105(0.99); C269(1.16); C348(1.13) | LDD0547 | [11] |
| LDCM0231 | AC114 | HCT 116 | C101(1.01); C105(1.17); C269(1.00); C348(0.90) | LDD0548 | [11] |
| LDCM0232 | AC115 | HCT 116 | C101(1.00); C105(1.23); C269(0.91); C348(0.86) | LDD0549 | [11] |
| LDCM0233 | AC116 | HCT 116 | C101(1.14); C105(1.22); C269(1.06); C348(0.88) | LDD0550 | [11] |
| LDCM0234 | AC117 | HCT 116 | C101(1.07); C105(1.14); C269(0.91); C348(0.97) | LDD0551 | [11] |
| LDCM0235 | AC118 | HCT 116 | C101(1.08); C105(1.10); C269(0.91); C348(0.99) | LDD0552 | [11] |
| LDCM0236 | AC119 | HCT 116 | C101(1.03); C105(1.31); C269(0.98); C348(0.97) | LDD0553 | [11] |
| LDCM0237 | AC12 | HCT 116 | C101(1.25); C105(0.87); C348(1.27); C434(1.15) | LDD0554 | [11] |
| LDCM0238 | AC120 | HCT 116 | C101(1.01); C105(1.21); C269(1.02); C348(0.94) | LDD0555 | [11] |
| LDCM0239 | AC121 | HCT 116 | C101(1.04); C105(1.25); C269(1.27); C348(1.12) | LDD0556 | [11] |
| LDCM0240 | AC122 | HCT 116 | C101(1.00); C105(1.26); C269(1.19); C348(1.00) | LDD0557 | [11] |
| LDCM0241 | AC123 | HCT 116 | C101(0.94); C105(1.02); C269(0.99); C348(1.07) | LDD0558 | [11] |
| LDCM0242 | AC124 | HCT 116 | C101(1.01); C105(1.12); C269(1.19); C348(1.05) | LDD0559 | [11] |
| LDCM0243 | AC125 | HCT 116 | C101(0.94); C105(1.17); C269(1.09); C348(1.01) | LDD0560 | [11] |
| LDCM0244 | AC126 | HCT 116 | C101(1.00); C105(1.11); C269(1.21); C348(1.08) | LDD0561 | [11] |
| LDCM0245 | AC127 | HCT 116 | C101(1.06); C105(1.13); C269(1.08); C348(1.06) | LDD0562 | [11] |
| LDCM0246 | AC128 | HCT 116 | C101(0.56); C105(1.00); C348(0.92); C96(0.56) | LDD0563 | [11] |
| LDCM0247 | AC129 | HCT 116 | C101(1.00); C105(0.87); C348(1.35); C96(1.00) | LDD0564 | [11] |
| LDCM0249 | AC130 | HCT 116 | C101(0.70); C105(1.03); C348(1.09); C96(0.70) | LDD0566 | [11] |
| LDCM0250 | AC131 | HCT 116 | C101(0.99); C105(0.81); C348(1.06); C96(0.99) | LDD0567 | [11] |
| LDCM0251 | AC132 | HCT 116 | C101(1.10); C105(1.07); C348(1.41); C96(1.10) | LDD0568 | [11] |
| LDCM0252 | AC133 | HCT 116 | C101(0.85); C105(1.05); C348(1.29); C96(0.85) | LDD0569 | [11] |
| LDCM0253 | AC134 | HCT 116 | C101(0.70); C105(1.05); C348(0.89); C96(0.70) | LDD0570 | [11] |
| LDCM0254 | AC135 | HCT 116 | C101(0.66); C105(1.00); C348(1.12); C96(0.66) | LDD0571 | [11] |
| LDCM0255 | AC136 | HCT 116 | C101(0.75); C105(1.08); C348(1.03); C96(0.75) | LDD0572 | [11] |
| LDCM0256 | AC137 | HCT 116 | C101(0.97); C105(1.09); C348(1.20); C96(0.97) | LDD0573 | [11] |
| LDCM0257 | AC138 | HCT 116 | C101(0.85); C105(1.05); C348(0.82); C96(0.85) | LDD0574 | [11] |
| LDCM0258 | AC139 | HCT 116 | C101(0.84); C105(1.05); C348(0.99); C96(0.84) | LDD0575 | [11] |
| LDCM0259 | AC14 | HCT 116 | C101(1.33); C105(0.98); C348(1.19); C434(0.74) | LDD0576 | [11] |
| LDCM0260 | AC140 | HCT 116 | C101(0.79); C105(1.05); C348(0.75); C96(0.79) | LDD0577 | [11] |
| LDCM0261 | AC141 | HCT 116 | C101(0.78); C105(0.99); C348(0.87); C96(0.78) | LDD0578 | [11] |
| LDCM0262 | AC142 | HCT 116 | C101(1.05); C105(1.06); C348(1.16); C96(1.05) | LDD0579 | [11] |
| LDCM0263 | AC143 | HCT 116 | C101(0.93); C105(1.13); C236(1.29); C348(0.68) | LDD0580 | [11] |
| LDCM0264 | AC144 | HCT 116 | C348(0.65); C101(0.91); C236(1.07); C105(1.36) | LDD0581 | [11] |
| LDCM0265 | AC145 | HCT 116 | C348(0.80); C101(0.85); C236(1.39); C105(1.42) | LDD0582 | [11] |
| LDCM0266 | AC146 | HCT 116 | C348(0.81); C101(0.93); C105(1.13); C236(1.46) | LDD0583 | [11] |
| LDCM0267 | AC147 | HCT 116 | C348(0.63); C101(0.87); C236(1.23); C105(1.30) | LDD0584 | [11] |
| LDCM0268 | AC148 | HCT 116 | C348(0.48); C101(0.80); C105(1.10); C236(3.34) | LDD0585 | [11] |
| LDCM0269 | AC149 | HCT 116 | C348(0.68); C101(0.90); C105(1.31); C236(2.08) | LDD0586 | [11] |
