Details of the Target
General Information of Target
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
A-EBA Probe Info |
 |
3.66 | LDD0215 | [1] | |
|
TH211 Probe Info |
 |
Y272(20.00); Y246(17.43); Y241(14.01); Y310(13.32) | LDD0260 | [2] | |
|
C-Sul Probe Info |
 |
8.42 | LDD0066 | [3] | |
|
AZ-9 Probe Info |
 |
D312(0.95); E36(1.21) | LDD2208 | [4] | |
|
ONAyne Probe Info |
 |
K86(0.00); K163(0.00) | LDD0273 | [5] | |
|
OPA-S-S-alkyne Probe Info |
 |
K63(1.66); K86(2.06); K163(3.88); K217(8.01) | LDD3494 | [6] | |
|
Probe 1 Probe Info |
 |
Y190(32.20); Y206(38.76); Y214(93.32); Y225(21.11) | LDD3495 | [7] | |
|
5E-2FA Probe Info |
 |
H275(0.00); H248(0.00); H128(0.00) | LDD2235 | [8] | |
|
AMP probe Probe Info |
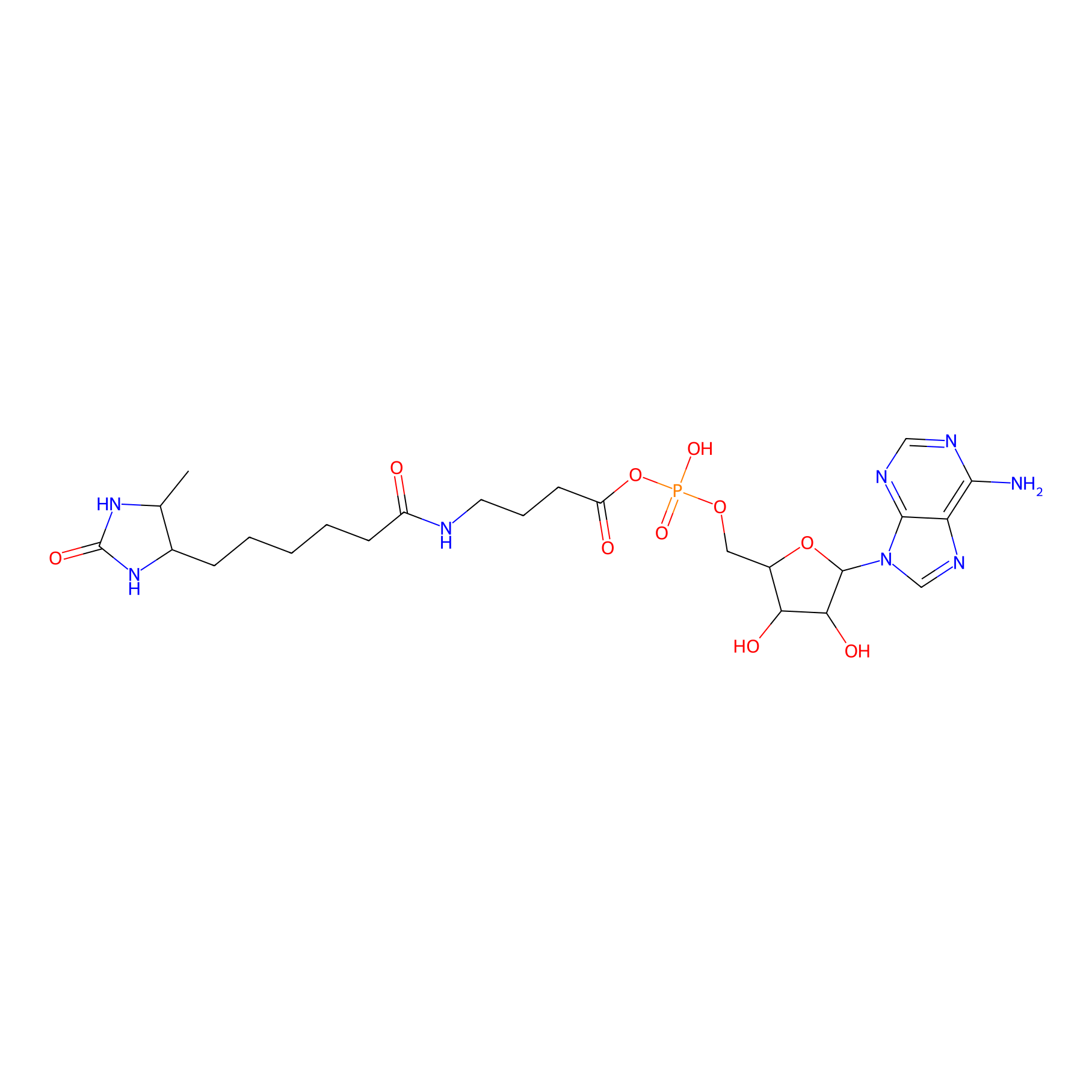 |
N.A. | LDD0200 | [9] | |
|
ATP probe Probe Info |
 |
K80(0.00); K86(0.00); K77(0.00); K30(0.00) | LDD0199 | [9] | |
|
m-APA Probe Info |
 |
H275(0.00); H128(0.00) | LDD2231 | [8] | |
|
ATP probe Probe Info |
 |
K86(0.00); K41(0.00) | LDD0035 | [10] | |
|
1d-yne Probe Info |
