Details of the Target
General Information of Target
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| IGROV1 | SNV: p.T73I | DBIA Probe Info | |||
| L428 | SNV: p.L428F | DBIA Probe Info | |||
| MOLT4 | SNV: p.Q372K | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
P8 Probe Info |
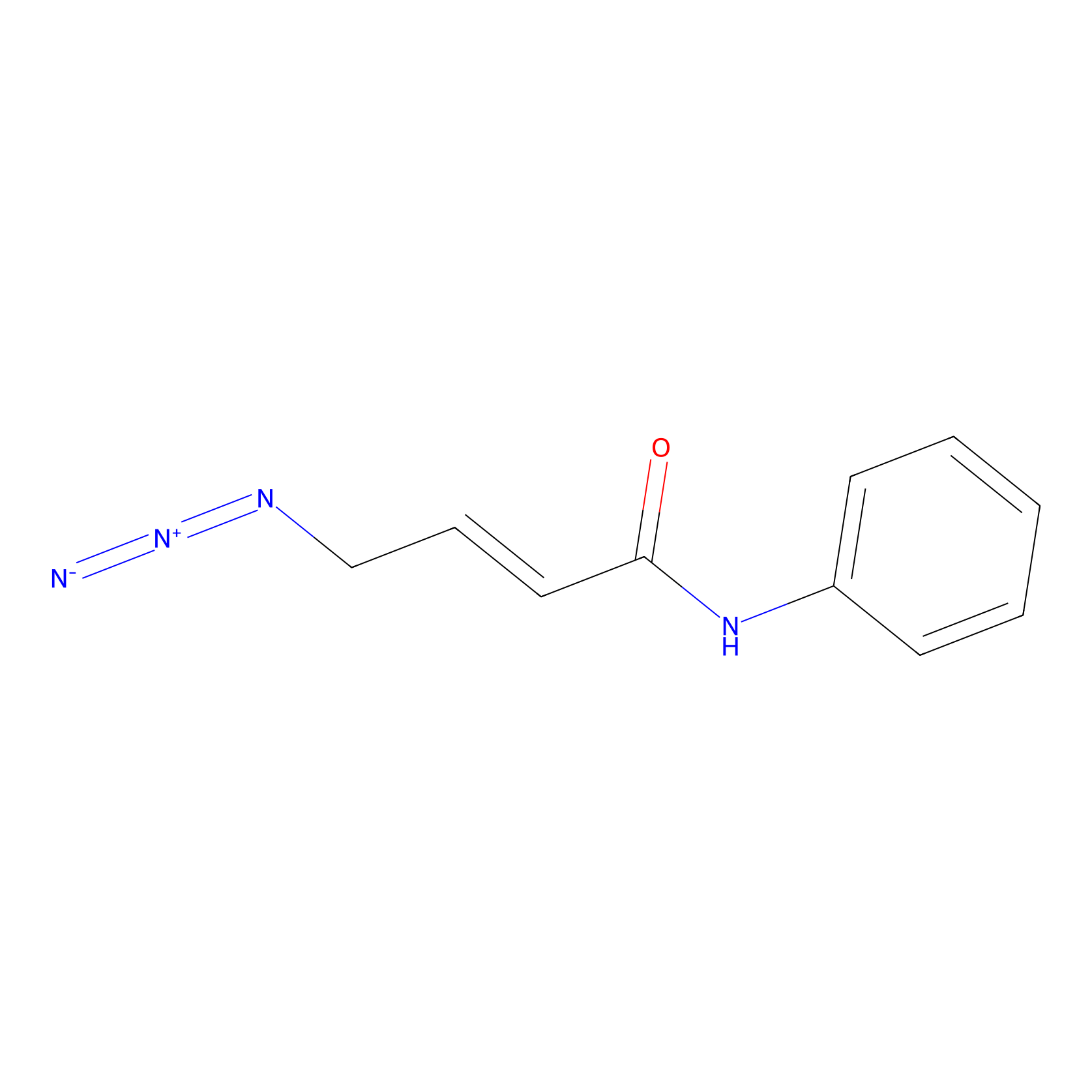 |
4.10 | LDD0455 | [1] | |
|
8RK64 Probe Info |
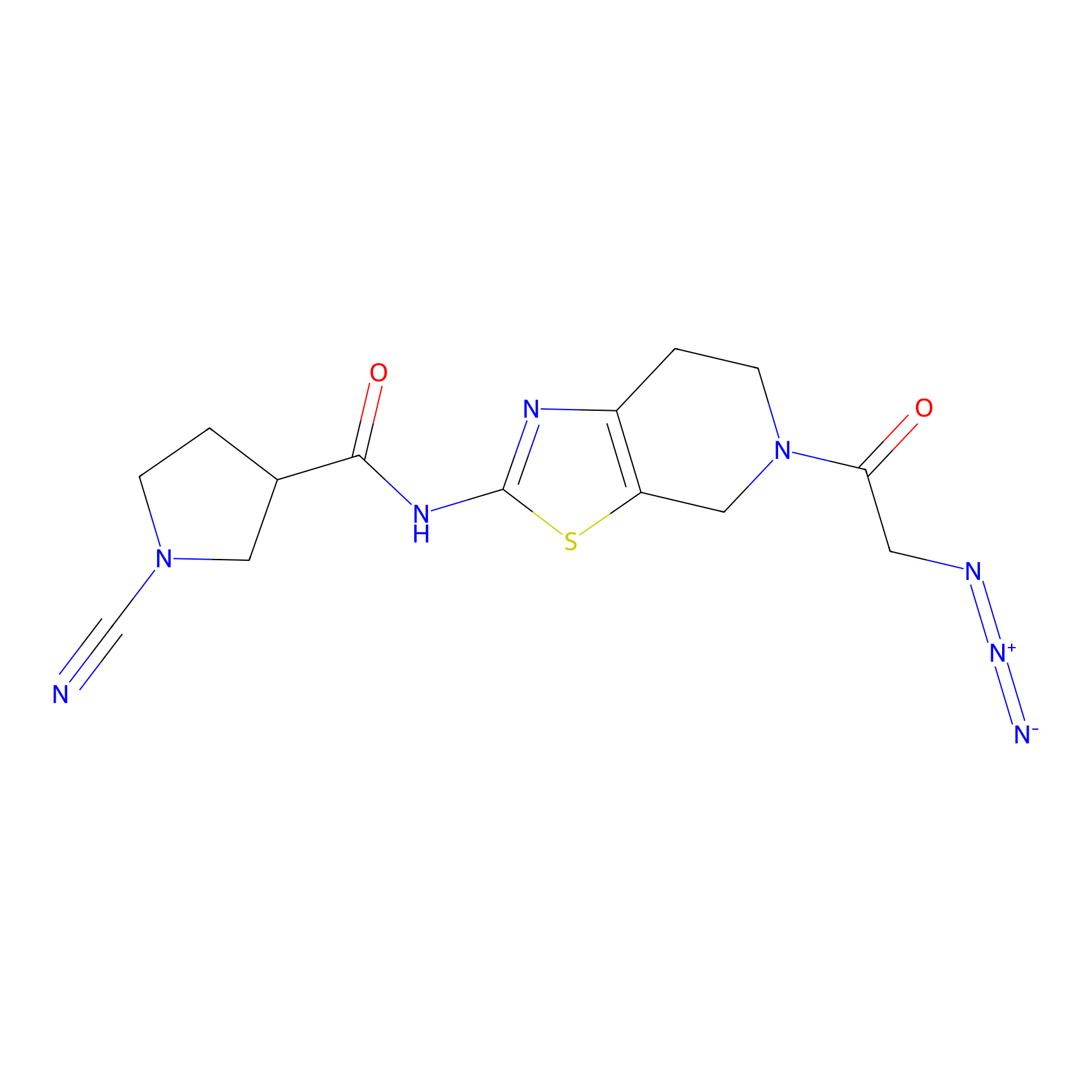 |
N.A. | LDD0039 | [2] | |
|
CHEMBL5175495 Probe Info |
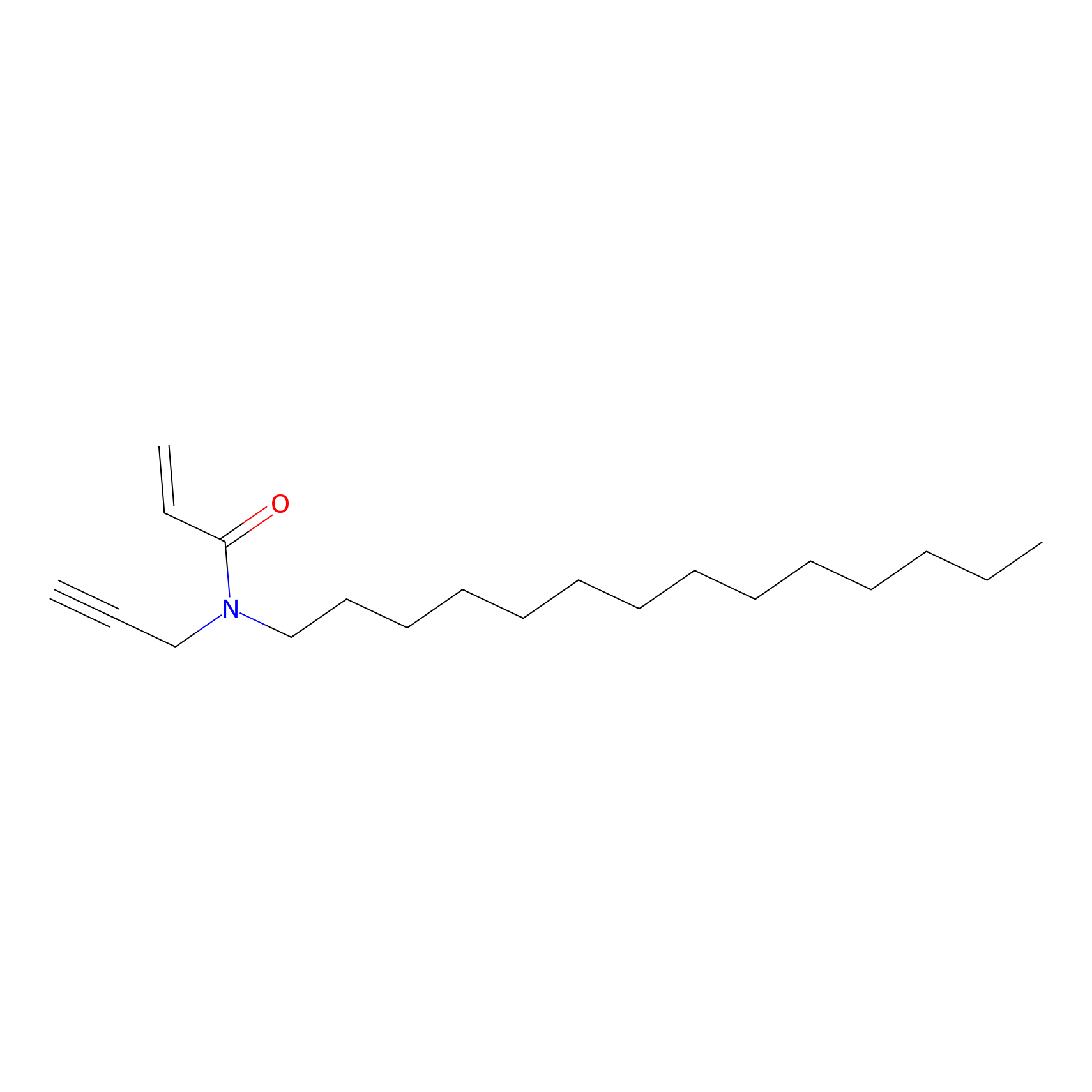 |
7.28 | LDD0196 | [3] | |
|
CY4 Probe Info |
 |
2.41 | LDD0244 | [4] | |
|
TH211 Probe Info |
 |
Y108(20.00); Y161(20.00); Y312(18.35); Y282(17.37) | LDD0257 | [5] | |
|
TH214 Probe Info |
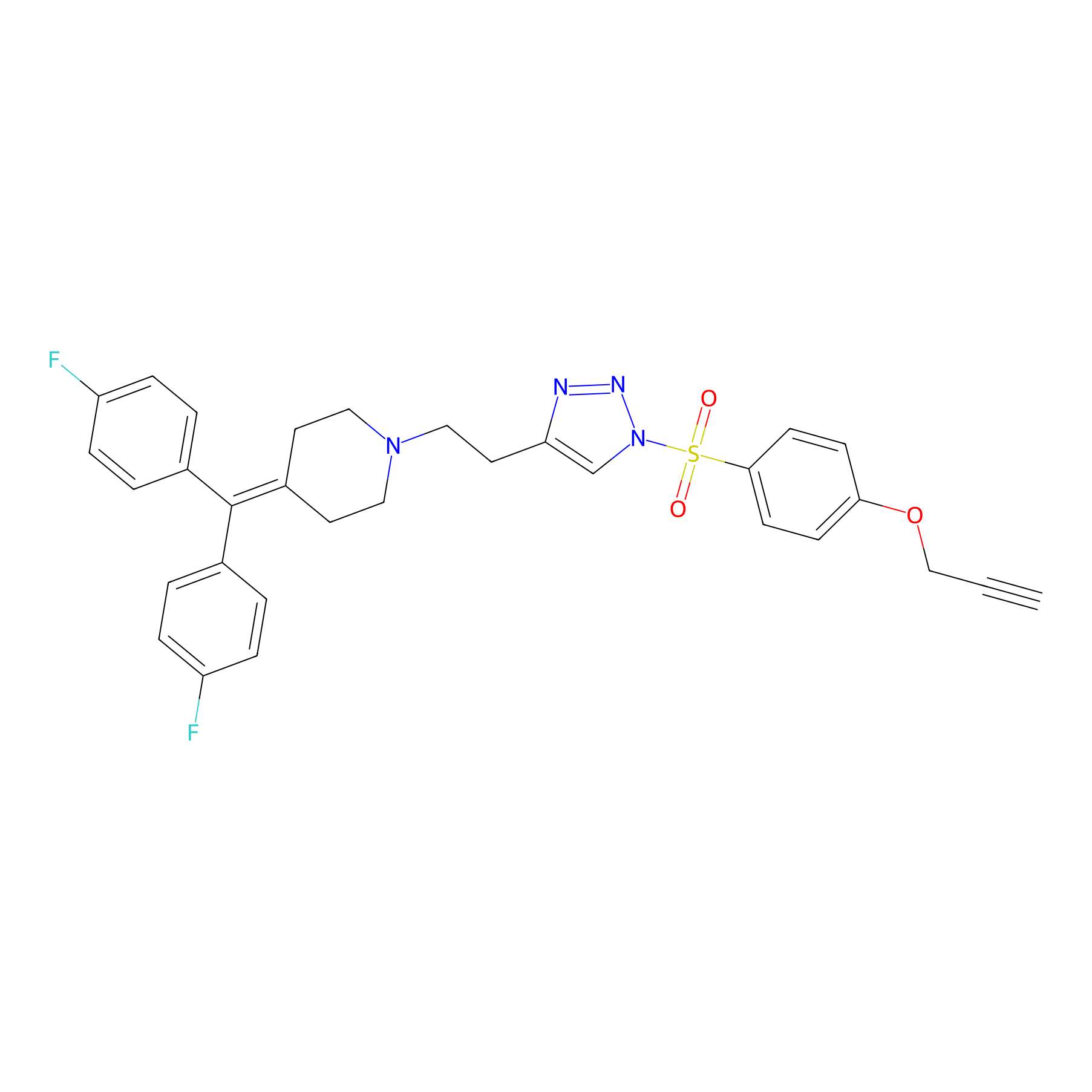 |
Y108(20.00); Y262(20.00); Y272(20.00); Y312(20.00) | LDD0258 | [5] | |
|
TH216 Probe Info |
 |
Y262(20.00); Y272(20.00); Y108(19.12); Y312(17.94) | LDD0259 | [5] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [6] | |
|
AZ-9 Probe Info |
 |