| LDCM0270 | AC15 | HCT 116 | C434(0.78); C105(1.03); C348(1.06); C96(1.12) | LDD0587 | [11] |
| LDCM0271 | AC150 | HCT 116 | C101(0.82); C348(0.85); C105(1.40); C236(1.50) | LDD0588 | [11] |
| LDCM0272 | AC151 | HCT 116 | C348(0.71); C101(0.92); C105(1.13); C236(1.78) | LDD0589 | [11] |
| LDCM0273 | AC152 | HCT 116 | C348(0.58); C101(0.93); C105(1.25); C236(1.36) | LDD0590 | [11] |
| LDCM0274 | AC153 | HCT 116 | C348(0.48); C101(0.86); C105(1.20); C236(2.65) | LDD0591 | [11] |
| LDCM0621 | AC154 | HCT 116 | C101(0.91); C105(1.34); C236(1.43); C348(0.68) | LDD2158 | [11] |
| LDCM0622 | AC155 | HCT 116 | C101(1.04); C105(1.31); C236(1.71); C348(0.82) | LDD2159 | [11] |
| LDCM0623 | AC156 | HCT 116 | C101(0.76); C105(1.17); C236(1.40); C348(1.02) | LDD2160 | [11] |
| LDCM0624 | AC157 | HCT 116 | C101(0.78); C105(1.09); C236(1.51); C348(0.68) | LDD2161 | [11] |
| LDCM0276 | AC17 | HCT 116 | C105(1.02); C348(1.04); C269(1.09); C101(1.16) | LDD0593 | [11] |
| LDCM0277 | AC18 | HCT 116 | C348(0.90); C434(1.05); C269(1.07); C101(1.14) | LDD0594 | [11] |
| LDCM0278 | AC19 | HCT 116 | C348(0.84); C269(0.96); C101(1.02); C96(1.02) | LDD0595 | [11] |
| LDCM0279 | AC2 | HCT 116 | C236(1.00); C348(1.03); C96(1.10); C105(1.11) | LDD0596 | [11] |
| LDCM0280 | AC20 | HCT 116 | C348(0.90); C269(0.94); C105(1.01); C434(1.02) | LDD0597 | [11] |
| LDCM0281 | AC21 | HCT 116 | C348(0.99); C269(1.02); C101(1.06); C96(1.06) | LDD0598 | [11] |
| LDCM0282 | AC22 | HCT 116 | C434(0.94); C105(0.99); C269(1.00); C348(1.00) | LDD0599 | [11] |
| LDCM0283 | AC23 | HCT 116 | C269(0.93); C434(0.95); C105(0.99); C348(0.99) | LDD0600 | [11] |
| LDCM0284 | AC24 | HCT 116 | C105(0.90); C348(0.99); C101(1.00); C96(1.00) | LDD0601 | [11] |
| LDCM0285 | AC25 | HCT 116 | C269(0.81); C105(0.94); C101(1.09); C348(1.13) | LDD0602 | [11] |
| LDCM0286 | AC26 | HCT 116 | C269(0.66); C348(0.93); C96(0.94); C101(0.95) | LDD0603 | [11] |
| LDCM0287 | AC27 | HCT 116 | C269(0.77); C348(0.91); C101(0.97); C105(0.99) | LDD0604 | [11] |
| LDCM0288 | AC28 | HCT 116 | C269(0.58); C348(0.85); C96(0.94); C101(0.96) | LDD0605 | [11] |
| LDCM0289 | AC29 | HCT 116 | C269(0.53); C96(0.76); C101(0.77); C348(0.83) | LDD0606 | [11] |
| LDCM0290 | AC3 | HCT 116 | C269(0.92); C236(0.97); C105(1.08); C348(1.11) | LDD0607 | [11] |
| LDCM0291 | AC30 | HCT 116 | C269(0.44); C348(0.73); C96(0.77); C101(0.81) | LDD0608 | [11] |
| LDCM0292 | AC31 | HCT 116 | C269(0.70); C348(0.84); C96(1.01); C101(1.05) | LDD0609 | [11] |
| LDCM0293 | AC32 | HCT 116 | C269(0.45); C348(0.67); C101(0.86); C96(0.94) | LDD0610 | [11] |
| LDCM0294 | AC33 | HCT 116 | C348(0.73); C269(0.73); C101(0.92); C105(0.94) | LDD0611 | [11] |
| LDCM0295 | AC34 | HCT 116 | C348(0.64); C269(0.76); C101(0.83); C96(0.87) | LDD0612 | [11] |
| LDCM0296 | AC35 | HCT 116 | C105(0.93); C269(1.23); C236(1.23); C101(1.31) | LDD0613 | [11] |
| LDCM0297 | AC36 | HCT 116 | C105(0.91); C236(1.03); C101(1.21); C269(1.30) | LDD0614 | [11] |
| LDCM0298 | AC37 | HCT 116 | C105(0.89); C236(1.04); C269(1.15); C101(1.36) | LDD0615 | [11] |
| LDCM0299 | AC38 | HCT 116 | C105(0.99); C96(0.99); C101(1.03); C236(1.05) | LDD0616 | [11] |
| LDCM0300 | AC39 | HCT 116 | C236(1.00); C105(1.03); C269(1.09); C96(1.17) | LDD0617 | [11] |
| LDCM0301 | AC4 | HCT 116 | C269(0.90); C236(0.90); C105(1.15); C96(1.16) | LDD0618 | [11] |
| LDCM0302 | AC40 | HCT 116 | C236(0.89); C269(0.91); C105(0.93); C101(1.05) | LDD0619 | [11] |
| LDCM0303 | AC41 | HCT 116 | C105(0.99); C236(1.03); C269(1.11); C101(1.21) | LDD0620 | [11] |
| LDCM0304 | AC42 | HCT 116 | C105(0.99); C236(1.07); C269(1.12); C101(1.12) | LDD0621 | [11] |
| LDCM0305 | AC43 | HCT 116 | C236(0.95); C101(0.99); C269(0.99); C105(1.00) | LDD0622 | [11] |
| LDCM0306 | AC44 | HCT 116 | C105(0.90); C236(1.02); C269(1.12); C101(1.24) | LDD0623 | [11] |