 |
K80(0.00); K63(0.00); K77(0.00) | LDD0356 | [11] | |
|
NHS Probe Info |
 |
K63(0.00); K30(0.00); K86(0.00); K9(0.00) | LDD0010 | [12] | |
|
OSF Probe Info |
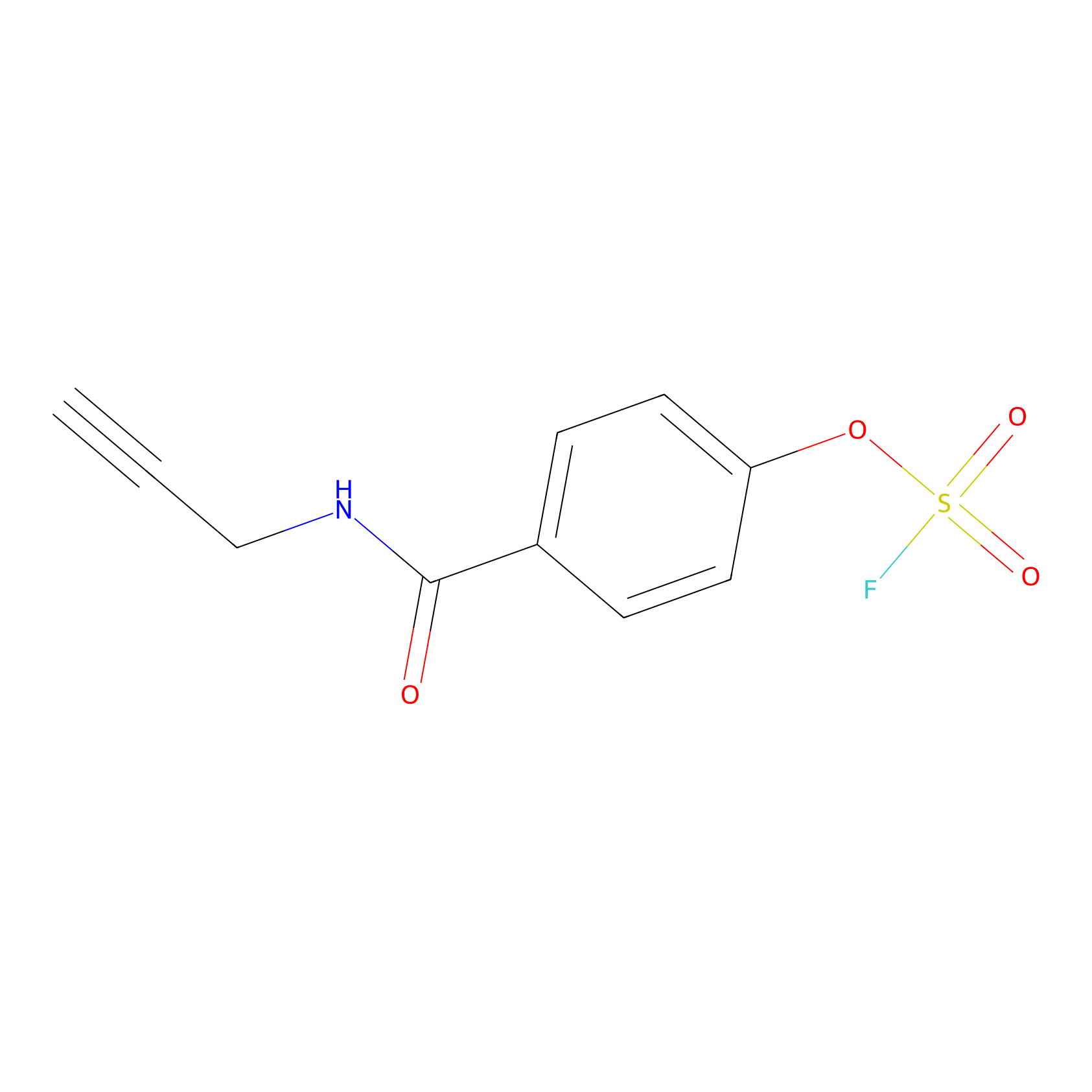 |
Y357(0.00); Y304(0.00); Y234(0.00); Y335(0.00) | LDD0029 | [13] | |
|
SF Probe Info |
 |
K63(0.00); Y313(0.00); Y288(0.00); Y243(0.00) | LDD0028 | [13] | |
|
STPyne Probe Info |
 |
K86(0.00); K217(0.00) | LDD0009 | [12] | |
|
1c-yne Probe Info |
 |
K80(0.00); K9(0.00) | LDD0228 | [11] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C040 Probe Info |