D46(1.01); D47(1.01); D345(1.15); E386(1.02) | LDD2208 | [7] | |
|
Probe 1 Probe Info |
 |
Y108(82.90); Y161(115.83); Y224(41.31); Y262(11.16) | LDD3495 | [8] | |
|
JZ128-DTB Probe Info |
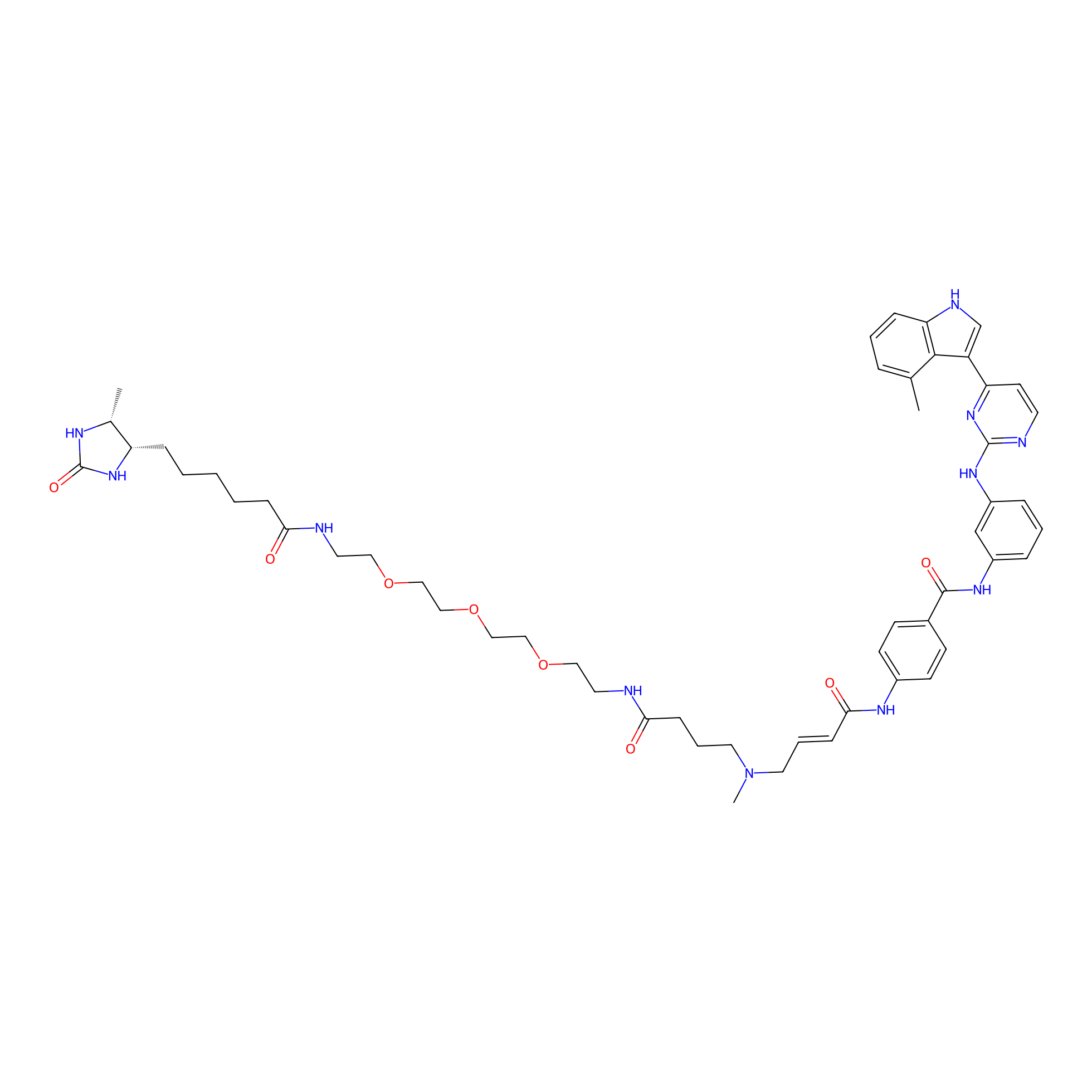 |
N.A. | LDD0462 | [9] | |
|
THZ1-DTB Probe Info |
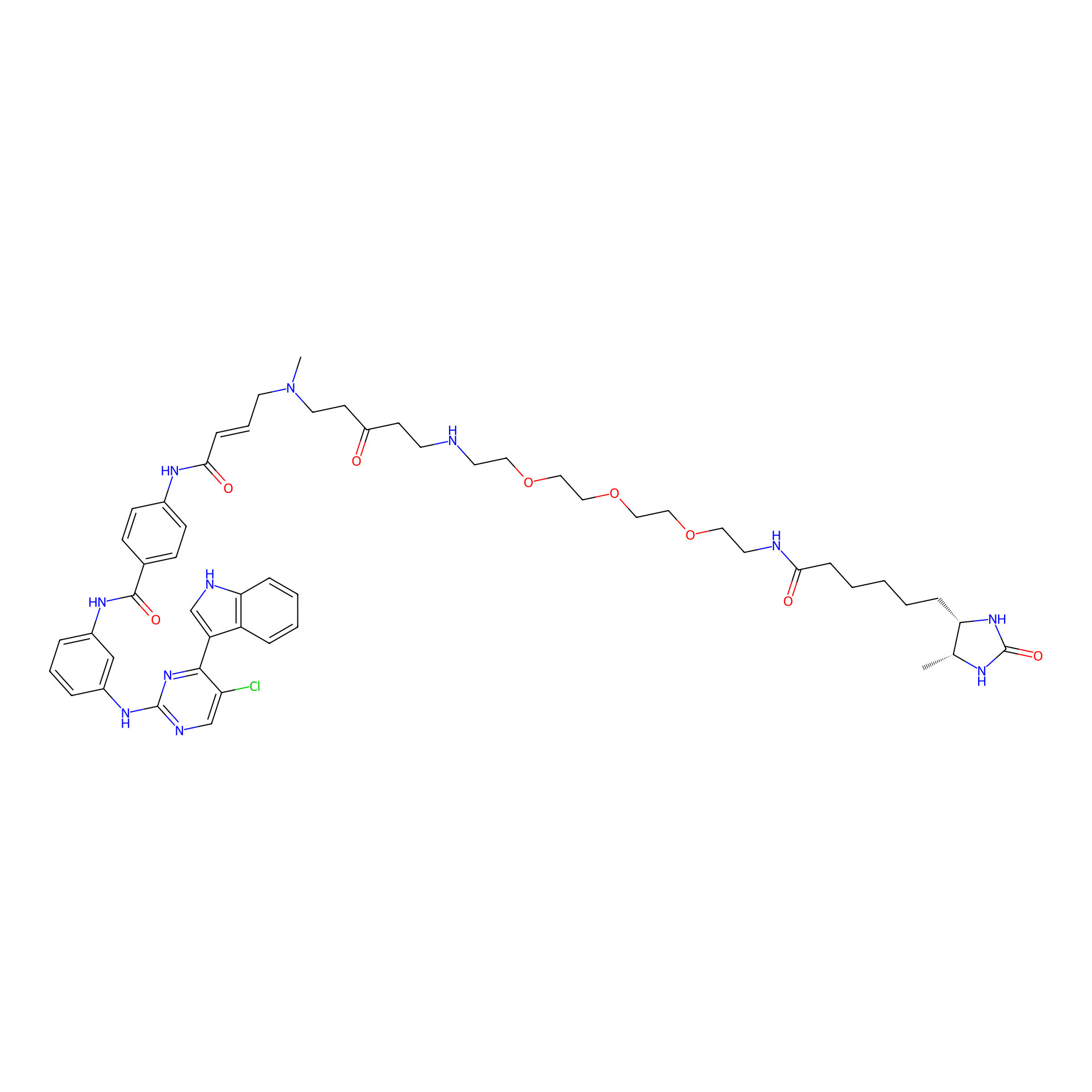 |
C315(1.09); C347(1.08); C213(1.10); C200(1.03) | LDD0460 | [9] | |
|
DA-P3 Probe Info |
 |
4.59 | LDD0183 | [10] | |
|
AHL-Pu-1 Probe Info |
 |
C295(7.28) | LDD0170 | [11] | |
|
HHS-482 Probe Info |
 |
Y103(0.88); Y108(0.87); Y161(0.88); Y224(0.87) | LDD0285 | [12] | |
|
HHS-475 Probe Info |
 |
Y408(0.57); Y103(0.64); Y399(0.65); Y262(0.67) | LDD0264 | [13] | |
|
HHS-465 Probe Info |
 |
Y103(10.00); Y108(10.00); Y161(7.71); Y224(10.00) | LDD2237 | [14] | |
|
DBIA Probe Info |
 |
C315(3.01); C316(3.01); C376(2.26); C347(1.58) | LDD0080 | [15] | |
|
AMP probe Probe Info |
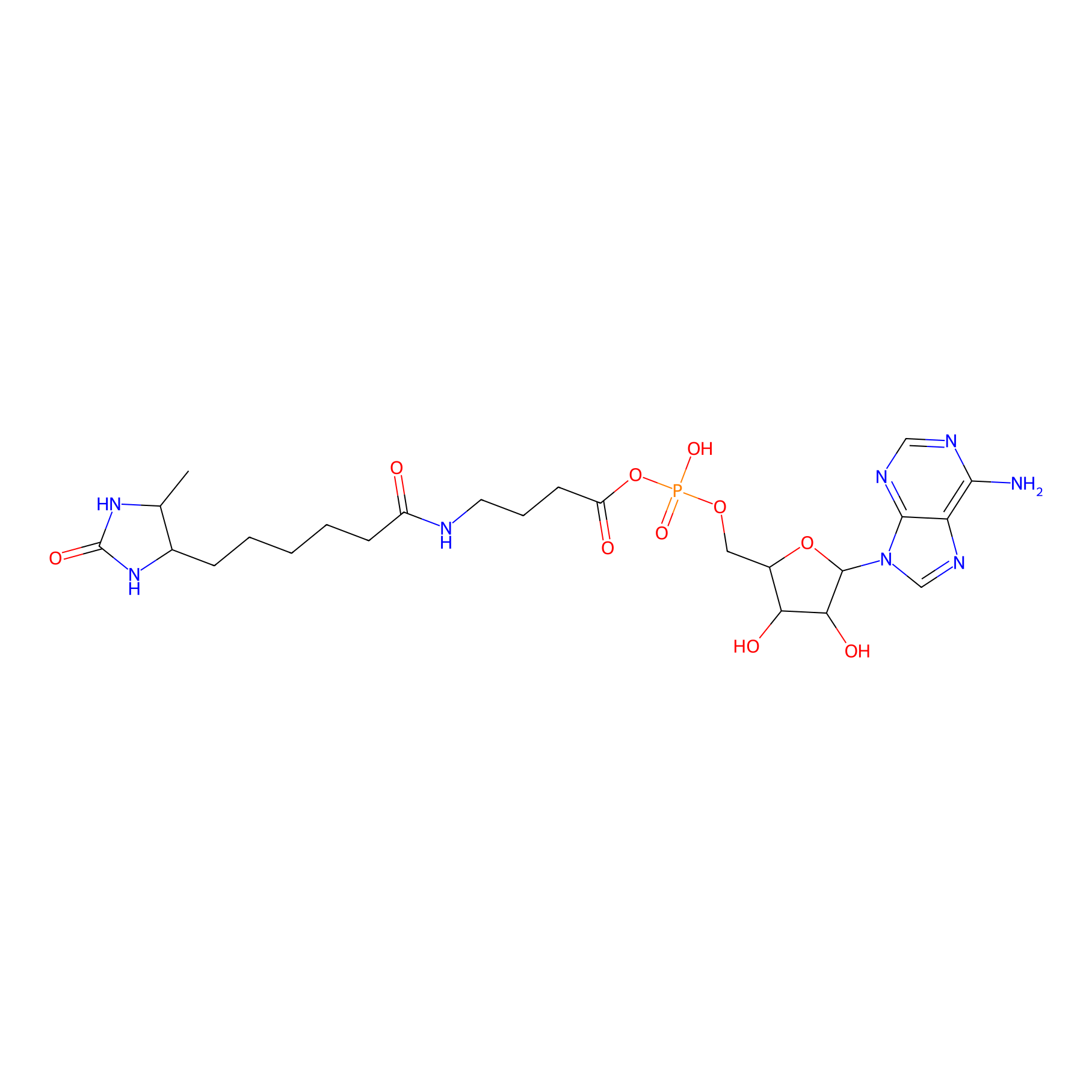 |
K60(0.00); K370(0.00); K336(0.00); K338(0.00) | LDD0200 | [16] | |
|