| LDCM0307 | AC45 | HCT 116 | C269(0.91); C236(0.93); C105(1.07); C101(1.45) | LDD0624 | [11] |
| LDCM0308 | AC46 | HCT 116 | C269(0.84); C348(0.91); C236(1.02); C105(1.11) | LDD0625 | [11] |
| LDCM0309 | AC47 | HCT 116 | C105(0.81); C269(0.86); C348(0.89); C236(0.96) | LDD0626 | [11] |
| LDCM0310 | AC48 | HCT 116 | C269(0.72); C105(0.84); C348(0.84); C236(0.98) | LDD0627 | [11] |
| LDCM0311 | AC49 | HCT 116 | C269(0.53); C348(0.53); C236(0.72); C105(0.85) | LDD0628 | [11] |
| LDCM0312 | AC5 | HCT 116 | C236(0.99); C269(1.07); C348(1.22); C105(1.26) | LDD0629 | [11] |
| LDCM0313 | AC50 | HCT 116 | C348(0.54); C269(0.56); C236(0.65); C105(0.88) | LDD0630 | [11] |
| LDCM0314 | AC51 | HCT 116 | C269(0.82); C105(0.87); C236(0.87); C348(1.41) | LDD0631 | [11] |
| LDCM0315 | AC52 | HCT 116 | C269(0.75); C236(0.88); C348(0.96); C105(1.12) | LDD0632 | [11] |
| LDCM0316 | AC53 | HCT 116 | C348(0.75); C269(0.79); C236(0.83); C105(0.86) | LDD0633 | [11] |
| LDCM0317 | AC54 | HCT 116 | C348(0.73); C269(0.76); C236(0.87); C105(0.91) | LDD0634 | [11] |
| LDCM0318 | AC55 | HCT 116 | C348(0.60); C236(0.63); C269(0.65); C105(0.87) | LDD0635 | [11] |
| LDCM0319 | AC56 | HCT 116 | C348(0.41); C236(0.48); C269(0.52); C105(0.91) | LDD0636 | [11] |
| LDCM0320 | AC57 | HCT 116 | C348(0.82); C269(0.91); C96(0.92); C236(1.01) | LDD0637 | [11] |
| LDCM0321 | AC58 | HCT 116 | C96(0.64); C348(0.67); C269(0.85); C101(0.91) | LDD0638 | [11] |
| LDCM0322 | AC59 | HCT 116 | C348(0.82); C269(0.83); C236(0.92); C96(0.92) | LDD0639 | [11] |
| LDCM0323 | AC6 | HCT 116 | C434(0.75); C348(0.92); C96(0.98); C101(1.11) | LDD0640 | [11] |
| LDCM0324 | AC60 | HCT 116 | C348(0.76); C96(0.92); C101(0.97); C236(1.05) | LDD0641 | [11] |
| LDCM0325 | AC61 | HCT 116 | C348(0.75); C96(0.75); C101(0.93); C236(1.06) | LDD0642 | [11] |
| LDCM0326 | AC62 | HCT 116 | C269(0.71); C236(0.72); C348(0.77); C96(1.00) | LDD0643 | [11] |
| LDCM0327 | AC63 | HCT 116 | C348(0.91); C269(0.92); C96(0.97); C236(1.16) | LDD0644 | [11] |
| LDCM0328 | AC64 | HCT 116 | C269(0.82); C348(0.86); C236(0.90); C105(1.13) | LDD0645 | [11] |
| LDCM0329 | AC65 | HCT 116 | C348(0.73); C96(0.75); C101(0.93); C269(1.16) | LDD0646 | [11] |
| LDCM0330 | AC66 | HCT 116 | C348(0.75); C96(0.92); C101(0.93); C269(1.15) | LDD0647 | [11] |
| LDCM0331 | AC67 | HCT 116 | C269(0.45); C236(0.59); C348(0.63); C101(0.91) | LDD0648 | [11] |
| LDCM0332 | AC68 | HCT 116 | C348(0.86); C105(0.90); C236(0.91); C269(0.98) | LDD0649 | [11] |
| LDCM0333 | AC69 | HCT 116 | C236(0.69); C269(0.84); C105(0.84); C348(0.94) | LDD0650 | [11] |
| LDCM0334 | AC7 | HCT 116 | C434(0.89); C105(0.99); C96(0.99); C348(1.11) | LDD0651 | [11] |
| LDCM0335 | AC70 | HCT 116 | C236(0.63); C269(0.69); C105(0.80); C348(0.85) | LDD0652 | [11] |
| LDCM0336 | AC71 | HCT 116 | C236(0.84); C269(0.85); C105(0.87); C96(1.02) | LDD0653 | [11] |
| LDCM0337 | AC72 | HCT 116 | C236(0.83); C105(0.84); C348(0.88); C269(0.89) | LDD0654 | [11] |
| LDCM0338 | AC73 | HCT 116 | C236(0.47); C348(0.66); C269(0.71); C105(0.75) | LDD0655 | [11] |
| LDCM0339 | AC74 | HCT 116 | C236(0.43); C269(0.63); C105(0.64); C348(0.67) | LDD0656 | [11] |
| LDCM0340 | AC75 | HCT 116 | C236(0.41); C269(0.64); C348(0.77); C105(0.81) | LDD0657 | [11] |
| LDCM0341 | AC76 | HCT 116 | C236(0.76); C348(0.82); C105(0.85); C96(0.95) | LDD0658 | [11] |
| LDCM0342 | AC77 | HCT 116 | C236(0.65); C348(0.78); C269(0.82); C105(0.83) | LDD0659 | [11] |
| LDCM0343 | AC78 | HCT 116 | C105(0.84); C236(0.88); C348(0.96); C269(1.01) | LDD0660 | [11] |
| LDCM0344 | AC79 | HCT 116 | C236(0.90); C269(0.95); C105(0.96); C348(1.02) | LDD0661 | [11] |
| LDCM0345 | AC8 | HCT 116 | C434(0.83); C348(1.00); C105(1.01); C96(1.08) | LDD0662 | [11] |