 |
6.59 | LDD1740 | [14] | |
|
C041 Probe Info |
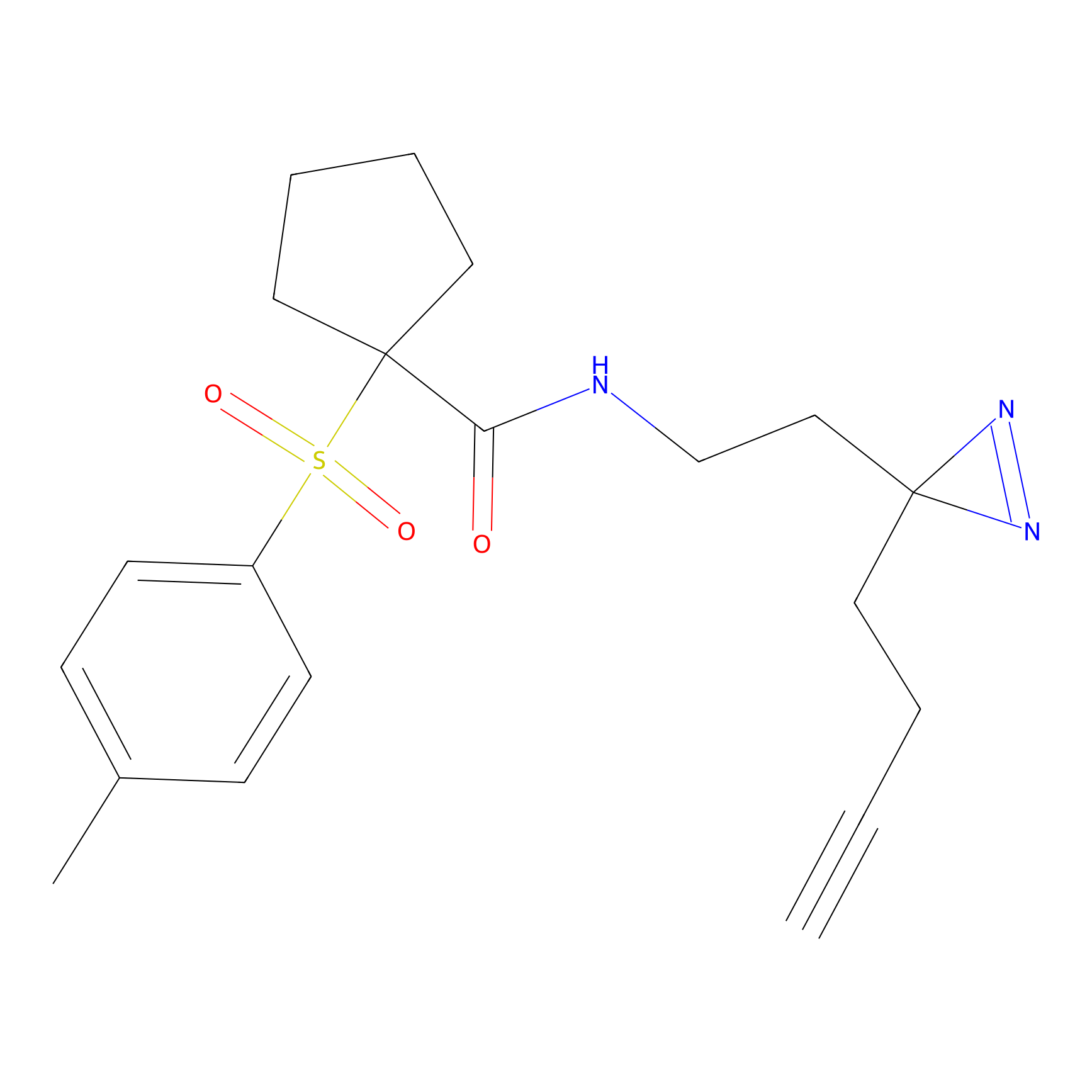 |
6.82 | LDD1741 | [14] | |
|
C063 Probe Info |
 |
12.73 | LDD1760 | [14] | |
|
C087 Probe Info |
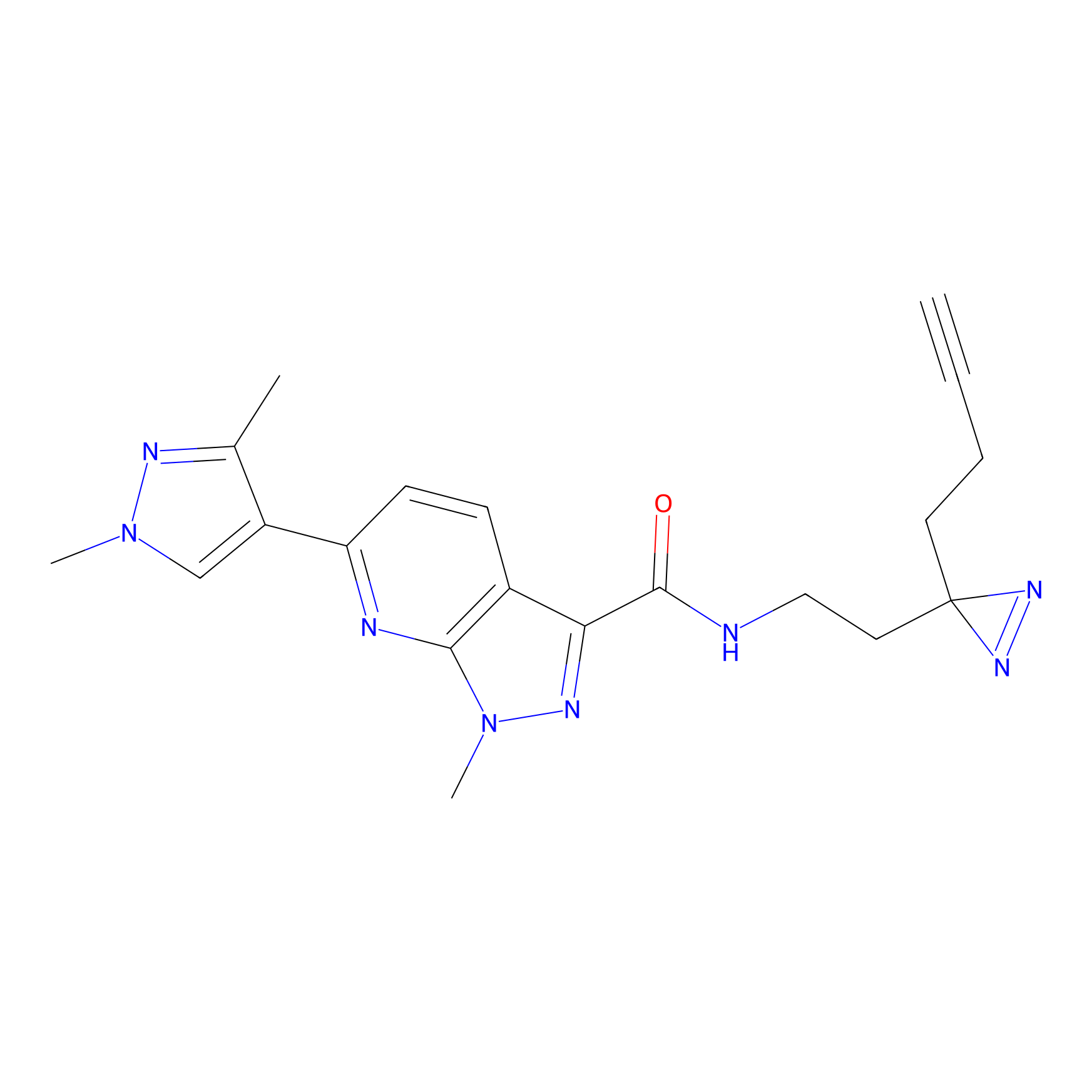 |
9.32 | LDD1779 | [14] | |
|
C139 Probe Info |
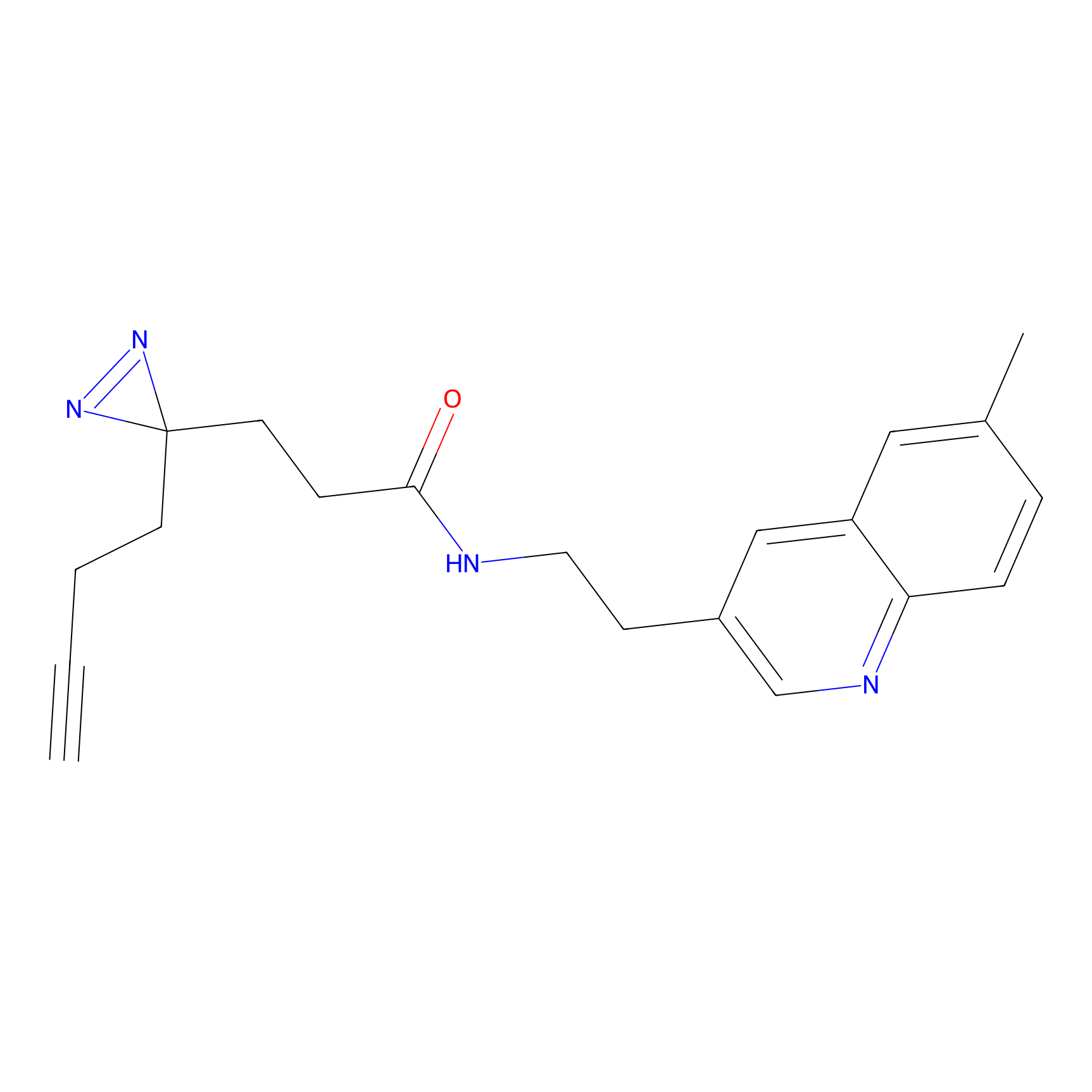 |
10.63 | LDD1821 | [14] | |
|
C145 Probe Info |
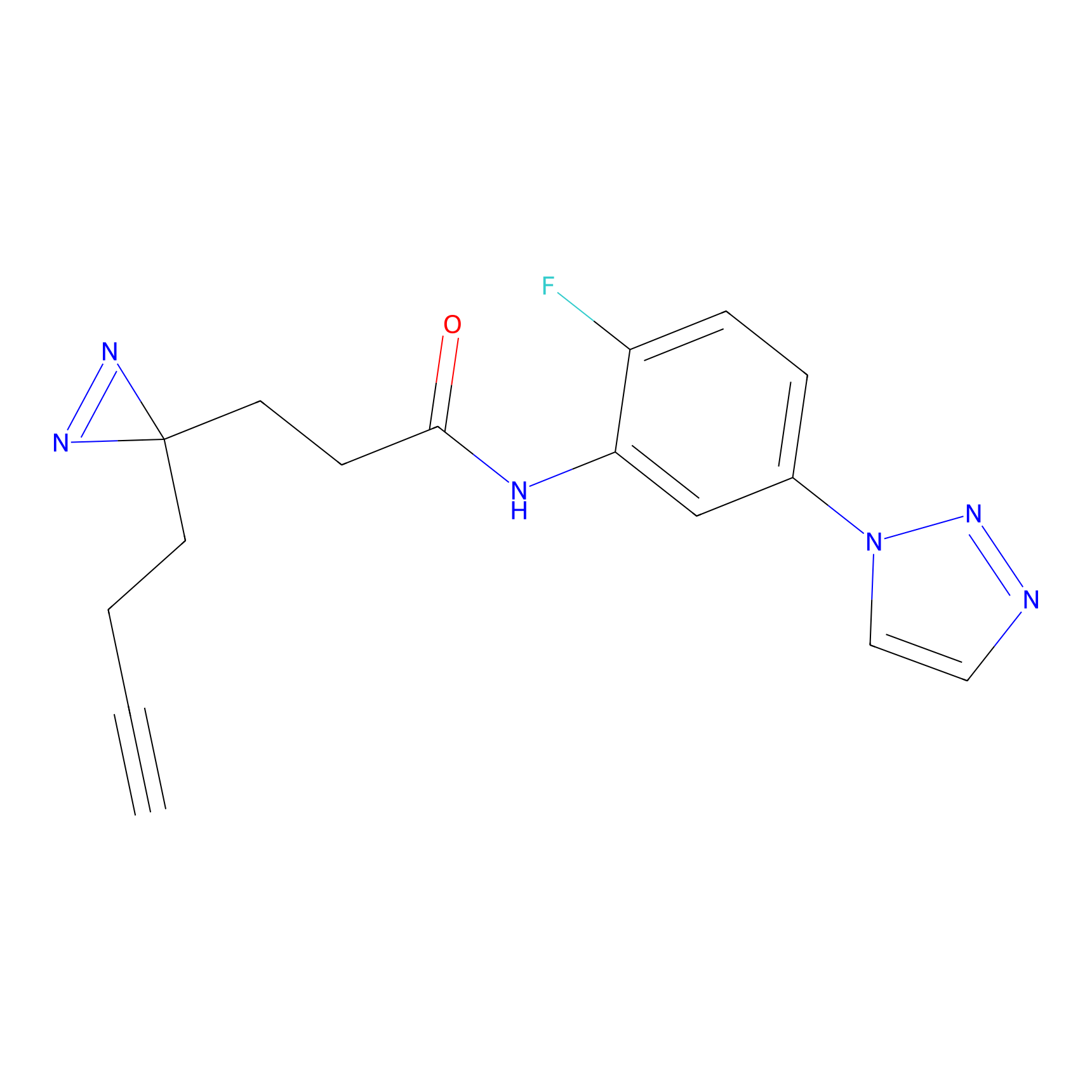 |
8.75 | LDD1827 | [14] | |
|
C165 Probe Info |
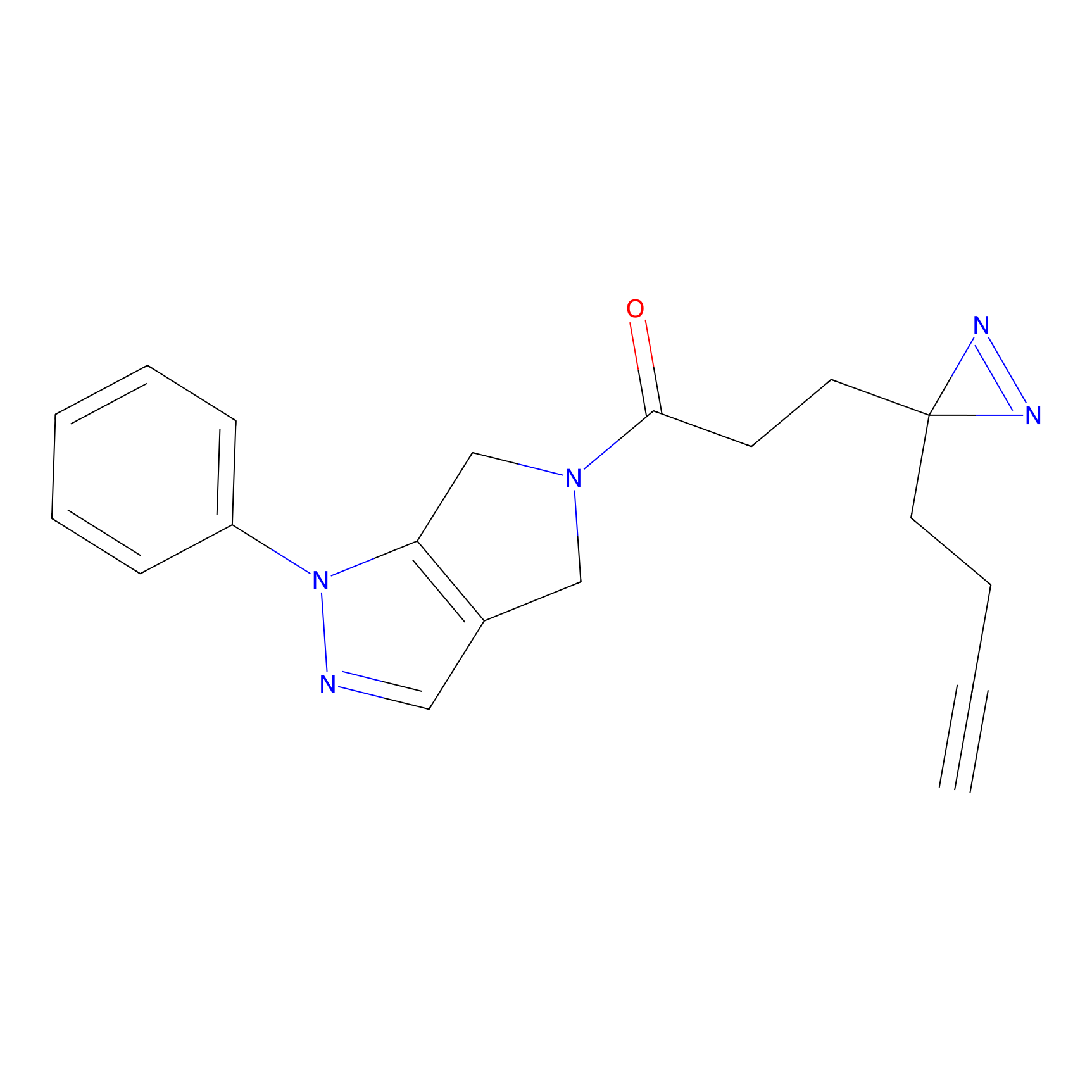 |
