Details of the Target
General Information of Target
| Target ID | LDTP03499 | |||||
|---|---|---|---|---|---|---|
| Target Name | Elongation factor 1-delta (EEF1D) | |||||
| Gene Name | EEF1D | |||||
| Gene ID | 1936 | |||||
| Synonyms |
EF1D; Elongation factor 1-delta; EF-1-delta; Antigen NY-CO-4 |
|||||
| 3D Structure | ||||||
| Sequence |
MATNFLAHEKIWFDKFKYDDAERRFYEQMNGPVAGASRQENGASVILRDIARARENIQKS
LAGSSGPGASSGTSGDHGELVVRIASLEVENQSLRGVVQELQQAISKLEARLNVLEKSSP GHRATAPQTQHVSPMRQVEPPAKKPATPAEDDEDDDIDLFGSDNEEEDKEAAQLREERLR QYAEKKAKKPALVAKSSILLDVKPWDDETDMAQLEACVRSIQLDGLVWGASKLVPVGYGI RKLQIQCVVEDDKVGTDLLEEEITKFEEHVQSVDIAAFNKI |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
EF-1-beta/EF-1-delta family
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
[Isoform 1]: EF-1-beta and EF-1-delta stimulate the exchange of GDP bound to EF-1-alpha to GTP, regenerating EF-1-alpha for another round of transfer of aminoacyl-tRNAs to the ribosome.; [Isoform 2]: Regulates induction of heat-shock-responsive genes through association with heat shock transcription factors and direct DNA-binding at heat shock promoter elements (HSE).
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