| LDCM0346 | AC80 | HCT 116 | C348(0.81); C236(0.82); C105(0.83); C269(1.00) | LDD0663 | [11] |
| LDCM0347 | AC81 | HCT 116 | C236(0.77); C269(0.87); C105(0.89); C96(1.00) | LDD0664 | [11] |
| LDCM0348 | AC82 | HCT 116 | C236(0.30); C269(0.56); C348(0.61); C105(0.62) | LDD0665 | [11] |
| LDCM0349 | AC83 | HCT 116 | C348(0.64); C105(0.87); C269(0.90); C101(0.92) | LDD0666 | [11] |
| LDCM0350 | AC84 | HCT 116 | C348(0.60); C96(0.82); C105(0.88); C101(0.90) | LDD0667 | [11] |
| LDCM0351 | AC85 | HCT 116 | C348(0.77); C269(0.84); C101(0.87); C96(0.87) | LDD0668 | [11] |
| LDCM0352 | AC86 | HCT 116 | C348(0.82); C269(0.90); C101(0.98); C96(0.99) | LDD0669 | [11] |
| LDCM0353 | AC87 | HCT 116 | C269(0.79); C348(0.90); C105(0.93); C96(1.00) | LDD0670 | [11] |
| LDCM0354 | AC88 | HCT 116 | C269(0.78); C348(0.82); C105(0.96); C101(0.99) | LDD0671 | [11] |
| LDCM0355 | AC89 | HCT 116 | C348(0.66); C269(0.81); C105(0.96); C101(0.99) | LDD0672 | [11] |
| LDCM0357 | AC90 | HCT 116 | C269(0.73); C105(1.00); C101(1.07); C96(1.09) | LDD0674 | [11] |
| LDCM0358 | AC91 | HCT 116 | C348(0.64); C105(0.91); C96(0.91); C101(0.96) | LDD0675 | [11] |
| LDCM0359 | AC92 | HCT 116 | C348(0.69); C96(0.88); C101(0.90); C105(0.92) | LDD0676 | [11] |
| LDCM0360 | AC93 | HCT 116 | C269(0.89); C105(0.93); C96(0.96); C101(0.98) | LDD0677 | [11] |
| LDCM0361 | AC94 | HCT 116 | C105(0.88); C348(0.94); C269(0.97); C96(1.06) | LDD0678 | [11] |
| LDCM0362 | AC95 | HCT 116 | C269(0.85); C348(0.85); C96(0.86); C105(0.90) | LDD0679 | [11] |
| LDCM0363 | AC96 | HCT 116 | C348(0.81); C105(0.89); C96(0.96); C101(1.06) | LDD0680 | [11] |
| LDCM0364 | AC97 | HCT 116 | C348(0.66); C269(0.73); C105(1.01); C96(1.02) | LDD0681 | [11] |
| LDCM0365 | AC98 | HCT 116 | C434(0.09); C269(0.53); C348(0.72); C105(0.74) | LDD0682 | [11] |
| LDCM0366 | AC99 | HCT 116 | C269(0.66); C434(0.68); C348(0.77); C105(0.95) | LDD0683 | [11] |
| LDCM0248 | AKOS034007472 | HCT 116 | C101(0.99); C105(1.05); C348(1.26); C434(1.18) | LDD0565 | [11] |
| LDCM0356 | AKOS034007680 | HCT 116 | C434(0.77); C105(0.95); C96(1.06); C348(1.07) | LDD0673 | [11] |
| LDCM0275 | AKOS034007705 | HCT 116 | C434(0.37); C348(0.71); C96(1.04); C101(1.10) | LDD0592 | [11] |
| LDCM0020 | ARS-1620 | HCC44 | C269(1.14); C105(1.07); C348(1.02); C101(1.00) | LDD0078 | [11] |
| LDCM0630 | CCW28-3 | 231MFP | C105(1.35) | LDD2214 | [32] |
| LDCM0108 | Chloroacetamide | HeLa | C105(0.00); C311(0.00) | LDD0222 | [25] |
| LDCM0632 | CL-Sc | Hep-G2 | C269(20.00); C434(2.29) | LDD2227 | [19] |
| LDCM0367 | CL1 | HCT 116 | C434(1.01); C236(1.03); C105(1.08); C348(1.08) | LDD0684 | [11] |
| LDCM0368 | CL10 | HCT 116 | C434(0.40); C348(1.00); C236(1.09); C105(1.21) | LDD0685 | [11] |
| LDCM0369 | CL100 | HCT 116 | C269(0.91); C96(1.05); C236(1.07); C348(1.08) | LDD0686 | [11] |
| LDCM0370 | CL101 | HCT 116 | C434(0.75); C348(0.93); C96(1.06); C105(1.13) | LDD0687 | [11] |
| LDCM0371 | CL102 | HCT 116 | C105(1.00); C434(1.05); C96(1.07); C101(1.20) | LDD0688 | [11] |
| LDCM0372 | CL103 | HCT 116 | C105(0.86); C434(0.91); C101(1.06); C348(1.06) | LDD0689 | [11] |
| LDCM0373 | CL104 | HCT 116 | C434(0.75); C105(0.91); C101(0.98); C96(1.11) | LDD0690 | [11] |
| LDCM0374 | CL105 | HCT 116 | C348(0.97); C434(1.18); C105(1.23); C269(1.31) | LDD0691 | [11] |
| LDCM0375 | CL106 | HCT 116 | C348(0.76); C434(1.09); C269(1.15); C105(1.16) | LDD0692 | [11] |
| LDCM0376 | CL107 | HCT 116 | C348(0.94); C434(1.11); C101(1.23); C96(1.23) | LDD0693 | [11] |
| LDCM0377 | CL108 | HCT 116 | C348(0.74); C434(1.11); C105(1.16); C101(1.32) | LDD0694 | [11] |
| LDCM0378 | CL109 | HCT 116 | C348(0.90); C101(1.07); C96(1.07); C105(1.08) | LDD0695 | [11] |
| LDCM0379 | CL11 | HCT 116 | C434(0.50); C236(0.86); C105(1.02); C348(1.27) | LDD0696 | [11] |