ATP probe Probe Info |
 |
K60(0.00); K370(0.00); K336(0.00); K338(0.00) | LDD0199 | [16] | |
|
N1 Probe Info |
 |
C347(0.00); C315(0.00); D345(0.00); K352(0.00) | LDD0245 | [4] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C376(0.00); C295(0.00); C4(0.00); C129(0.00) | LDD0038 | [17] | |
|
IA-alkyne Probe Info |
 |
C295(0.00); C347(0.00); C376(0.00); C129(0.00) | LDD0032 | [18] | |
|
IPIAA_H Probe Info |
 |
C129(0.00); C376(0.00); C295(0.00); C347(0.00) | LDD0030 | [19] | |
|
Lodoacetamide azide Probe Info |
 |
C376(0.00); C295(0.00); C315(0.00); C316(0.00) | LDD0037 | [17] | |
|
2PCA Probe Info |
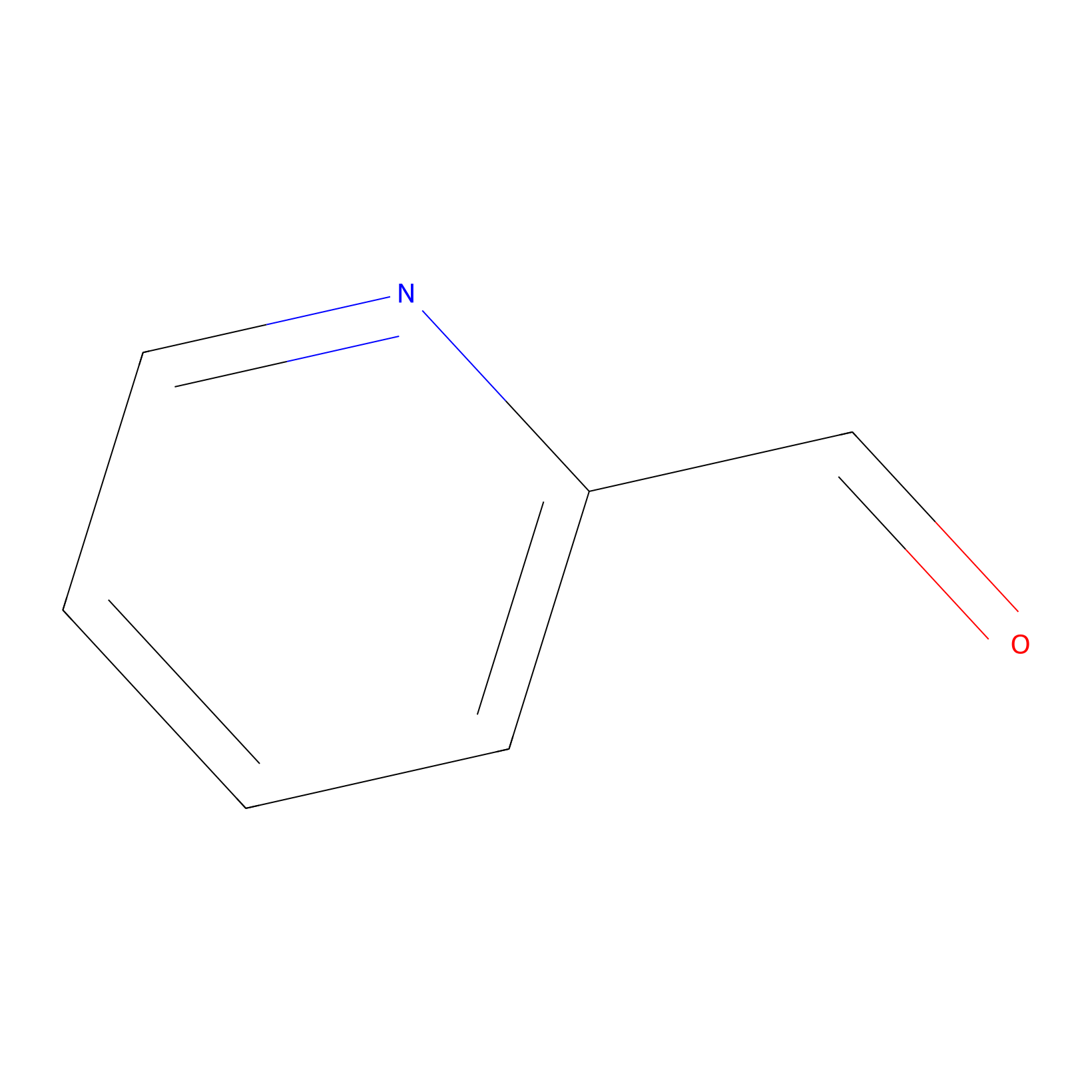 |
K352(0.00); R64(0.00); R264(0.00) | LDD0034 | [20] | |
|
NAIA_4 Probe Info |
 |
C20(0.00); C129(0.00); C213(0.00); C347(0.00) | LDD2226 | [21] | |
|
WYneO Probe Info |
 |
N.A. | LDD0022 | [22] | |
|
aHNE Probe Info |
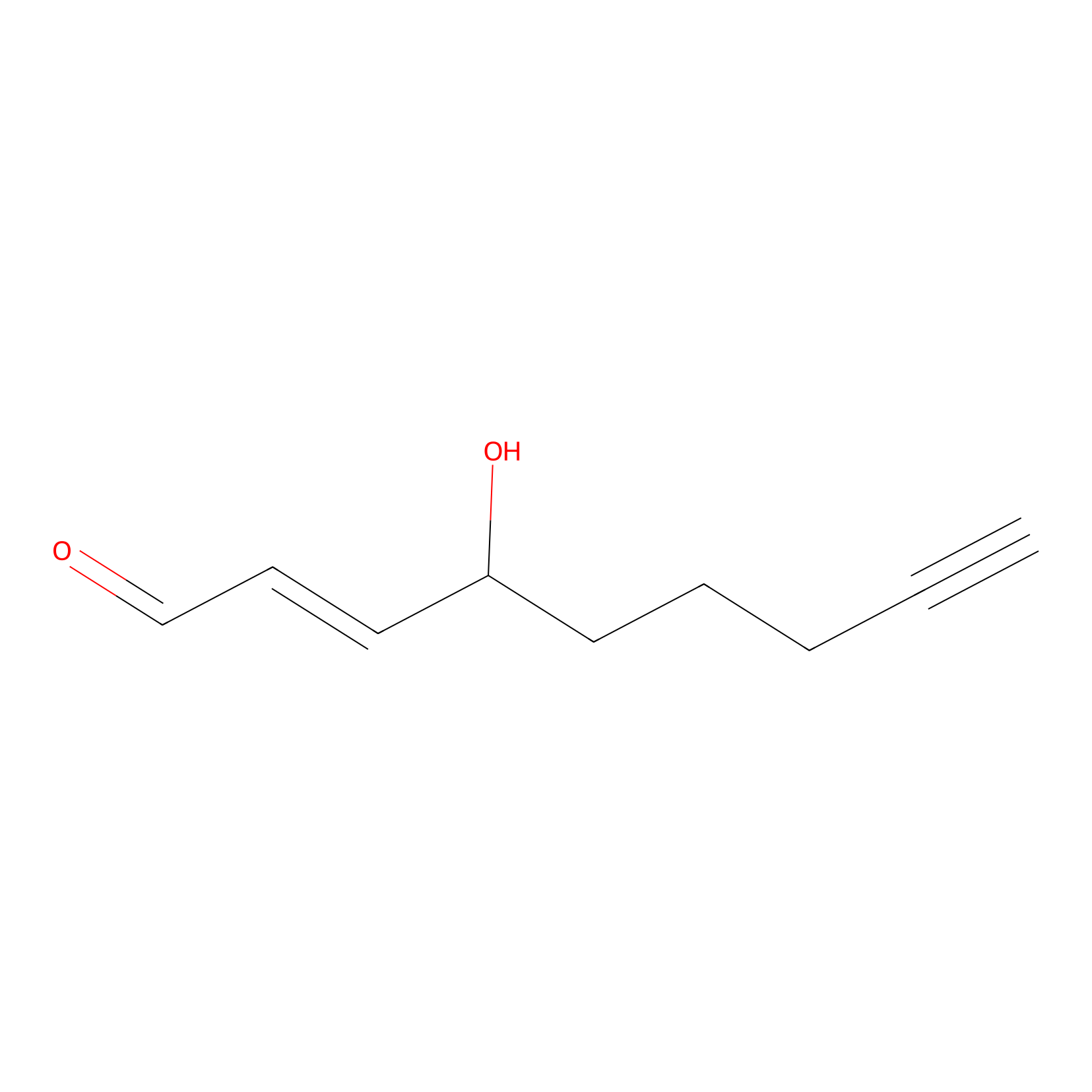 |
C347(0.00); C295(0.00) | LDD0001 | [22] | |
|
1d-yne Probe Info |
 |
K352(0.00); K60(0.00); K370(0.00); K96(0.00) | LDD0356 | [23] | |