P2 Probe Info |
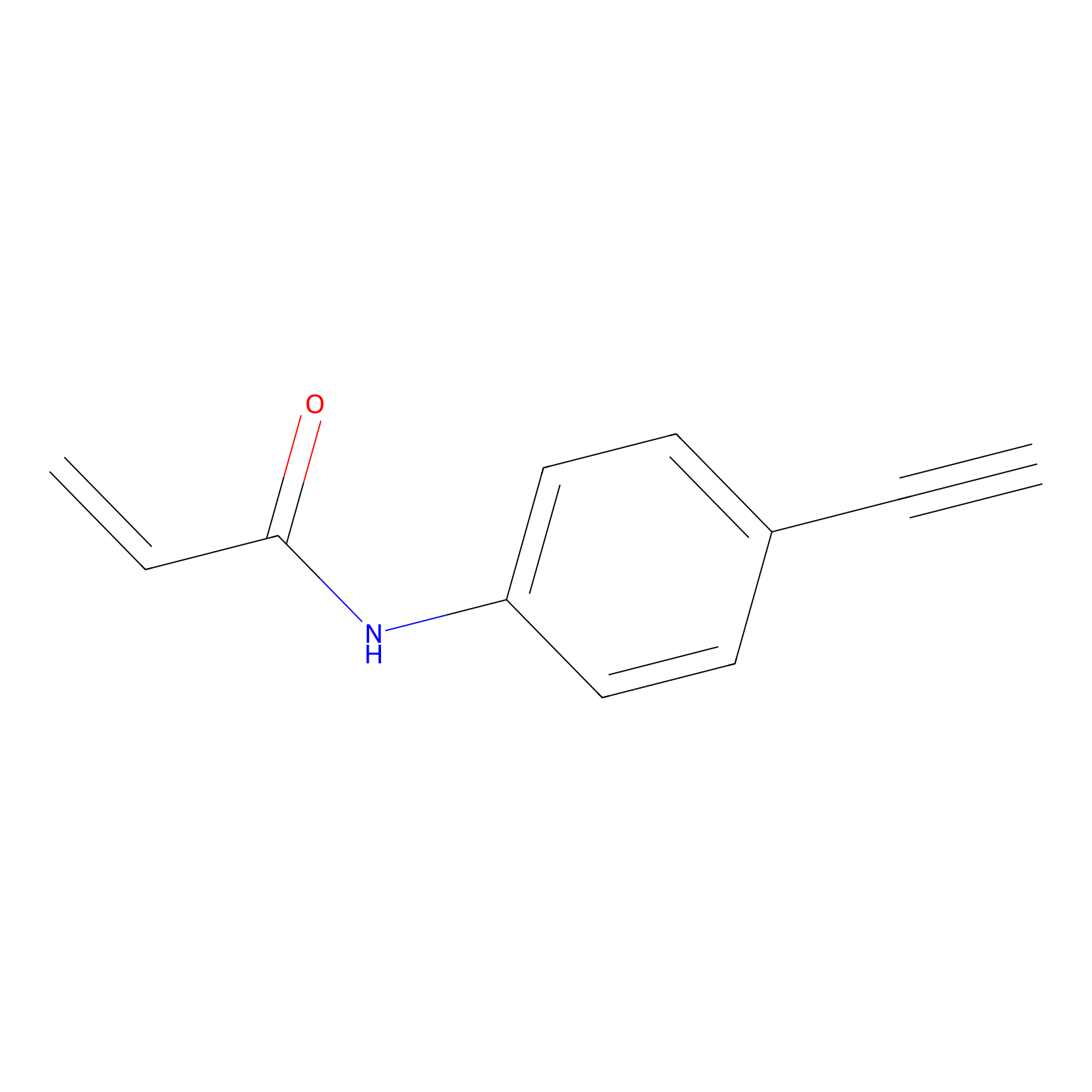 |
10.00 | LDD0449 | [1] | |
|
P3 Probe Info |
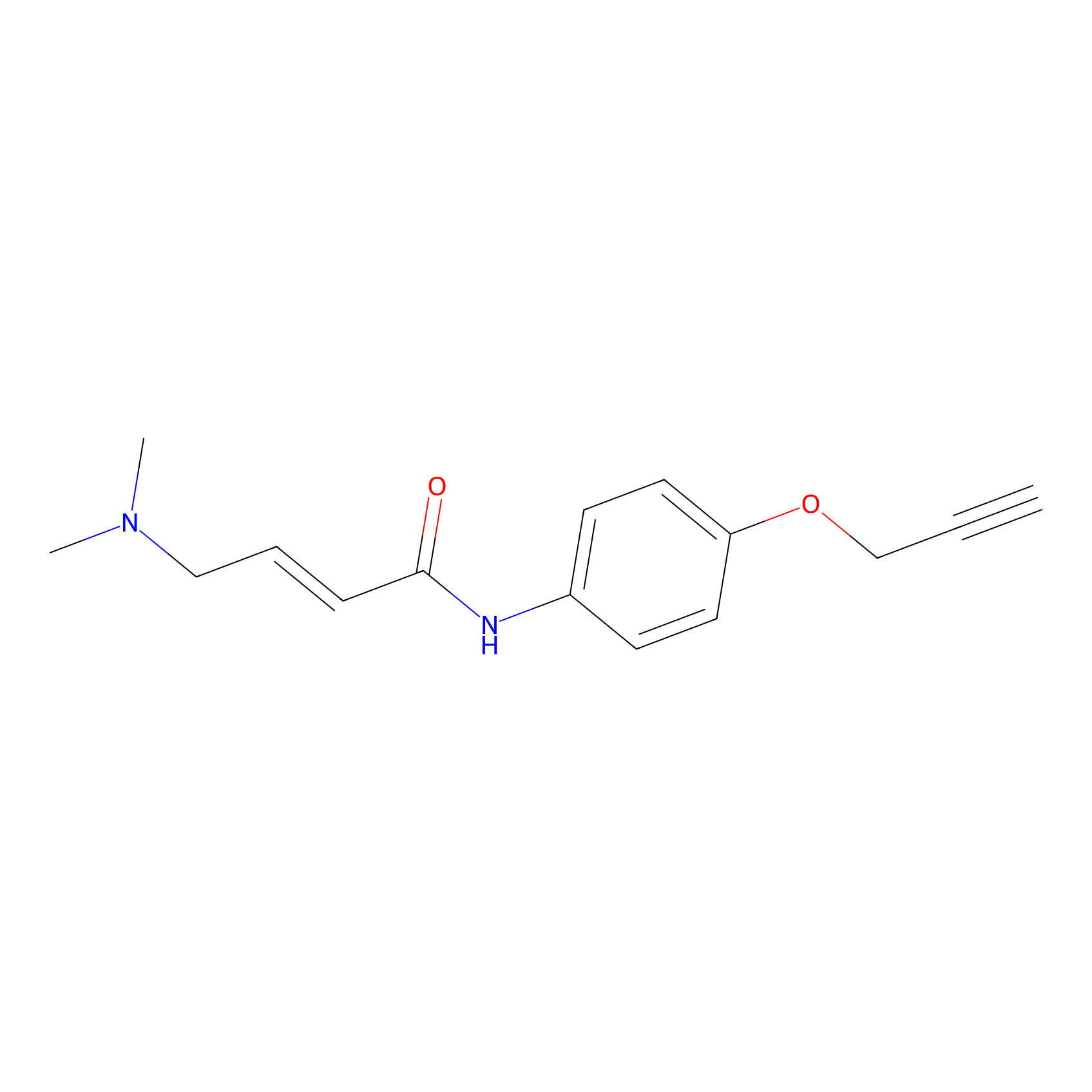 |
10.00 | LDD0450 | [1] | |
|
P8 Probe Info |
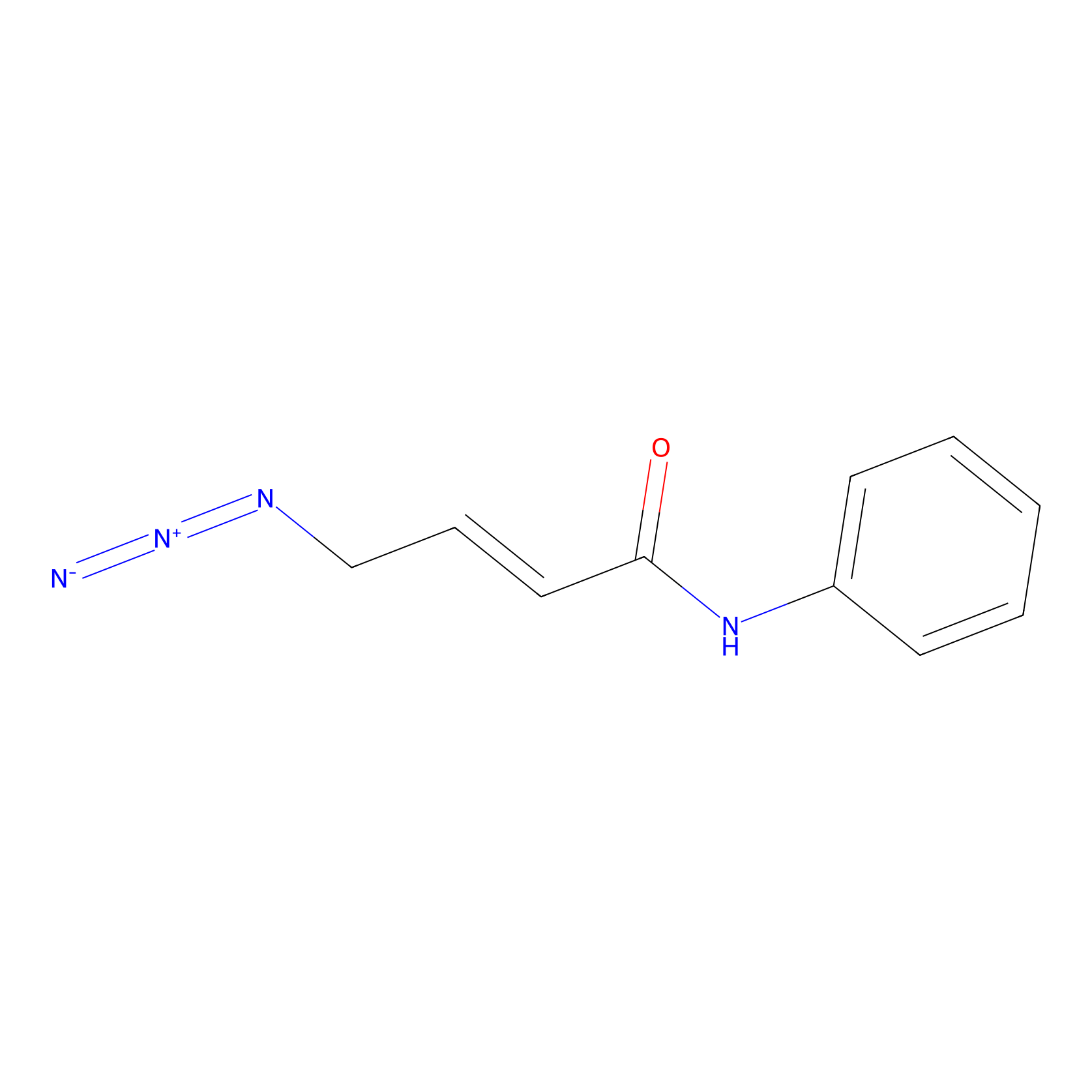 |
10.00 | LDD0451 | [1] | |
|
A-EBA Probe Info |
 |
3.31 | LDD0215 | [2] | |
|
ILS-1 Probe Info |
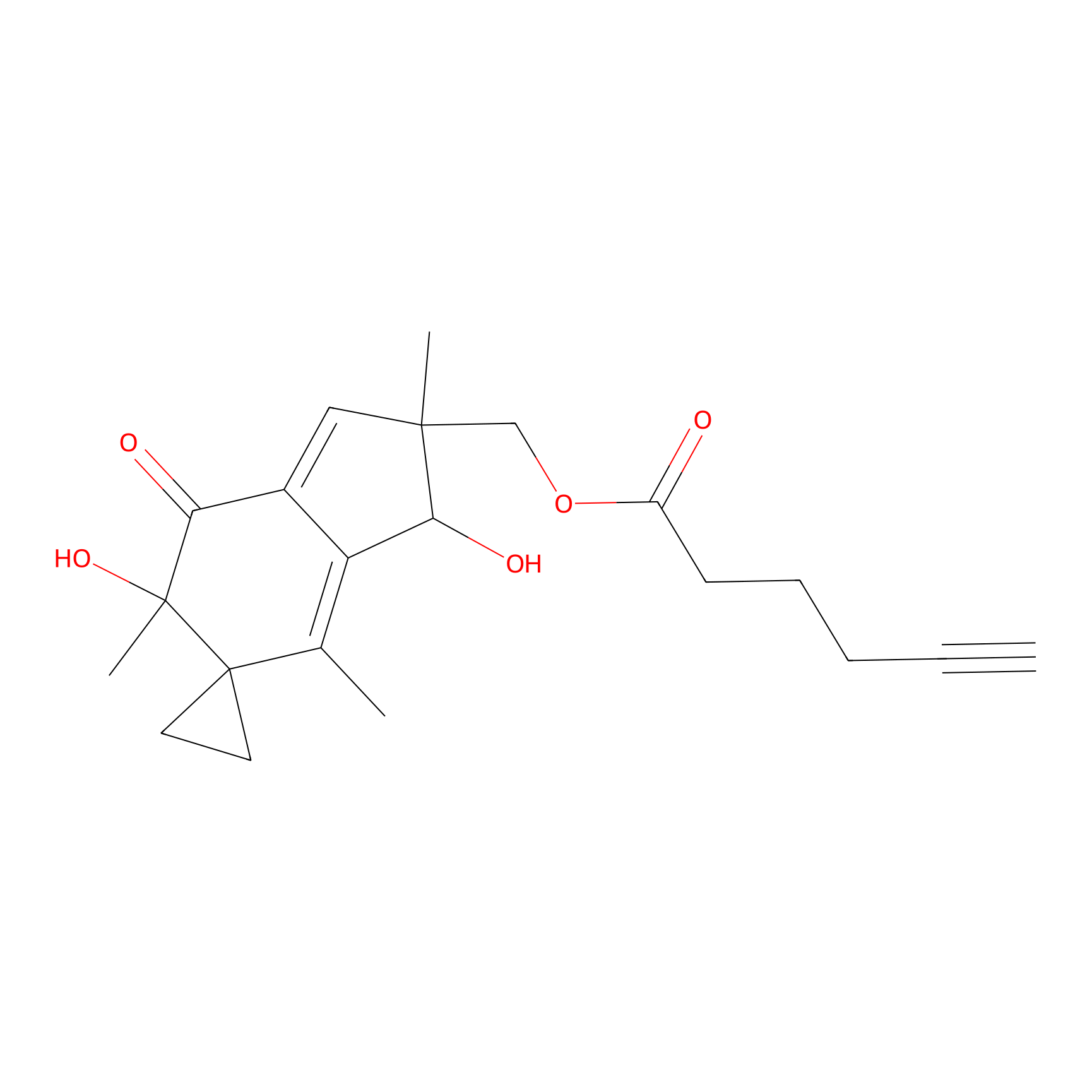 |
2.16 | LDD0415 | [3] | |
|
CY-1 Probe Info |
 |
100.00 | LDD0243 | [4] | |
|
CY4 Probe Info |
 |
100.00 | LDD0244 | [4] | |
|
N1 Probe Info |
 |
100.00 | LDD0242 | [4] | |
|
C-Sul Probe Info |
 |
2.59 | LDD0066 | [5] | |