| LDCM0380 | CL110 | HCT 116 | C348(0.73); C269(0.84); C101(0.88); C96(0.88) | LDD0697 | [11] |
| LDCM0381 | CL111 | HCT 116 | C269(0.81); C348(0.85); C434(0.92); C101(0.98) | LDD0698 | [11] |
| LDCM0382 | CL112 | HCT 116 | C269(0.69); C105(0.85); C96(0.90); C101(0.90) | LDD0699 | [11] |
| LDCM0383 | CL113 | HCT 116 | C269(0.48); C348(0.73); C101(0.84); C96(0.88) | LDD0700 | [11] |
| LDCM0384 | CL114 | HCT 116 | C269(0.52); C101(0.85); C96(0.89); C348(0.91) | LDD0701 | [11] |
| LDCM0385 | CL115 | HCT 116 | C269(0.50); C96(0.82); C348(0.86); C101(0.86) | LDD0702 | [11] |
| LDCM0386 | CL116 | HCT 116 | C269(0.72); C101(0.88); C348(0.90); C96(0.90) | LDD0703 | [11] |
| LDCM0387 | CL117 | HCT 116 | C269(0.61); C236(0.67); C101(0.84); C96(0.92) | LDD0704 | [11] |
| LDCM0388 | CL118 | HCT 116 | C105(0.96); C236(1.00); C101(1.18); C269(1.20) | LDD0705 | [11] |
| LDCM0389 | CL119 | HCT 116 | C236(0.89); C105(0.95); C269(1.07); C101(1.14) | LDD0706 | [11] |
| LDCM0390 | CL12 | HCT 116 | C434(0.53); C348(0.93); C236(1.01); C105(1.05) | LDD0707 | [11] |
| LDCM0391 | CL120 | HCT 116 | C101(0.99); C105(1.00); C96(1.03); C236(1.12) | LDD0708 | [11] |
| LDCM0392 | CL121 | HCT 116 | C348(0.71); C269(0.81); C236(1.00); C105(1.21) | LDD0709 | [11] |
| LDCM0393 | CL122 | HCT 116 | C348(0.66); C269(0.67); C236(0.68); C105(0.97) | LDD0710 | [11] |
| LDCM0394 | CL123 | HCT 116 | C269(0.73); C348(0.74); C105(0.77); C236(0.78) | LDD0711 | [11] |
| LDCM0395 | CL124 | HCT 116 | C348(0.60); C269(0.63); C236(0.71); C105(1.12) | LDD0712 | [11] |
| LDCM0396 | CL125 | HCT 116 | C96(0.70); C348(0.94); C101(0.97); C236(1.12) | LDD0713 | [11] |
| LDCM0397 | CL126 | HCT 116 | C96(0.68); C348(0.80); C101(0.83); C236(1.09) | LDD0714 | [11] |
| LDCM0398 | CL127 | HCT 116 | C96(0.81); C348(0.89); C101(0.98); C269(0.98) | LDD0715 | [11] |
| LDCM0399 | CL128 | HCT 116 | C348(0.72); C96(0.89); C236(0.95); C101(0.97) | LDD0716 | [11] |
| LDCM0400 | CL13 | HCT 116 | C434(0.52); C105(0.95); C236(1.28); C348(1.28) | LDD0717 | [11] |
| LDCM0401 | CL14 | HCT 116 | C434(0.84); C105(0.97); C236(1.20); C348(1.25) | LDD0718 | [11] |
| LDCM0402 | CL15 | HCT 116 | C434(0.43); C105(0.87); C348(1.26); C236(1.34) | LDD0719 | [11] |
| LDCM0403 | CL16 | HCT 116 | C269(0.74); C348(0.75); C236(0.98); C101(1.03) | LDD0720 | [11] |
| LDCM0404 | CL17 | HCT 116 | C269(0.80); C101(0.87); C236(1.05); C96(1.11) | LDD0721 | [11] |
| LDCM0405 | CL18 | HCT 116 | C101(0.81); C269(0.95); C96(1.01); C348(1.03) | LDD0722 | [11] |
| LDCM0406 | CL19 | HCT 116 | C101(0.76); C236(0.93); C269(0.96); C96(0.98) | LDD0723 | [11] |
| LDCM0407 | CL2 | HCT 116 | C434(1.03); C348(1.08); C105(1.13); C236(1.26) | LDD0724 | [11] |
| LDCM0408 | CL20 | HCT 116 | C236(0.84); C101(0.88); C269(0.90); C96(1.03) | LDD0725 | [11] |
| LDCM0409 | CL21 | HCT 116 | C269(0.73); C236(0.73); C101(0.86); C105(1.17) | LDD0726 | [11] |
| LDCM0410 | CL22 | HCT 116 | C269(0.56); C236(0.65); C101(1.09); C96(1.14) | LDD0727 | [11] |
| LDCM0411 | CL23 | HCT 116 | C348(0.77); C101(0.87); C96(0.89); C236(0.93) | LDD0728 | [11] |
| LDCM0412 | CL24 | HCT 116 | C269(0.62); C236(0.80); C348(0.86); C96(1.06) | LDD0729 | [11] |
| LDCM0413 | CL25 | HCT 116 | C101(0.99); C105(1.20); C236(0.95); C269(0.79) | LDD0730 | [11] |
| LDCM0414 | CL26 | HCT 116 | C101(1.03); C105(1.19); C236(0.89); C269(0.83) | LDD0731 | [11] |
| LDCM0415 | CL27 | HCT 116 | C101(0.93); C105(1.23); C236(0.96); C269(0.93) | LDD0732 | [11] |
| LDCM0416 | CL28 | HCT 116 | C101(0.93); C105(1.11); C236(0.85); C269(0.93) | LDD0733 | [11] |
| LDCM0417 | CL29 | HCT 116 | C101(0.91); C105(1.19); C236(0.75); C269(0.70) | LDD0734 | [11] |
| LDCM0418 | CL3 | HCT 116 | C105(1.04); C236(1.39); C348(1.17); C434(0.87) | LDD0735 | [11] |