|
Compound 10 Probe Info |
 |
C295(0.00); C315(0.00); C316(0.00); C347(0.00) | LDD2216 | [24] | |
|
Compound 11 Probe Info |
 |
C295(0.00); C315(0.00); C316(0.00); C347(0.00) | LDD2213 | [24] | |
|
IPM Probe Info |
 |
C316(0.00); C295(0.00); C315(0.00); C376(0.00) | LDD0005 | [22] | |
|
NHS Probe Info |
 |
K60(0.00); K304(0.00); K394(0.00); K370(0.00) | LDD0010 | [22] | |
|
OSF Probe Info |
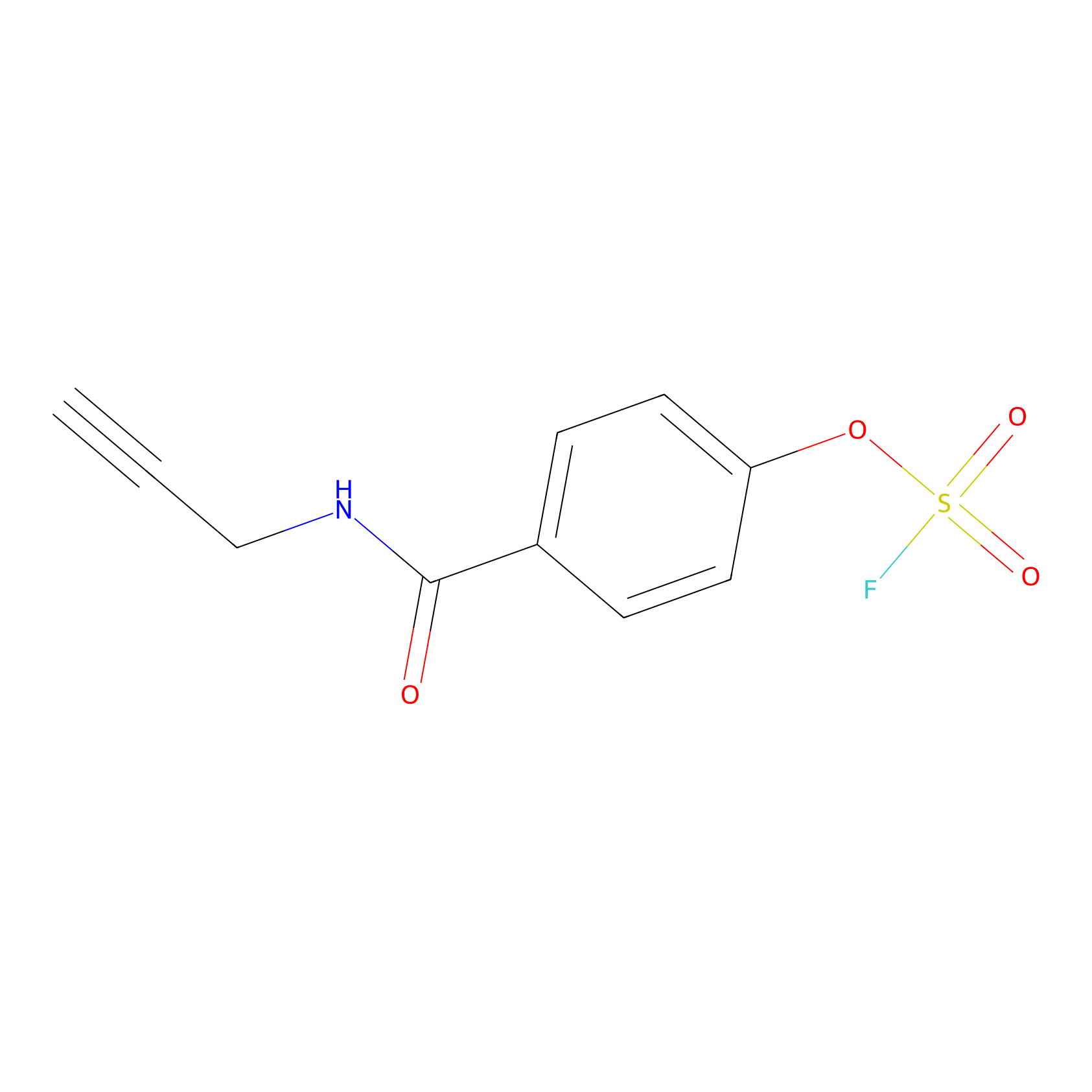 |
Y272(0.00); H107(0.00); Y161(0.00); H266(0.00) | LDD0029 | [25] | |
|
PF-06672131 Probe Info |
 |
C295(0.00); C347(0.00); C376(0.00) | LDD0017 | [26] | |
|
SF Probe Info |
 |
Y399(0.00); Y272(0.00); K336(0.00); K401(0.00) | LDD0028 | [25] | |
|
VSF Probe Info |
 |
C295(0.00); C376(0.00) | LDD0007 | [22] | |
|
YN-1 Probe Info |
 |
N.A. | LDD0447 | [6] | |
|
Phosphinate-6 Probe Info |
 |
C315(0.00); C25(0.00); C347(0.00); C305(0.00) | LDD0018 | [27] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [28] | |
|
1c-yne Probe Info |
 |