|
TH211 Probe Info |
 |
Y26(10.00); Y18(8.23) | LDD0257 | [6] | |
|
TH214 Probe Info |
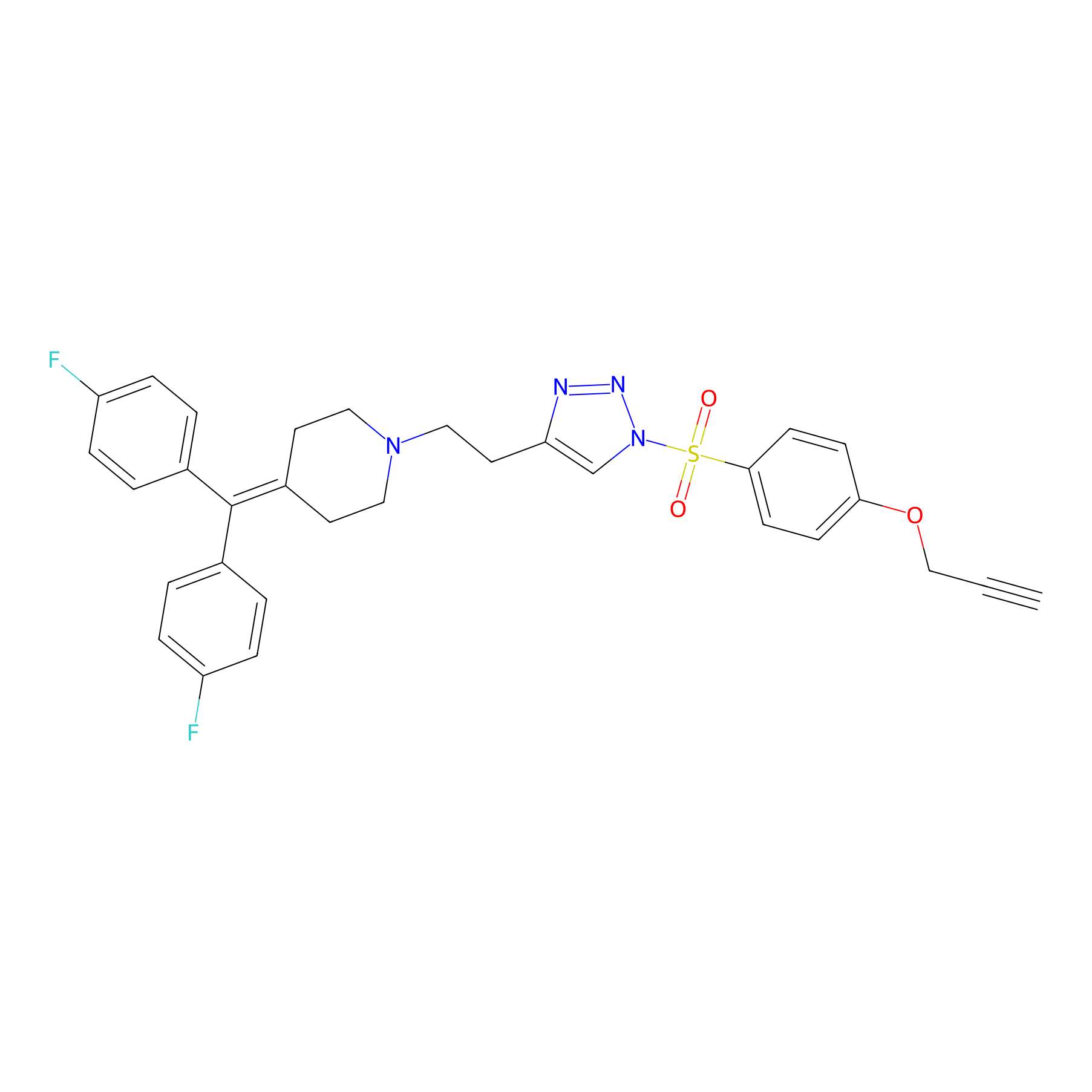 |
Y18(20.00); Y26(20.00) | LDD0258 | [6] | |
|
TH216 Probe Info |
 |
Y18(20.00); Y26(12.71) | LDD0259 | [6] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [7] | |
|
AZ-9 Probe Info |
 |
E100(0.88); E90(0.94); D224(0.95); E79(0.92) | LDD2208 | [8] | |
|
ONAyne Probe Info |
 |
N.A. | LDD0273 | [9] | |
|
Probe 1 Probe Info |
 |
Y18(270.12); Y26(41.10) | LDD3495 | [10] | |
|
m-APA Probe Info |
 |
7.32 | LDD0403 | [11] | |
|
DBIA Probe Info |
 |