| LDCM0419 | CL30 | HCT 116 | C101(0.92); C105(1.13); C236(0.92); C269(0.92) | LDD0736 | [11] |
| LDCM0420 | CL31 | HCT 116 | C101(1.08); C105(0.98); C236(0.97); C269(0.83) | LDD0737 | [11] |
| LDCM0421 | CL32 | HCT 116 | C101(1.04); C105(1.09); C236(0.94); C269(0.74) | LDD0738 | [11] |
| LDCM0422 | CL33 | HCT 116 | C101(0.98); C105(1.04); C236(0.86); C269(0.79) | LDD0739 | [11] |
| LDCM0423 | CL34 | HCT 116 | C101(0.95); C105(0.92); C236(0.69); C269(0.52) | LDD0740 | [11] |
| LDCM0424 | CL35 | HCT 116 | C101(0.90); C105(0.99); C236(0.75); C269(0.71) | LDD0741 | [11] |
| LDCM0425 | CL36 | HCT 116 | C101(1.00); C105(0.94); C236(0.82); C269(0.85) | LDD0742 | [11] |
| LDCM0426 | CL37 | HCT 116 | C101(0.92); C105(0.94); C236(0.78); C269(0.74) | LDD0743 | [11] |
| LDCM0428 | CL39 | HCT 116 | C101(1.12); C105(1.00); C236(0.79); C269(0.70) | LDD0745 | [11] |
| LDCM0429 | CL4 | HCT 116 | C105(0.95); C236(1.02); C348(1.14); C434(0.85) | LDD0746 | [11] |
| LDCM0430 | CL40 | HCT 116 | C101(1.06); C105(0.98); C236(0.72); C269(0.85) | LDD0747 | [11] |
| LDCM0431 | CL41 | HCT 116 | C101(1.08); C105(0.94); C236(0.93); C269(0.83) | LDD0748 | [11] |
| LDCM0432 | CL42 | HCT 116 | C101(1.03); C105(0.92); C236(0.54); C269(0.43) | LDD0749 | [11] |
| LDCM0433 | CL43 | HCT 116 | C101(1.04); C105(0.96); C236(0.71); C269(0.53) | LDD0750 | [11] |
| LDCM0434 | CL44 | HCT 116 | C101(1.09); C105(0.93); C236(0.78); C269(0.70) | LDD0751 | [11] |
| LDCM0435 | CL45 | HCT 116 | C101(1.01); C105(0.91); C236(0.54); C269(0.42) | LDD0752 | [11] |
| LDCM0436 | CL46 | HCT 116 | C105(1.06); C348(0.87); C434(1.33) | LDD0753 | [11] |
| LDCM0437 | CL47 | HCT 116 | C105(1.01); C348(0.81); C434(1.26) | LDD0754 | [11] |
| LDCM0438 | CL48 | HCT 116 | C105(0.94); C348(1.10); C434(1.27) | LDD0755 | [11] |
| LDCM0439 | CL49 | HCT 116 | C105(0.98); C348(0.97); C434(1.10) | LDD0756 | [11] |
| LDCM0440 | CL5 | HCT 116 | C105(0.97); C236(1.31); C348(1.19); C434(0.86) | LDD0757 | [11] |
| LDCM0441 | CL50 | HCT 116 | C105(1.04); C348(1.06); C434(1.40) | LDD0758 | [11] |
| LDCM0442 | CL51 | HCT 116 | C105(1.16); C348(1.17); C434(1.31) | LDD0759 | [11] |
| LDCM0443 | CL52 | HCT 116 | C105(1.01); C348(0.91); C434(1.31) | LDD0760 | [11] |
| LDCM0444 | CL53 | HCT 116 | C105(0.96); C348(0.90); C434(1.29) | LDD0761 | [11] |
| LDCM0445 | CL54 | HCT 116 | C105(1.10); C348(0.84); C434(1.24) | LDD0762 | [11] |
| LDCM0446 | CL55 | HCT 116 | C105(1.00); C348(1.14); C434(1.22) | LDD0763 | [11] |
| LDCM0447 | CL56 | HCT 116 | C105(1.04); C348(0.97); C434(1.05) | LDD0764 | [11] |
| LDCM0448 | CL57 | HCT 116 | C105(0.95); C348(0.94); C434(1.29) | LDD0765 | [11] |
| LDCM0449 | CL58 | HCT 116 | C105(0.92); C348(0.86); C434(1.33) | LDD0766 | [11] |
| LDCM0450 | CL59 | HCT 116 | C105(0.97); C348(1.07); C434(1.20) | LDD0767 | [11] |
| LDCM0451 | CL6 | HCT 116 | C105(0.98); C236(1.45); C348(1.19); C434(0.72) | LDD0768 | [11] |
| LDCM0452 | CL60 | HCT 116 | C105(1.03); C348(0.99); C434(1.12) | LDD0769 | [11] |
| LDCM0453 | CL61 | HCT 116 | C101(1.21); C105(1.10); C236(0.96); C348(1.11) | LDD0770 | [11] |
| LDCM0454 | CL62 | HCT 116 | C101(0.91); C105(1.07); C236(0.88); C348(0.93) | LDD0771 | [11] |
| LDCM0455 | CL63 | HCT 116 | C101(0.98); C105(1.28); C236(0.91); C348(1.06) | LDD0772 | [11] |
| LDCM0456 | CL64 | HCT 116 | C101(1.01); C105(1.00); C236(0.95); C348(1.11) | LDD0773 | [11] |
| LDCM0457 | CL65 | HCT 116 | C101(1.05); C105(0.99); C236(0.97); C348(1.17) | LDD0774 | [11] |
| LDCM0458 | CL66 | HCT 116 | C101(1.02); C105(1.15); C236(0.79); C348(1.13) | LDD0775 | [11] |
| LDCM0459 | CL67 | HCT 116 | C101(1.04); C105(1.17); C236(1.00); C348(1.14) | LDD0776 | [11] |
| LDCM0460 | CL68 | HCT 116 | C101(1.17); C105(1.09); C236(1.02); C348(1.15) | LDD0777 | [11] |
| LDCM0461 | CL69 | HCT 116 | C101(1.01); C105(0.99); C236(1.09); C348(0.78) | LDD0778 | [11] |