18.51 | LDD1845 | [14] | |
|
C232 Probe Info |
 |
47.50 | LDD1905 | [14] | |
|
C302 Probe Info |
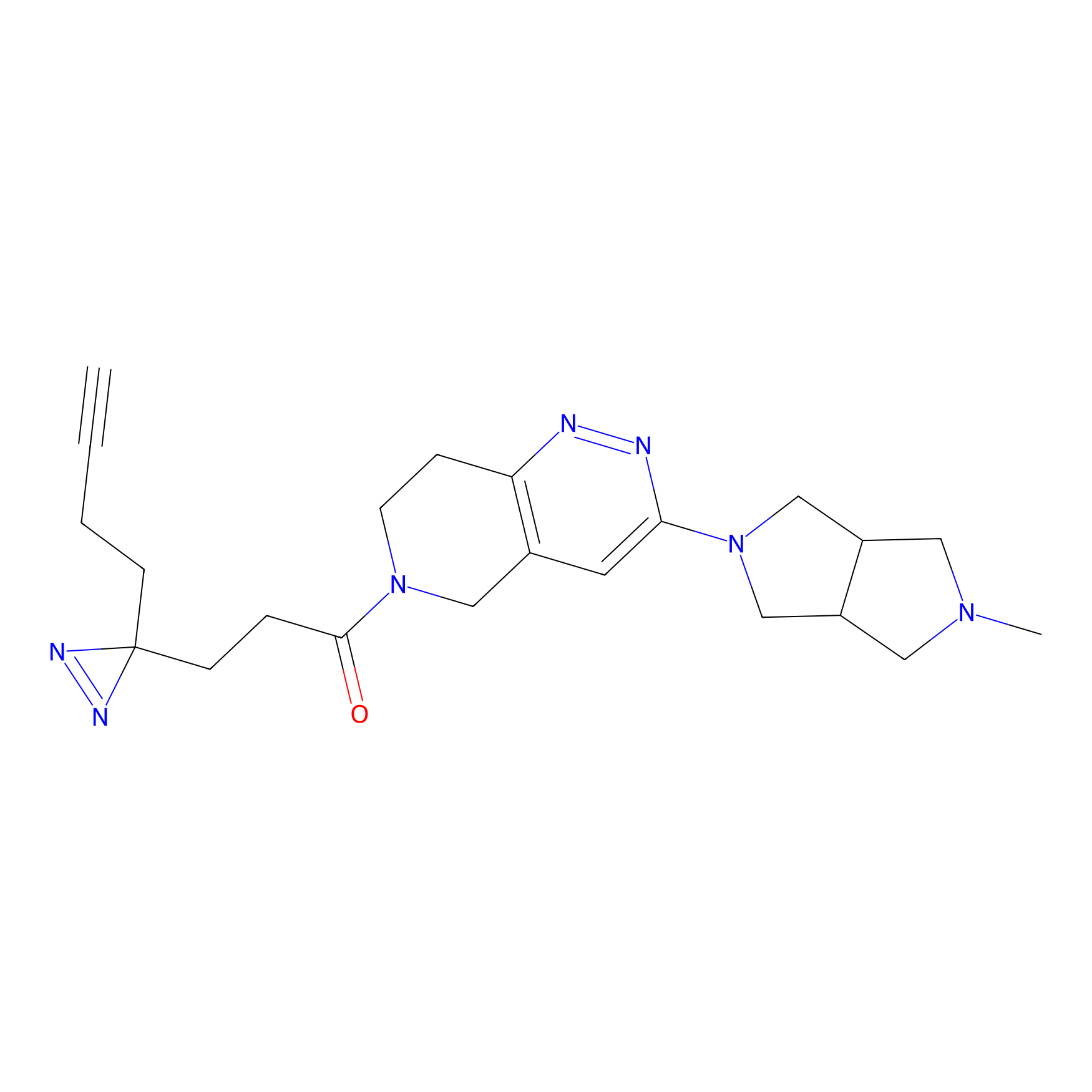 |
5.70 | LDD1971 | [14] | |
|
C313 Probe Info |
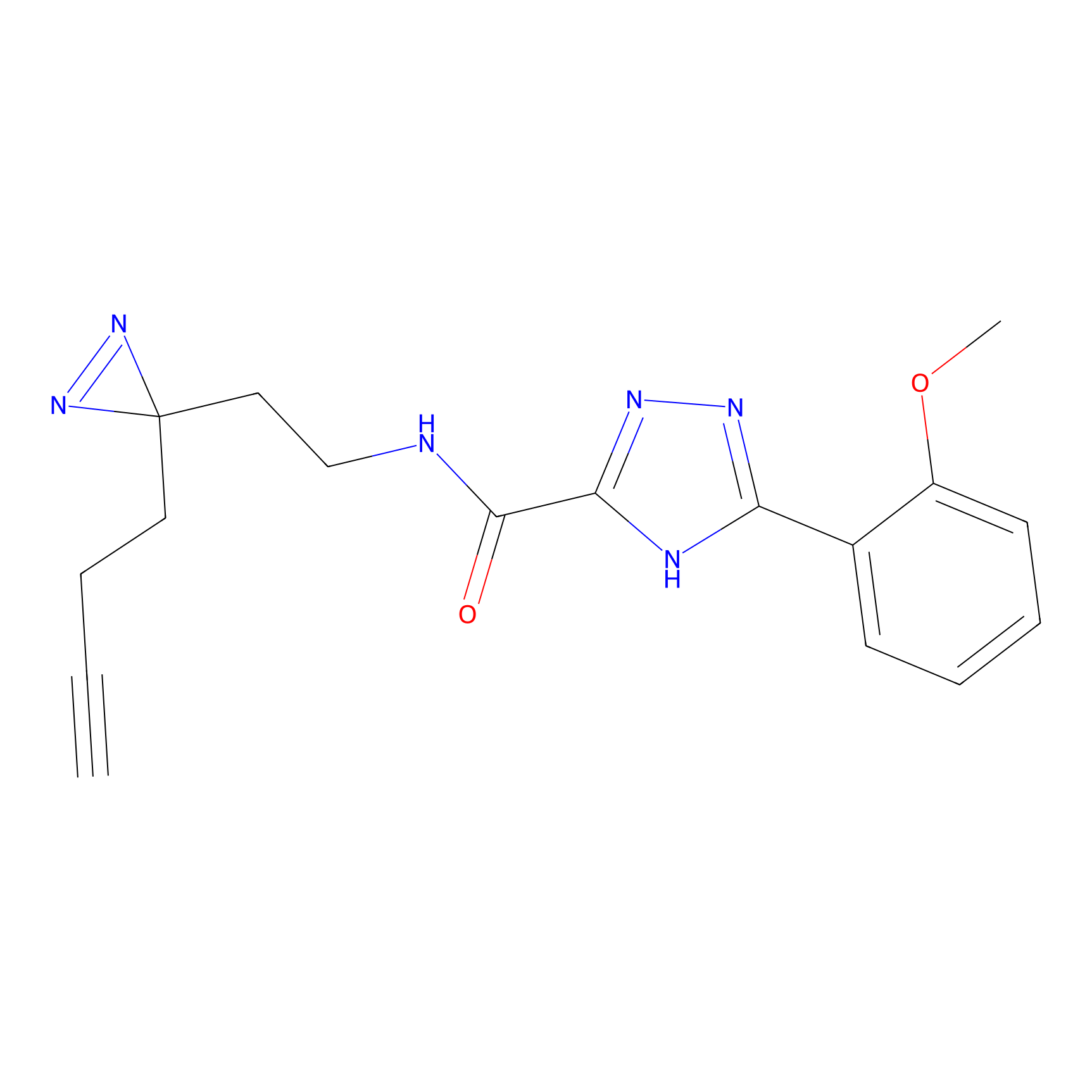 |
14.62 | LDD1980 | [14] | |
|
C314 Probe Info |
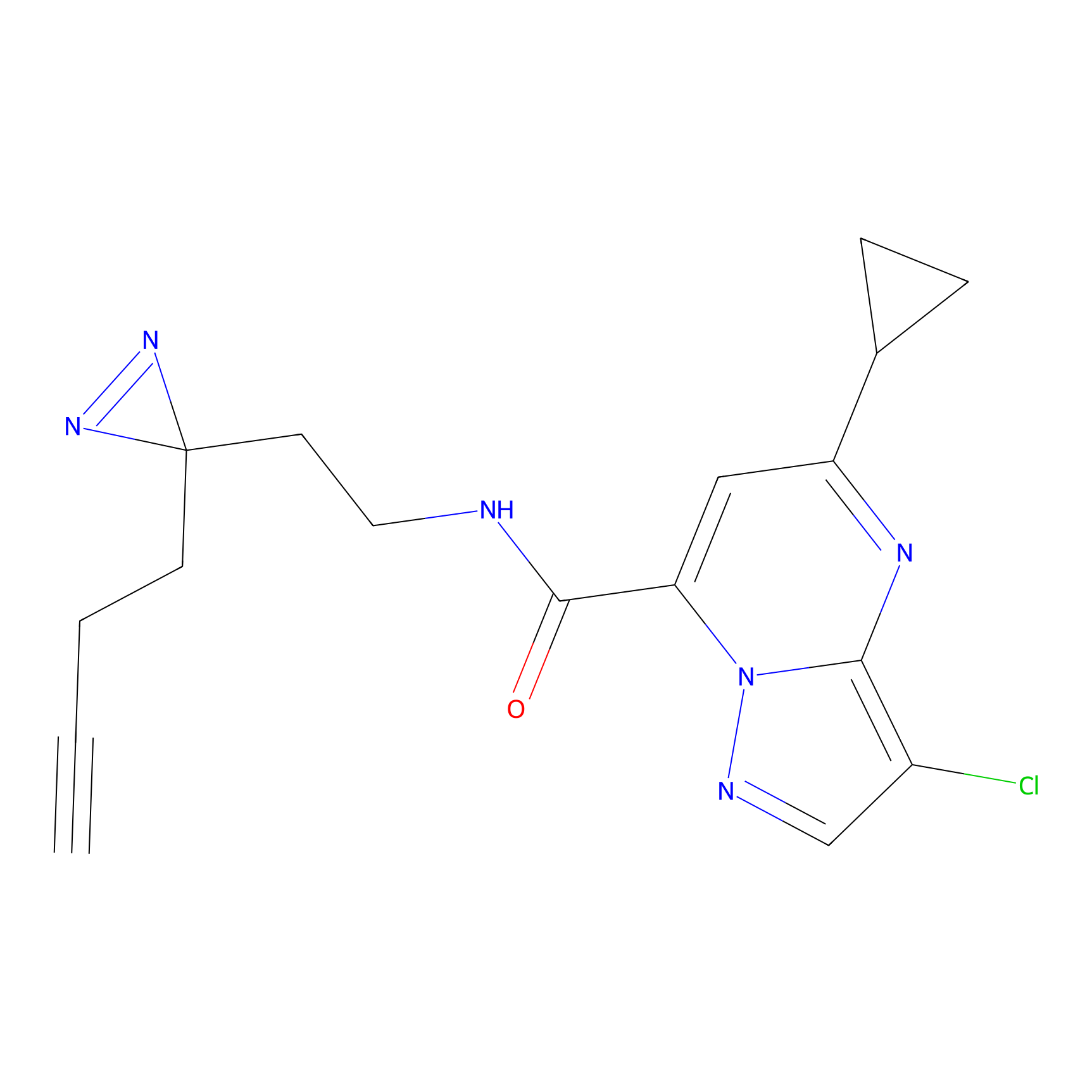 |
11.16 | LDD1981 | [14] | |
|
C338 Probe Info |
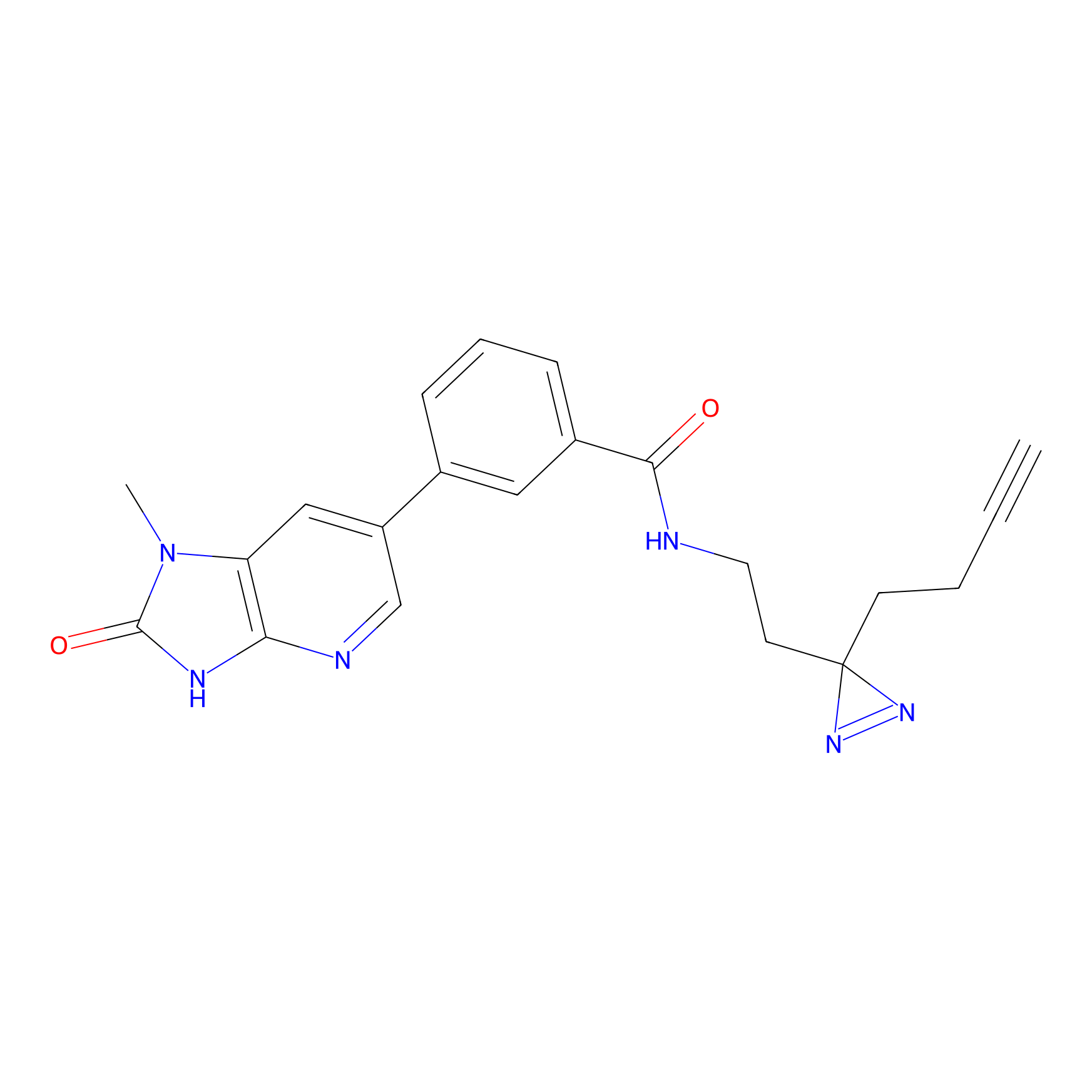 |
19.03 | LDD2001 | [14] | |
|
C346 Probe Info |
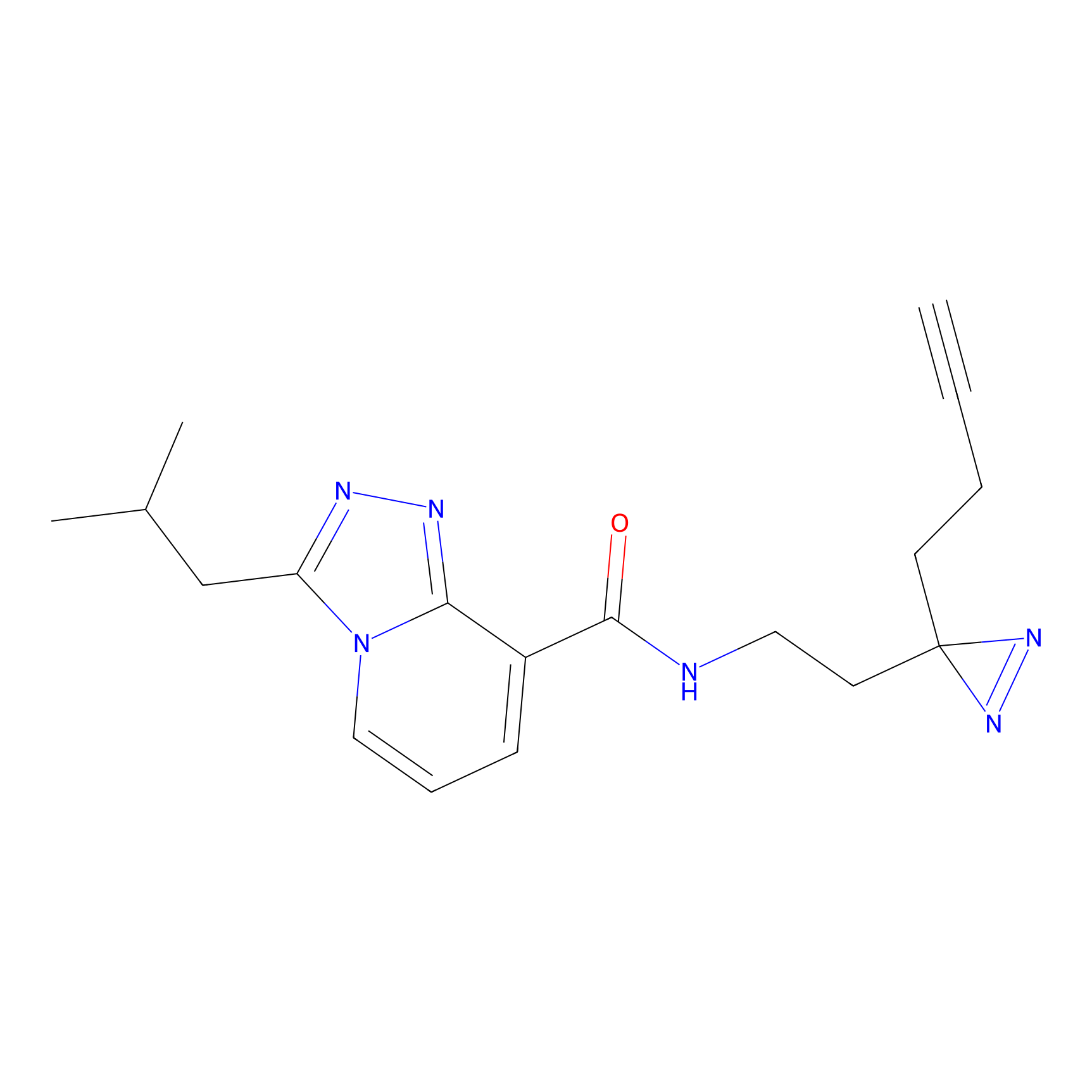 |