C613(0.96) | LDD3310 | [12] | |
|
JZ128-DTB Probe Info |
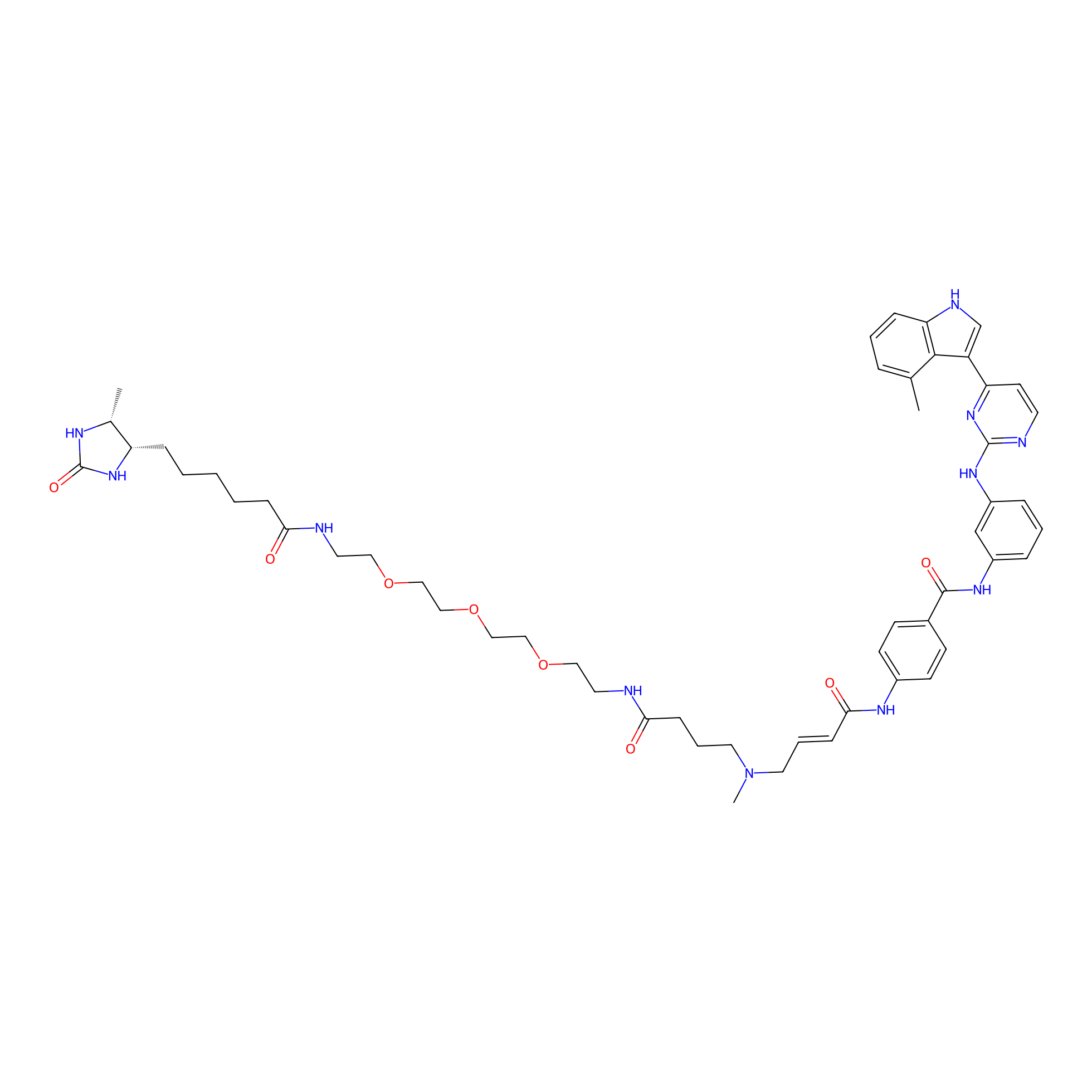 |
N.A. | LDD0462 | [13] | |
|
BTD Probe Info |
 |
C247(0.63) | LDD2145 | [14] | |
|
AHL-Pu-1 Probe Info |
 |
C217(2.33) | LDD0168 | [15] | |
|
HHS-482 Probe Info |
 |
Y18(0.95); Y182(1.01); Y238(0.44); Y26(1.01) | LDD0285 | [16] | |
|
HHS-475 Probe Info |
 |
Y18(0.81); Y26(0.82); Y182(0.86); Y238(3.92) | LDD0264 | [17] | |
|
HHS-465 Probe Info |
 |
Y18(10.00) | LDD2237 | [18] | |
|
5E-2FA Probe Info |
 |
H131(0.00); H8(0.00); H269(0.00); H77(0.00) | LDD2235 | [19] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C247(0.00); C217(0.00) | LDD0038 | [20] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0032 | [21] | |
|
IPIAA_H Probe Info |
 |
N.A. | LDD0030 | [22] | |
|
IPIAA_L Probe Info |
 |
N.A. | LDD0031 | [22] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [20] | |
|
ATP probe Probe Info |
 |
K189(0.00); K59(0.00); K15(0.00); K107(0.00) | LDD0035 | [23] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [24] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [25] | |
|
1d-yne Probe Info |
 |
N.A. | LDD0356 | [26] | |
|
IPM Probe Info |
 |
N.A. | LDD2156 | [27] | |
|
NHS Probe Info |
 |
K59(0.00); K117(0.00); K17(0.00) | LDD0010 | [25] | |
|
OSF Probe Info |
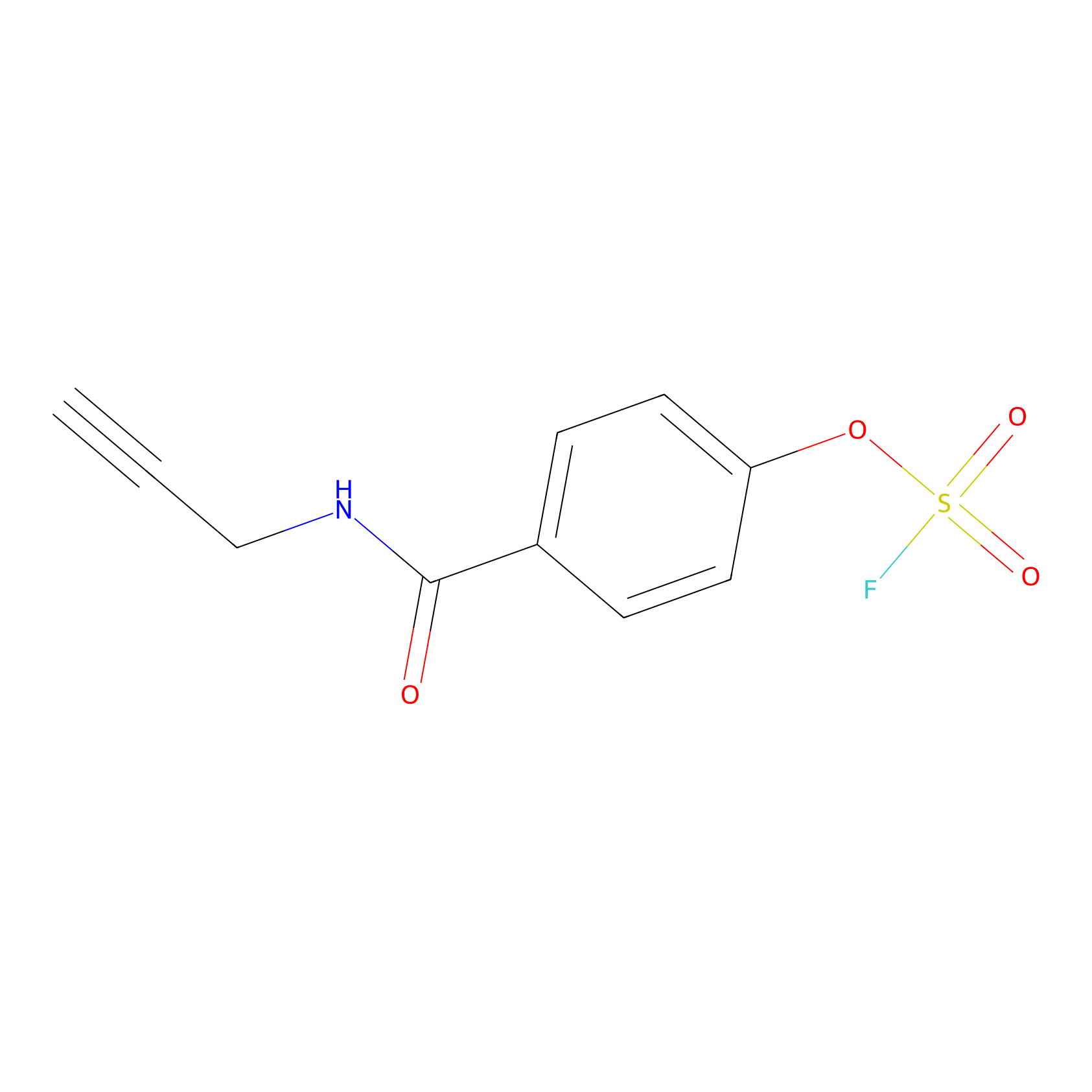 |
N.A. | LDD0029 | [28] | |
|
PF-06672131 Probe Info |
 |
N.A. | LDD0152 | [29] | |
|
SF Probe Info |
 |
Y18(0.00); K189(0.00); K117(0.00); K188(0.00) | LDD0028 | [28] | |
|
STPyne Probe Info |
 |
K117(0.00); K59(0.00) | LDD0009 | [25] | |
|
Ox-W18 Probe Info |
 |
W12(0.00); W228(0.00) | LDD2175 | [30] | |
|
1c-yne Probe Info |
 |
K59(0.00); K15(0.00); K232(0.00); K10(0.00) | LDD0228 | [26] | |
|
Acrolein Probe Info |
 |