| LDCM0462 | CL7 | HCT 116 | C105(1.14); C236(1.23); C348(1.26); C434(0.59) | LDD0779 | [11] |
| LDCM0463 | CL70 | HCT 116 | C101(1.01); C105(1.04); C236(0.97); C348(0.94) | LDD0780 | [11] |
| LDCM0464 | CL71 | HCT 116 | C101(1.11); C105(1.04); C236(0.87); C348(1.00) | LDD0781 | [11] |
| LDCM0465 | CL72 | HCT 116 | C101(1.02); C105(0.98); C236(0.90); C348(1.01) | LDD0782 | [11] |
| LDCM0466 | CL73 | HCT 116 | C101(0.89); C105(1.03); C236(0.92); C348(1.09) | LDD0783 | [11] |
| LDCM0467 | CL74 | HCT 116 | C101(1.07); C105(1.05); C236(0.87); C348(0.99) | LDD0784 | [11] |
| LDCM0469 | CL76 | HCT 116 | C105(0.96); C236(1.55); C269(1.07); C348(0.80) | LDD0786 | [11] |
| LDCM0470 | CL77 | HCT 116 | C105(0.82); C236(0.66); C269(0.86); C348(0.78) | LDD0787 | [11] |
| LDCM0471 | CL78 | HCT 116 | C105(0.93); C236(1.05); C269(1.07); C348(0.76) | LDD0788 | [11] |
| LDCM0472 | CL79 | HCT 116 | C105(1.03); C236(0.80); C269(0.95); C348(0.68) | LDD0789 | [11] |
| LDCM0473 | CL8 | HCT 116 | C105(1.01); C236(0.92); C348(0.97); C434(0.46) | LDD0790 | [11] |
| LDCM0474 | CL80 | HCT 116 | C105(0.85); C236(1.62); C269(0.97); C348(0.88) | LDD0791 | [11] |
| LDCM0475 | CL81 | HCT 116 | C105(0.92); C236(1.17); C269(1.06); C348(0.87) | LDD0792 | [11] |
| LDCM0476 | CL82 | HCT 116 | C105(0.90); C236(0.75); C269(0.73); C348(0.59) | LDD0793 | [11] |
| LDCM0477 | CL83 | HCT 116 | C105(0.96); C236(0.85); C269(0.87); C348(0.66) | LDD0794 | [11] |
| LDCM0478 | CL84 | HCT 116 | C105(0.85); C236(0.52); C269(0.61); C348(0.54) | LDD0795 | [11] |
| LDCM0479 | CL85 | HCT 116 | C105(0.95); C236(1.44); C269(1.29); C348(0.93) | LDD0796 | [11] |
| LDCM0480 | CL86 | HCT 116 | C105(0.91); C236(1.03); C269(1.08); C348(1.18) | LDD0797 | [11] |
| LDCM0481 | CL87 | HCT 116 | C105(0.84); C236(1.36); C269(1.05); C348(0.92) | LDD0798 | [11] |
| LDCM0482 | CL88 | HCT 116 | C105(0.83); C236(0.79); C269(0.89); C348(0.72) | LDD0799 | [11] |
| LDCM0483 | CL89 | HCT 116 | C105(0.80); C236(0.64); C269(0.48); C348(0.49) | LDD0800 | [11] |
| LDCM0484 | CL9 | HCT 116 | C105(1.14); C236(1.02); C348(0.99); C434(0.75) | LDD0801 | [11] |
| LDCM0485 | CL90 | HCT 116 | C105(0.76); C236(1.30); C269(1.19); C348(1.12) | LDD0802 | [11] |
| LDCM0486 | CL91 | HCT 116 | C101(0.93); C105(1.18); C236(0.77); C269(1.03) | LDD0803 | [11] |
| LDCM0487 | CL92 | HCT 116 | C101(1.31); C105(1.12); C236(0.98); C269(0.90) | LDD0804 | [11] |
| LDCM0488 | CL93 | HCT 116 | C101(1.36); C105(1.03); C236(0.85); C269(0.80) | LDD0805 | [11] |
| LDCM0489 | CL94 | HCT 116 | C101(1.07); C105(0.95); C236(0.82); C269(0.85) | LDD0806 | [11] |
| LDCM0490 | CL95 | HCT 116 | C101(1.21); C105(1.13); C236(0.84); C269(0.92) | LDD0807 | [11] |
| LDCM0491 | CL96 | HCT 116 | C101(1.11); C105(1.16); C236(0.84); C269(0.80) | LDD0808 | [11] |
| LDCM0492 | CL97 | HCT 116 | C101(1.09); C105(1.25); C236(1.07); C269(0.95) | LDD0809 | [11] |
| LDCM0493 | CL98 | HCT 116 | C101(1.03); C105(1.27); C236(1.08); C269(1.00) | LDD0810 | [11] |
| LDCM0494 | CL99 | HCT 116 | C101(1.09); C105(1.24); C236(1.03); C269(0.87) | LDD0811 | [11] |
| LDCM0495 | E2913 | HEK-293T | C105(0.93); C269(1.07); C434(1.14); C101(1.11) | LDD1698 | [33] |
| LDCM0625 | F8 | Ramos | C434(0.73); C105(1.10); C269(1.06); C311(15.53) | LDD2187 | [34] |
| LDCM0572 | Fragment10 | Ramos | C434(0.85); C105(0.93); C269(0.23); C311(0.37) | LDD2189 | [34] |
| LDCM0573 | Fragment11 | Ramos | C434(0.65); C105(0.10); C269(4.06); C311(6.82) | LDD2190 | [34] |
| LDCM0574 | Fragment12 | Ramos | C434(0.60); C105(0.57); C269(0.29); C311(0.61) | LDD2191 | [34] |
| LDCM0575 | Fragment13 | Ramos | C434(0.83); C105(0.88); C269(0.90); C311(0.80) | LDD2192 | [34] |
| LDCM0576 | Fragment14 | Ramos | C434(0.55); C105(0.58); C269(1.17); C311(0.67) | LDD2193 | [34] |