10.63 | LDD2007 | [14] | |
|
C348 Probe Info |
 |
11.71 | LDD2009 | [14] | |
|
C353 Probe Info |
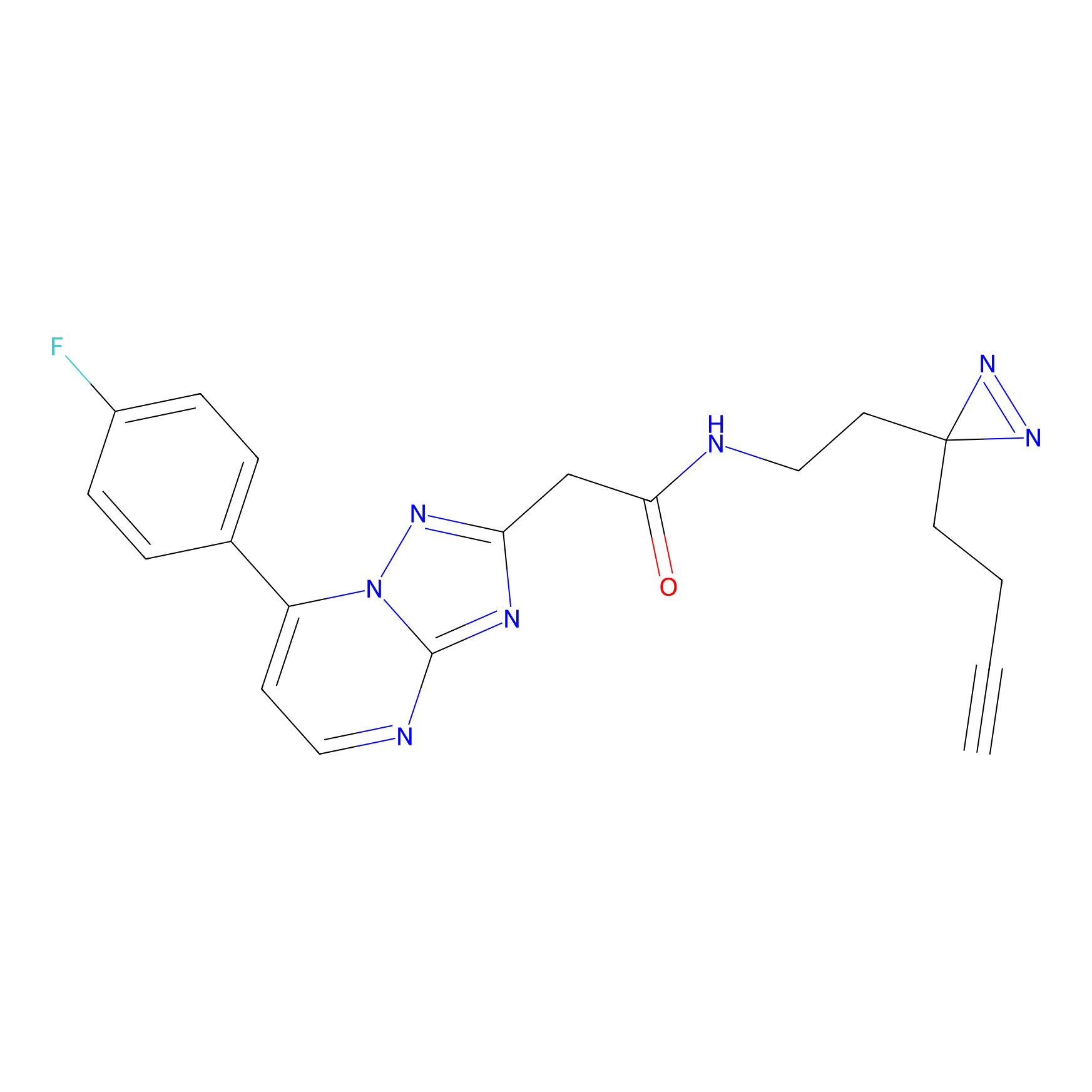 |
7.67 | LDD2014 | [14] | |
|
C355 Probe Info |
 |
32.22 | LDD2016 | [14] | |
|
C361 Probe Info |
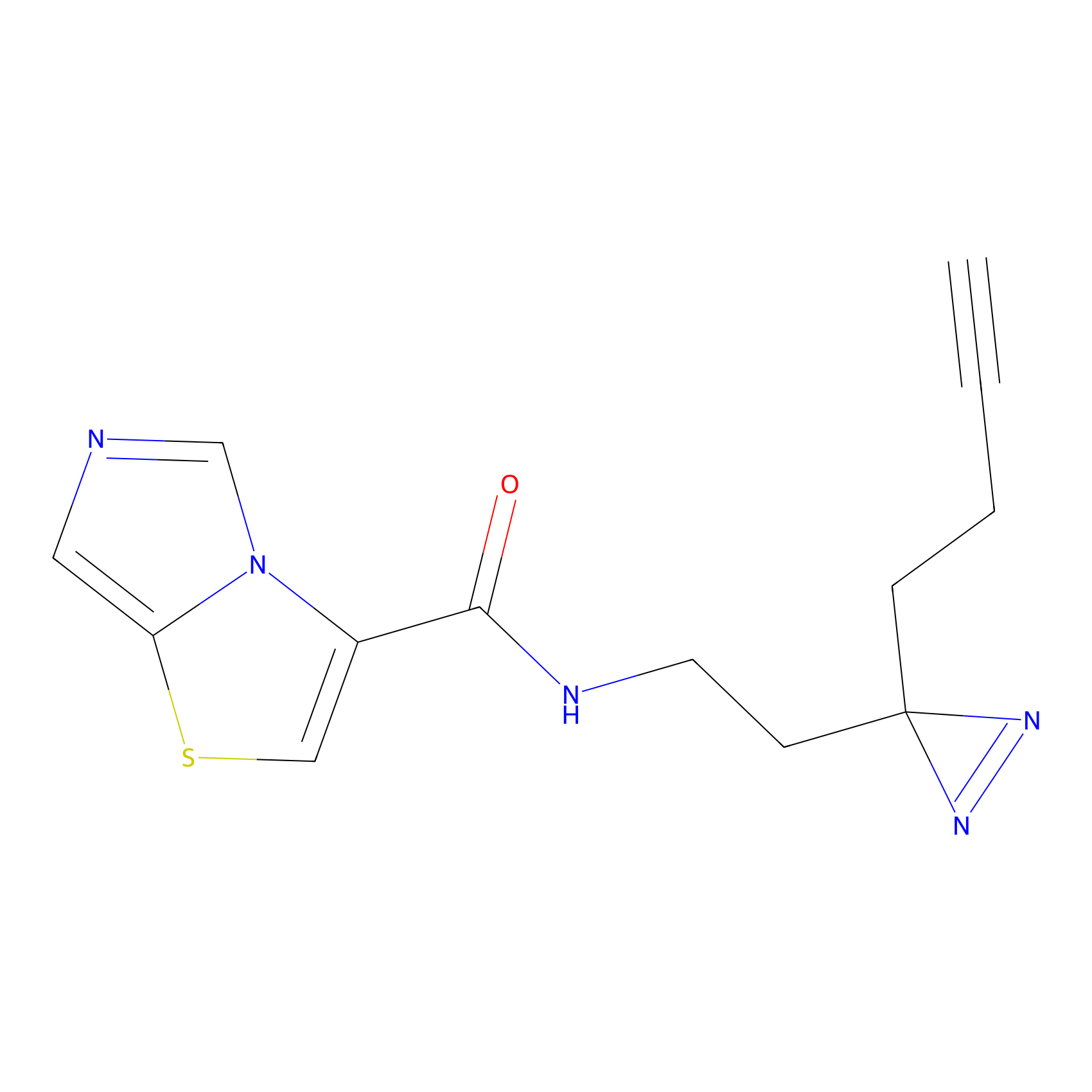 |
25.81 | LDD2022 | [14] | |
|
C390 Probe Info |
 |
21.11 | LDD2049 | [14] | |
|
C391 Probe Info |
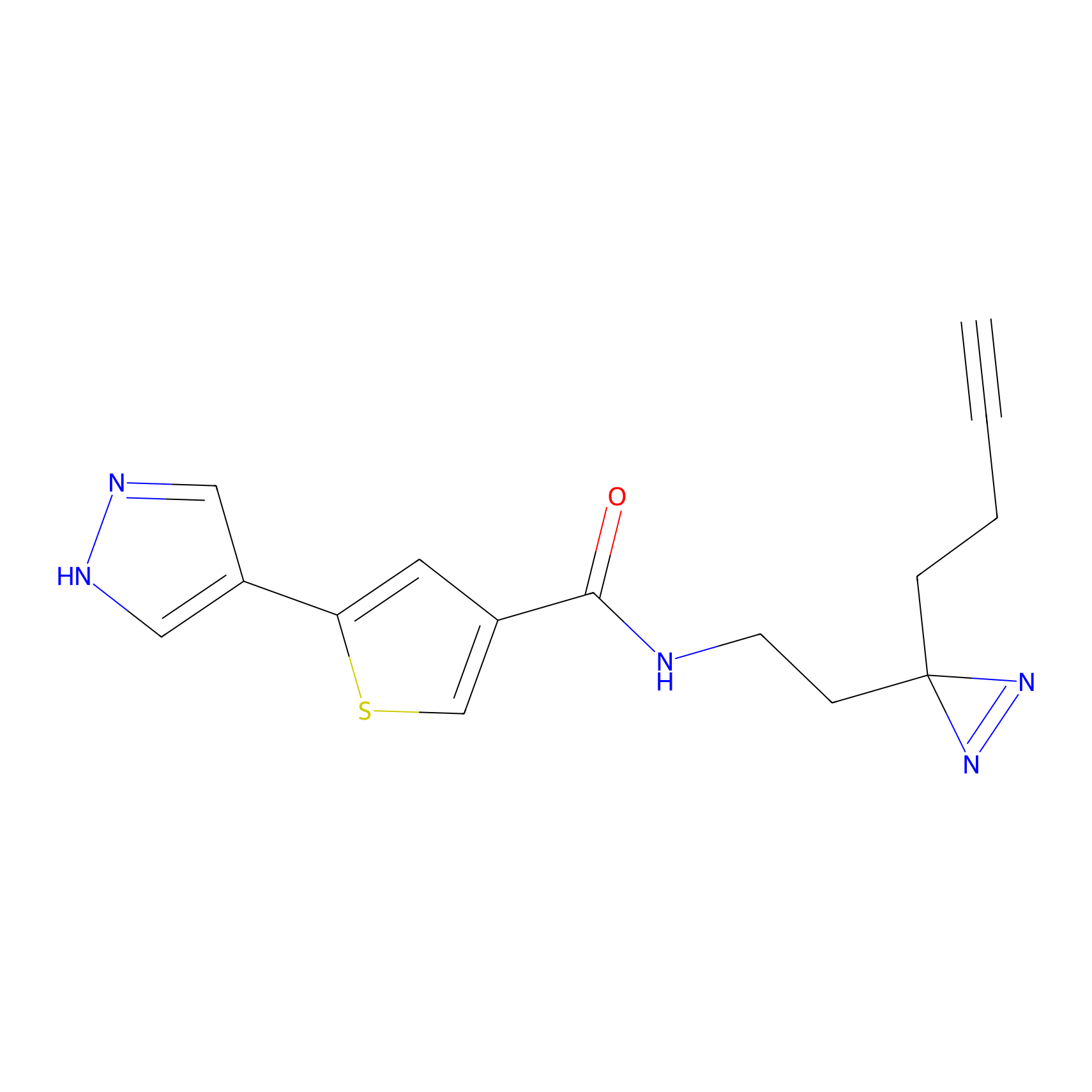 |
18.51 | LDD2050 | [14] | |
|
FFF probe11 Probe Info |
 |
13.55 | LDD0471 | [15] | |
|
FFF probe12 Probe Info |