K352(0.00); K394(0.00); K326(0.00); K112(0.00) | LDD0228 | [23] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [29] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [29] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [29] | |
|
W1 Probe Info |
 |
C347(0.00); C316(0.00); C295(0.00); C315(0.00) | LDD0236 | [30] | |
|
MPP-AC Probe Info |
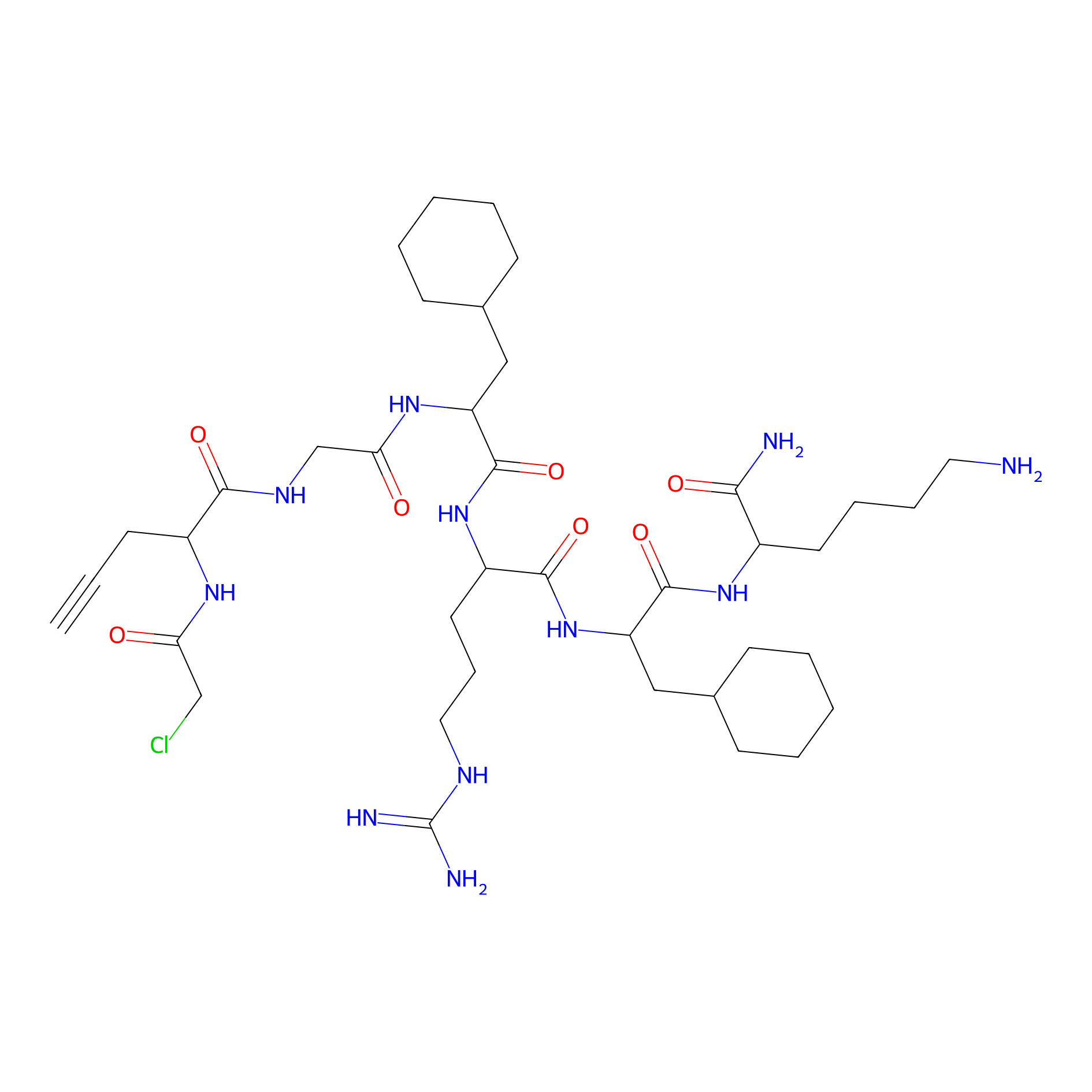 |
N.A. | LDD0428 | [31] | |
|
NAIA_5 Probe Info |
 |
C316(0.00); C315(0.00); C129(0.00); C347(0.00) | LDD2223 | [21] | |
|
TER-AC Probe Info |
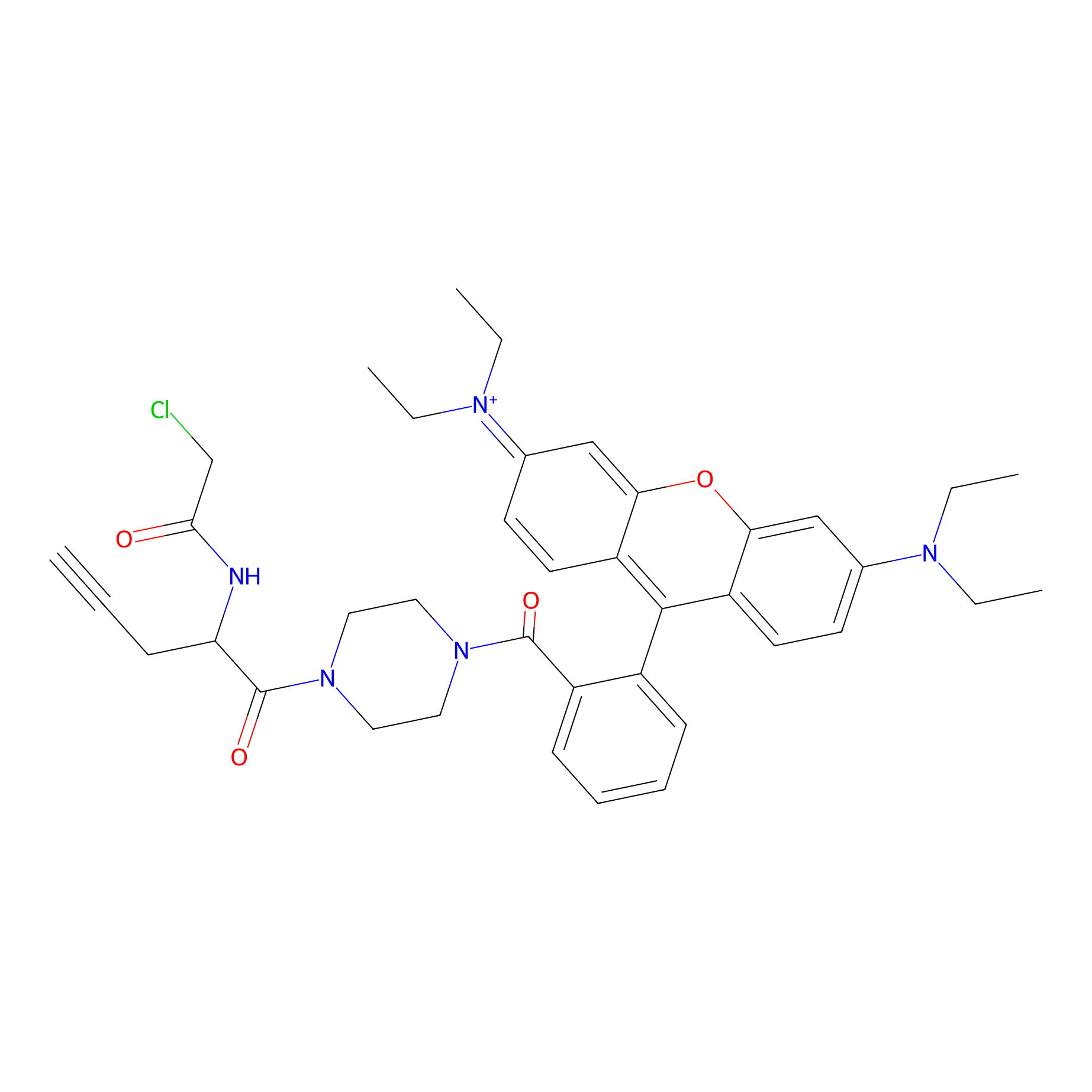 |
N.A. | LDD0426 | [31] | |
|
TPP-AC Probe Info |
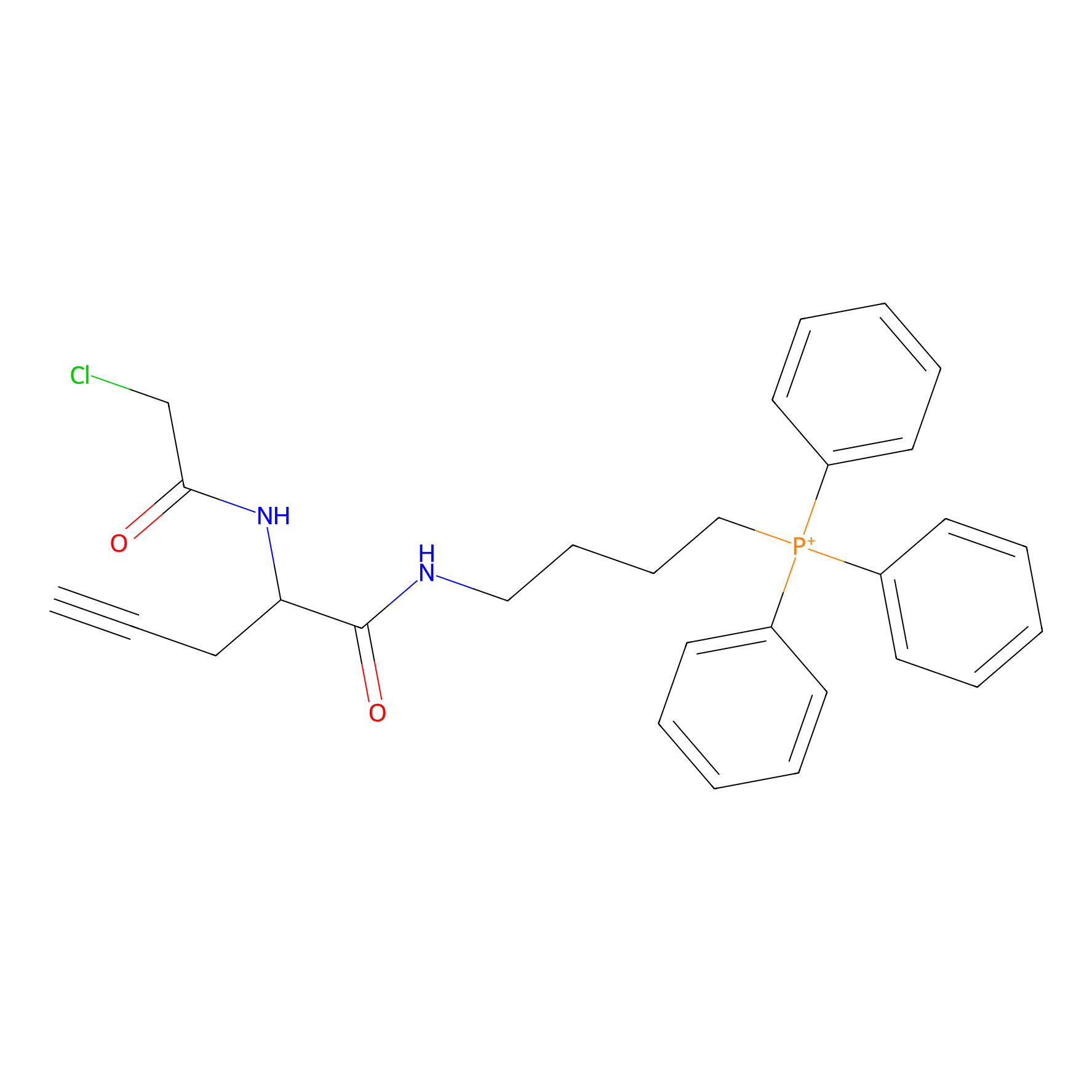 |
N.A. | LDD0427 | [31] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FFF probe11 Probe Info |
 |
20.00 | LDD0472 | [32] | |
|
FFF probe13 Probe Info |
 |
16.44 | LDD0476 | [32] | |
|
FFF probe3 Probe Info |
 |
20.00 | LDD0465 | [32] | |
|
Diazir Probe Info |
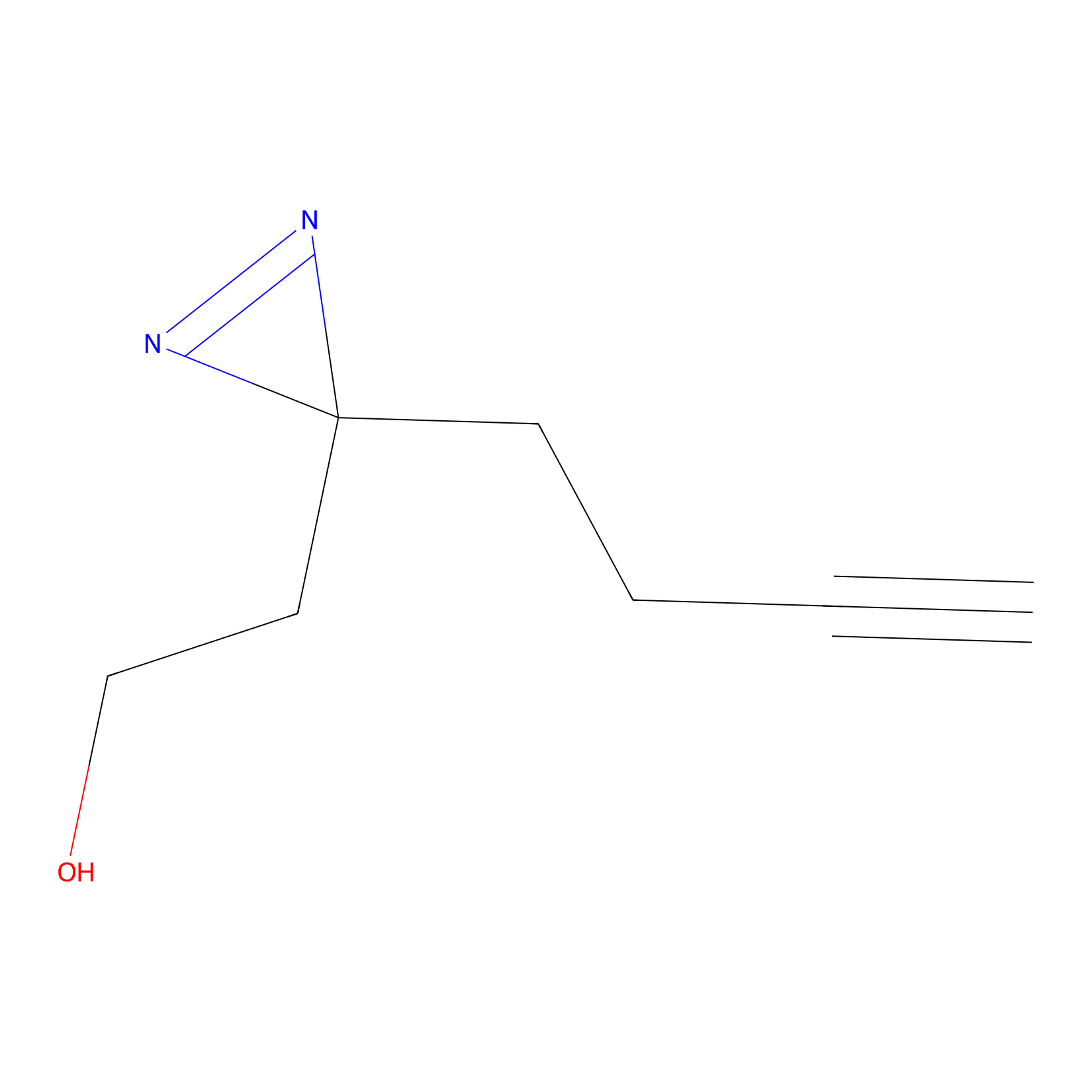 |