| LDCM0579 | Fragment20 | Ramos | C434(0.54); C105(0.51); C269(0.22); C311(0.29) | LDD2194 | [34] |
| LDCM0580 | Fragment21 | Ramos | C434(0.85); C105(0.88); C269(0.69); C311(1.10) | LDD2195 | [34] |
| LDCM0582 | Fragment23 | Ramos | C434(1.17); C105(1.06); C269(1.03); C311(0.70) | LDD2196 | [34] |
| LDCM0578 | Fragment27 | Ramos | C434(0.88); C105(0.89); C269(0.91); C311(0.72) | LDD2197 | [34] |
| LDCM0586 | Fragment28 | Ramos | C434(0.80); C105(1.33); C269(0.88); C311(0.67) | LDD2198 | [34] |
| LDCM0588 | Fragment30 | Ramos | C434(1.07); C105(0.98); C269(0.54); C311(1.32) | LDD2199 | [34] |
| LDCM0589 | Fragment31 | Ramos | C434(1.04); C105(0.94); C269(0.74); C311(1.29) | LDD2200 | [34] |
| LDCM0590 | Fragment32 | Ramos | C434(0.61); C105(0.56); C269(0.30); C311(0.32) | LDD2201 | [34] |
| LDCM0468 | Fragment33 | HCT 116 | C101(1.29); C105(1.00); C236(0.98); C348(1.18) | LDD0785 | [11] |
| LDCM0596 | Fragment38 | Ramos | C434(1.05); C105(0.79); C269(0.48); C311(0.78) | LDD2203 | [34] |
| LDCM0566 | Fragment4 | Ramos | C434(0.84); C105(1.11); C269(0.71); C311(1.18) | LDD2184 | [34] |
| LDCM0427 | Fragment51 | HCT 116 | C101(0.90); C105(0.89); C236(0.67); C269(0.49) | LDD0744 | [11] |
| LDCM0610 | Fragment52 | Ramos | C434(1.03); C105(1.05); C269(0.60); C311(0.69) | LDD2204 | [34] |
| LDCM0614 | Fragment56 | Ramos | C434(1.15); C105(1.14); C269(0.63); C311(0.89) | LDD2205 | [34] |
| LDCM0569 | Fragment7 | Ramos | C434(0.57); C105(0.77); C269(0.54); C311(0.45) | LDD2186 | [34] |
| LDCM0571 | Fragment9 | Ramos | C434(0.67); C105(0.69); C269(0.19); C311(0.31) | LDD2188 | [34] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [25] |
| LDCM0179 | JZ128 | PC-3 | N.A. | LDD0462 | [9] |
| LDCM0022 | KB02 | HEK-293T | C269(0.89); C105(1.00); C101(1.01) | LDD1492 | [33] |
| LDCM0023 | KB03 | HEK-293T | C269(1.12); C105(1.00); C101(1.07) | LDD1497 | [33] |
| LDCM0024 | KB05 | COLO792 | C105(1.12) | LDD3310 | [35] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C348(0.99) | LDD2121 | [31] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [25] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C348(0.70); C105(0.56) | LDD2089 | [31] |
| LDCM0503 | Nucleophilic fragment 14b | MDA-MB-231 | C269(0.22); C311(0.39) | LDD2096 | [31] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C348(1.04); C269(1.25); C105(1.10); C101(0.91) | LDD2099 | [31] |
| LDCM0523 | Nucleophilic fragment 24b | MDA-MB-231 | C348(1.74); C269(0.44); C311(0.46) | LDD2116 | [31] |
| LDCM0525 | Nucleophilic fragment 25b | MDA-MB-231 | C269(0.53); C311(0.56) | LDD2118 | [31] |
| LDCM0529 | Nucleophilic fragment 27b | MDA-MB-231 | C269(0.22); C311(0.55); C434(0.98) | LDD2122 | [31] |
| LDCM0531 | Nucleophilic fragment 28b | MDA-MB-231 | C348(1.64); C269(0.33); C311(0.37) | LDD2124 | [31] |
| LDCM0533 | Nucleophilic fragment 29b | MDA-MB-231 | C311(0.44) | LDD2126 | [31] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C348(0.38) | LDD2148 | [31] |
| LDCM0555 | Nucleophilic fragment 7b | MDA-MB-231 | C311(0.23) | LDD2149 | [31] |
| LDCM0557 | Nucleophilic fragment 8b | MDA-MB-231 | C269(0.28); C311(0.41) | LDD2151 | [31] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C105(1.39) | LDD2206 | [36] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C105(0.85) | LDD2207 | [36] |
| LDCM0131 | RA190 | MM1.R | C105(1.34); C434(1.30) | LDD0304 | [37] |
| LDCM0021 | THZ1 | HeLa S3 | C434(1.11); C269(1.09) | LDD0460 | [9] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Huntingtin (HTT) | Huntingtin family | P42858 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Zinc finger protein with KRAB and SCAN domains 3 (ZKSCAN3) | Krueppel C2H2-type zinc-finger protein family | Q9BRR0 | |||
Other
References