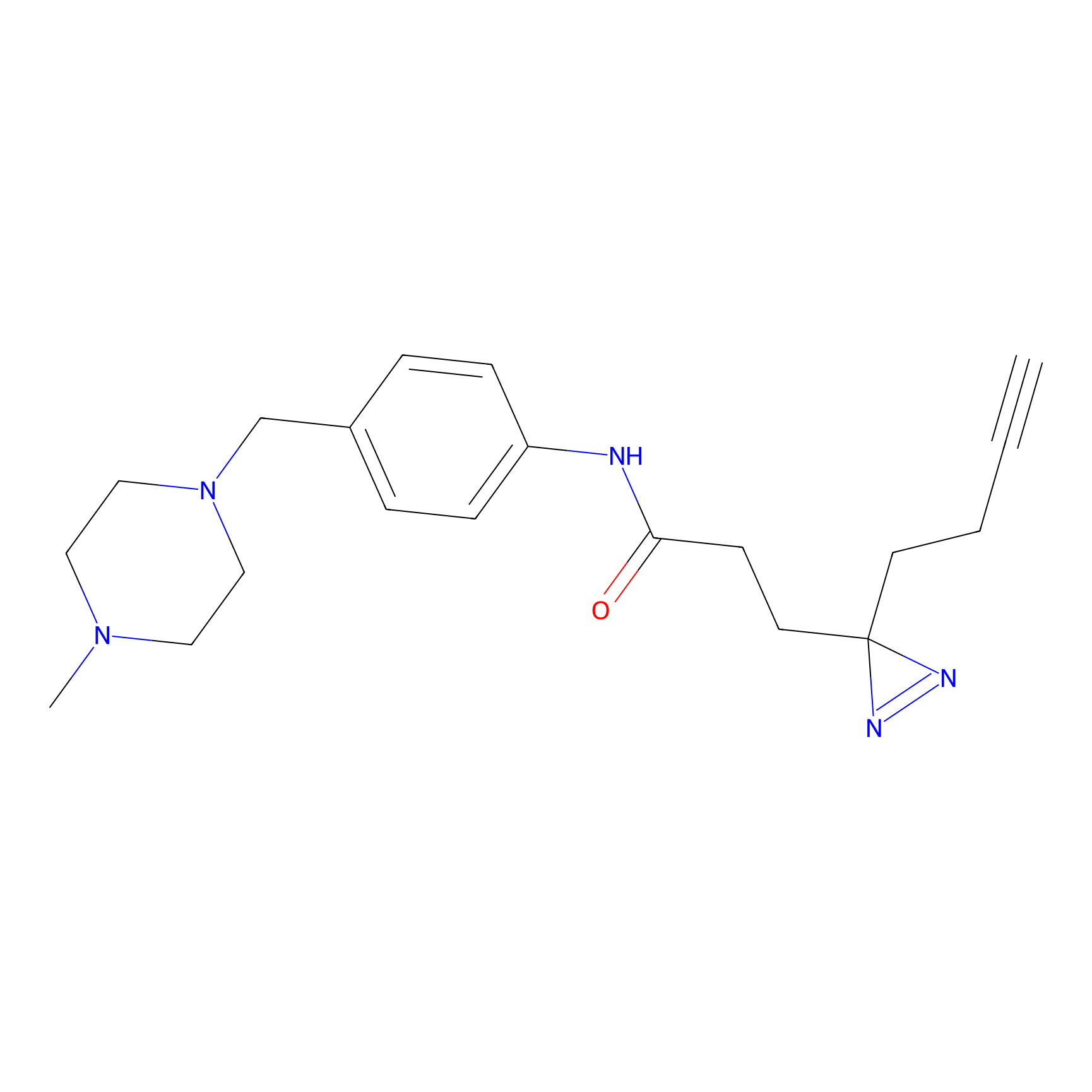 |
5.79 | LDD0473 | [15] | |
|
FFF probe13 Probe Info |
 |
18.11 | LDD0475 | [15] | |
|
FFF probe14 Probe Info |
 |
11.65 | LDD0477 | [15] | |
|
FFF probe2 Probe Info |
 |
17.95 | LDD0463 | [15] | |
|
FFF probe3 Probe Info |
 |
12.88 | LDD0464 | [15] | |
|
FFF probe6 Probe Info |
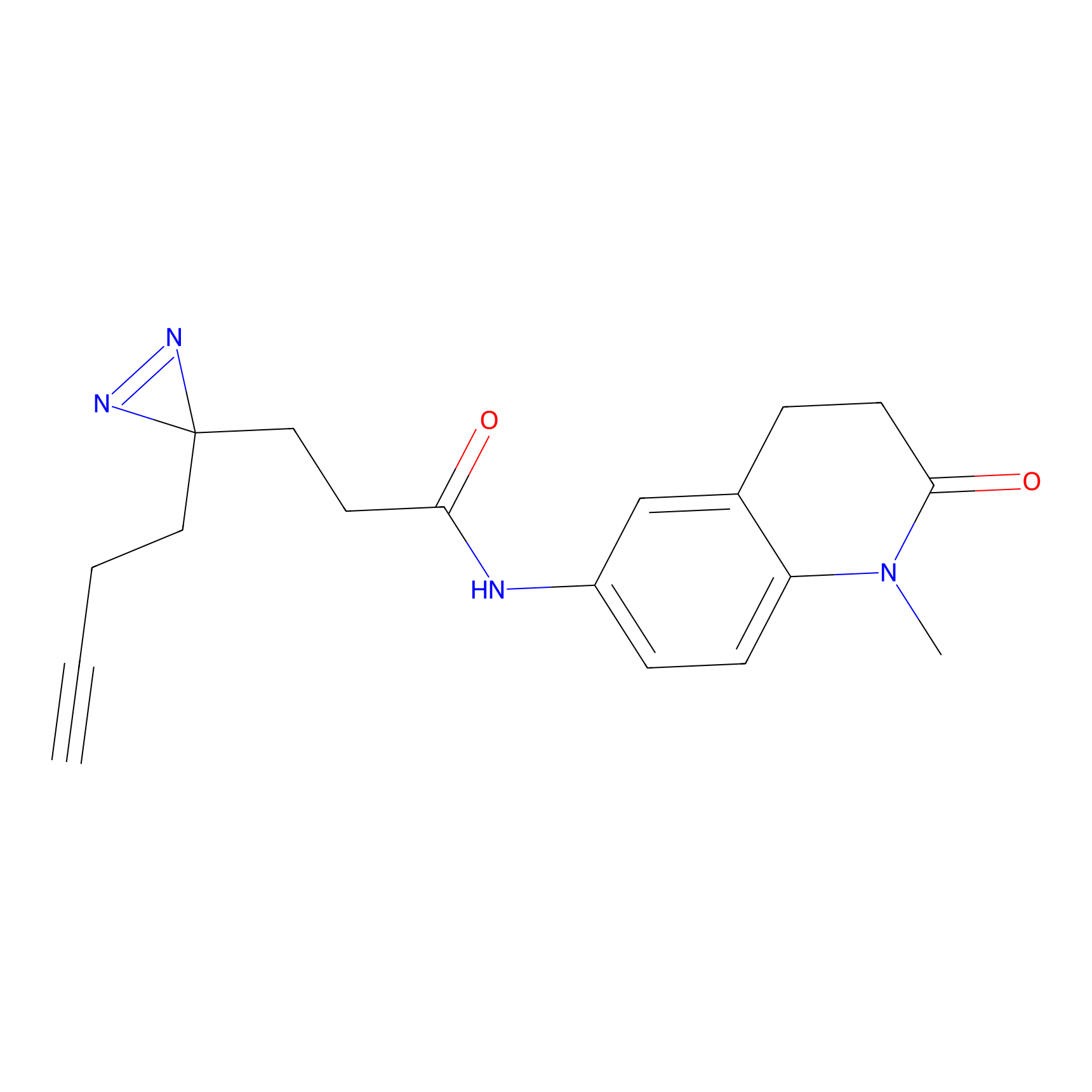 |
5.22 | LDD0467 | [15] | |
|
FFF probe9 Probe Info |
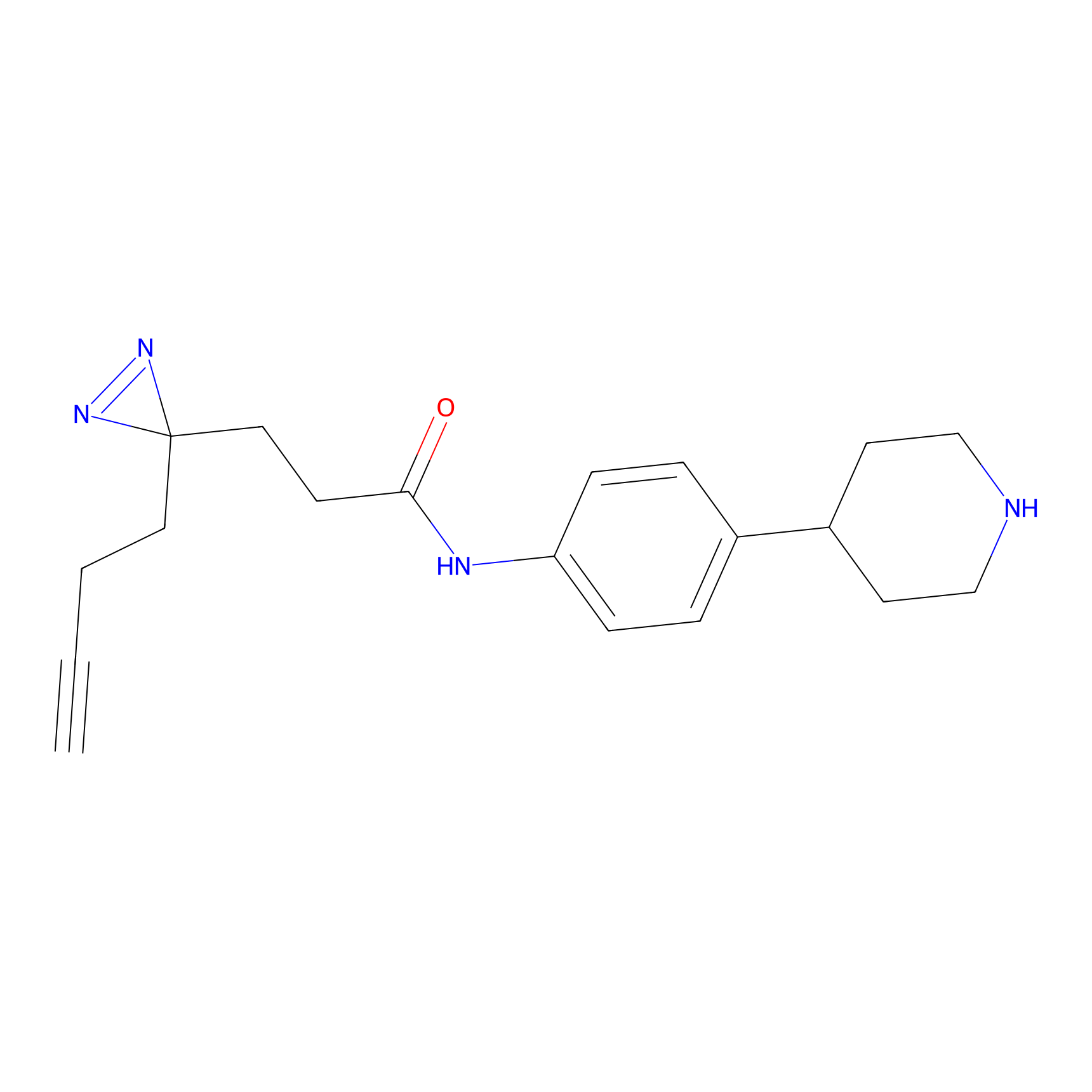 |
6.19 | LDD0470 | [15] | |
|
JN0003 Probe Info |
 |
11.71 | LDD0469 | [15] | |
|
OEA-DA Probe Info |
 |
7.36 | LDD0046 | [16] | |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Metal transporter CNNM3 (CNNM3) | ACDP family | Q8NE01 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Myb-related transcription factor, partner of profilin (MYPOP) | . | Q86VE0 | |||
Cytokine and receptor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| CKLF-like MARVEL transmembrane domain-containing protein 6 (CMTM6) | Chemokine-like factor family | Q9NX76 | |||
Other
References