E55(0.00); E77(0.00); Y357(0.00); E71(0.00) | LDD0011 | [22] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | DM93 | C295(7.28) | LDD0170 | [11] |
| LDCM0630 | CCW28-3 | 231MFP | C376(1.56); C295(1.50); C347(1.38) | LDD2214 | [33] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [29] |
| LDCM0632 | CL-Sc | Hep-G2 | C347(1.31); C315(1.00); C315(0.95); C347(0.88) | LDD2227 | [21] |
| LDCM0634 | CY-0357 | Hep-G2 | C129(1.18); C376(1.16); C347(0.86); C347(0.80) | LDD2228 | [21] |
| LDCM0031 | Epigallocatechin gallate | HEK-293T | 4.59 | LDD0183 | [10] |
| LDCM0116 | HHS-0101 | DM93 | Y408(0.57); Y103(0.64); Y399(0.65); Y262(0.67) | LDD0264 | [13] |
| LDCM0117 | HHS-0201 | DM93 | Y399(0.53); Y161(0.53); Y103(0.60); Y312(0.60) | LDD0265 | [13] |
| LDCM0118 | HHS-0301 | DM93 | Y312(0.52); Y262(0.52); Y399(0.61); Y272(0.64) | LDD0266 | [13] |
| LDCM0119 | HHS-0401 | DM93 | Y272(0.39); Y312(0.42); Y262(0.44); Y399(0.49) | LDD0267 | [13] |
| LDCM0120 | HHS-0701 | DM93 | Y103(0.40); Y272(0.47); Y262(0.52); Y312(0.53) | LDD0268 | [13] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [29] |
| LDCM0123 | JWB131 | DM93 | Y103(0.88); Y108(0.87); Y161(0.88); Y224(0.87) | LDD0285 | [12] |
| LDCM0124 | JWB142 | DM93 | Y103(0.60); Y108(0.59); Y161(0.60); Y224(0.61) | LDD0286 | [12] |
| LDCM0125 | JWB146 | DM93 | Y103(1.12); Y108(1.13); Y161(1.08); Y224(1.11) | LDD0287 | [12] |
| LDCM0126 | JWB150 | DM93 | Y103(2.30); Y108(2.21); Y161(2.31); Y224(2.46) | LDD0288 | [12] |
| LDCM0127 | JWB152 | DM93 | Y103(1.74); Y108(1.71); Y161(1.62); Y224(1.83) | LDD0289 | [12] |
| LDCM0128 | JWB198 | DM93 | Y103(1.06); Y108(1.06); Y161(1.04); Y224(1.02) | LDD0290 | [12] |
| LDCM0129 | JWB202 | DM93 | Y103(0.39); Y108(0.39); Y161(0.37); Y224(0.43) | LDD0291 | [12] |
| LDCM0130 | JWB211 | DM93 | Y103(0.91); Y108(0.89); Y161(0.90); Y224(0.89) | LDD0292 | [12] |
| LDCM0179 | JZ128 | PC-3 | N.A. | LDD0462 | [9] |
| LDCM0022 | KB02 | HCT 116 | C315(3.01); C316(3.01); C376(2.26); C347(1.58) | LDD0080 | [15] |
| LDCM0023 | KB03 | HCT 116 | C315(8.72); C316(8.72); C376(4.03); C347(3.83) | LDD0081 | [15] |
| LDCM0024 | KB05 | HCT 116 | C315(7.25); C316(7.25); C376(2.51); C347(1.58) | LDD0082 | [15] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [29] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C347(1.59) | LDD2206 | [34] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C347(0.96); C347(0.89); C347(0.84); C347(0.23) | LDD2207 | [34] |
| LDCM0021 | THZ1 | HeLa S3 | C315(1.09); C347(1.08); C213(1.10); C200(1.03) | LDD0460 | [9] |
| LDCM0110 | W12 | Hep-G2 | H107(2.12); Y108(2.23); C347(2.49) | LDD0237 | [30] |
| LDCM0111 | W14 | Hep-G2 | C347(1.30); H107(1.70) | LDD0238 | [30] |
| LDCM0112 | W16 | Hep-G2 | K338(0.63); K336(0.66); K370(0.71); Q372(0.71) | LDD0239 | [30] |
| LDCM0113 | W17 | Hep-G2 | C347(0.67); Y108(1.07) | LDD0240 | [30] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Amyloid-beta precursor protein (APP) | APP family | P05067 | |||
| Alpha-synuclein (SNCA) | Synuclein family | P37840 | |||
Other
References