H77(0.00); C217(0.00) | LDD0217 | [31] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [31] | |
|
Methacrolein Probe Info |
 |
H131(0.00); H77(0.00) | LDD0218 | [31] | |
|
W1 Probe Info |
 |
N.A. | LDD0236 | [32] | |
|
MPP-AC Probe Info |
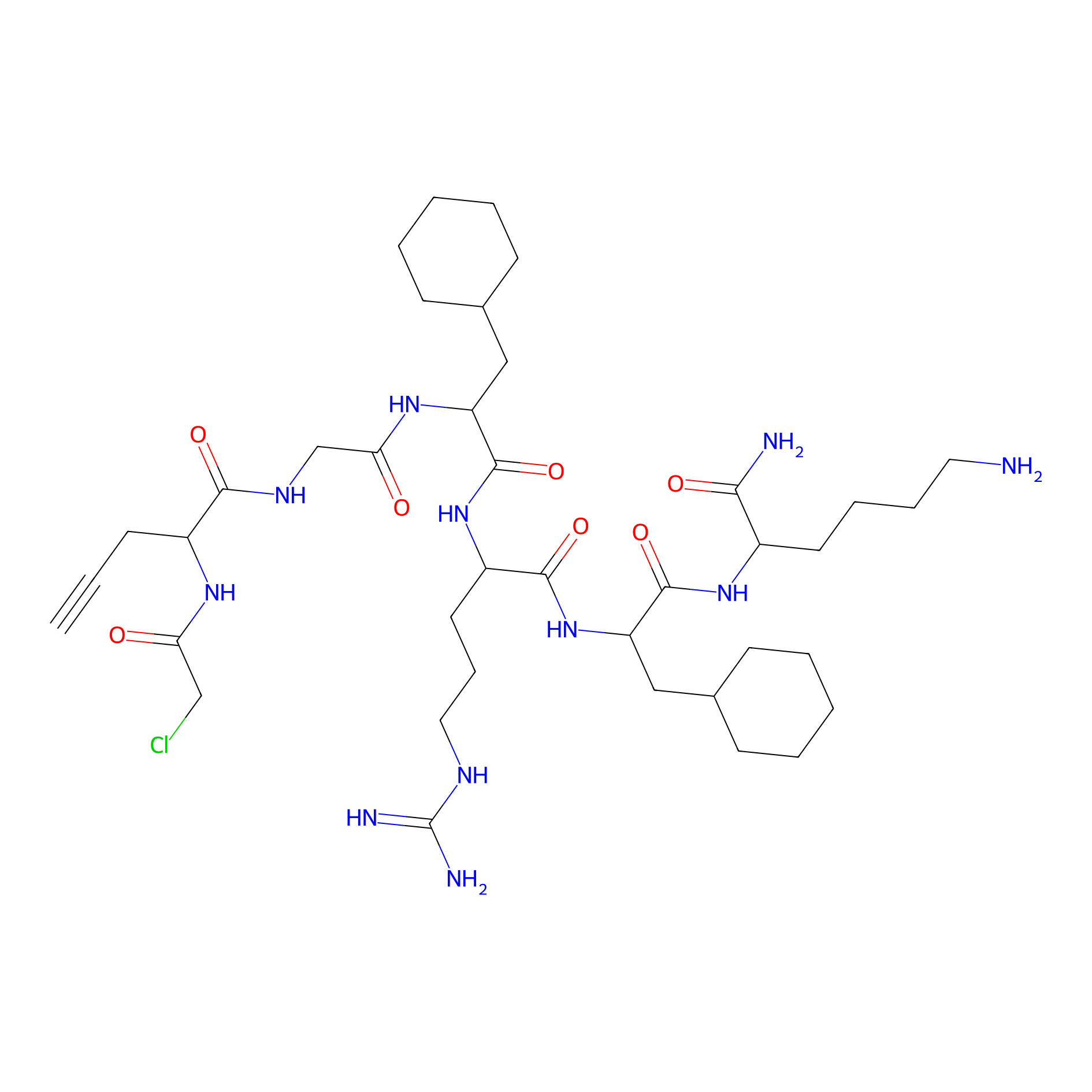 |
N.A. | LDD0428 | [33] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [34] | |
|
TER-AC Probe Info |
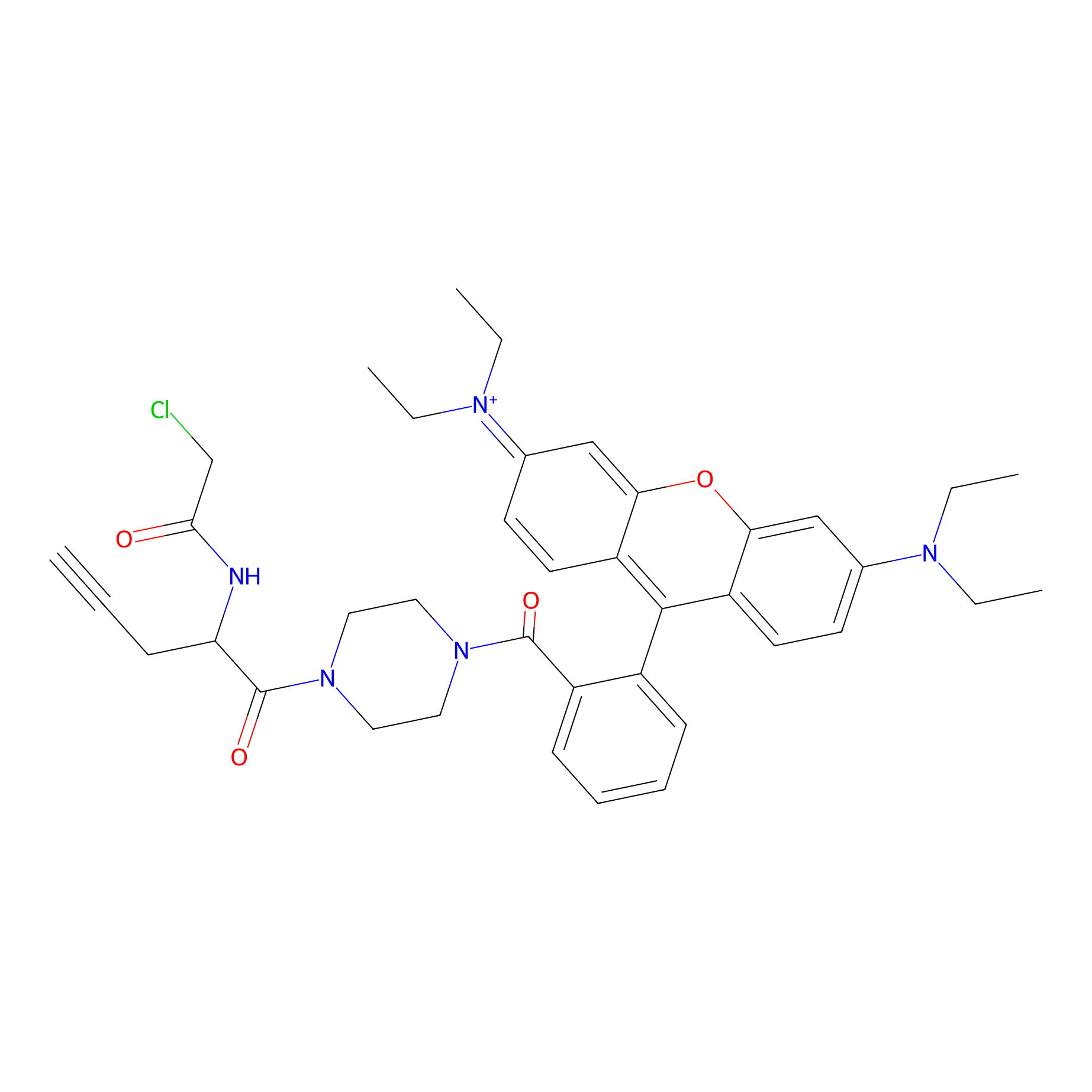 |
N.A. | LDD0426 | [33] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C003 Probe Info |
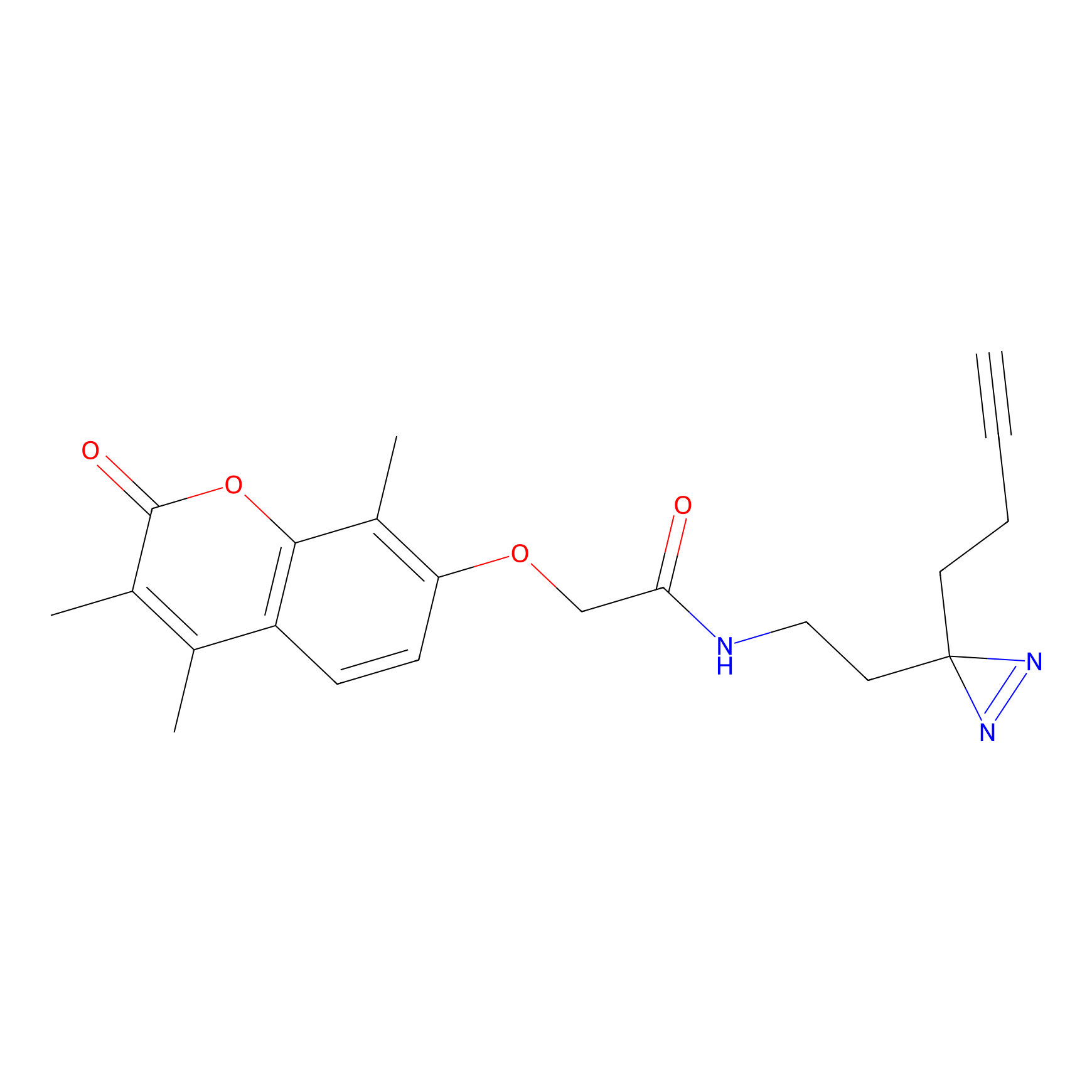 |
12.64 | LDD1713 | [35] | |
|
C301 Probe Info |
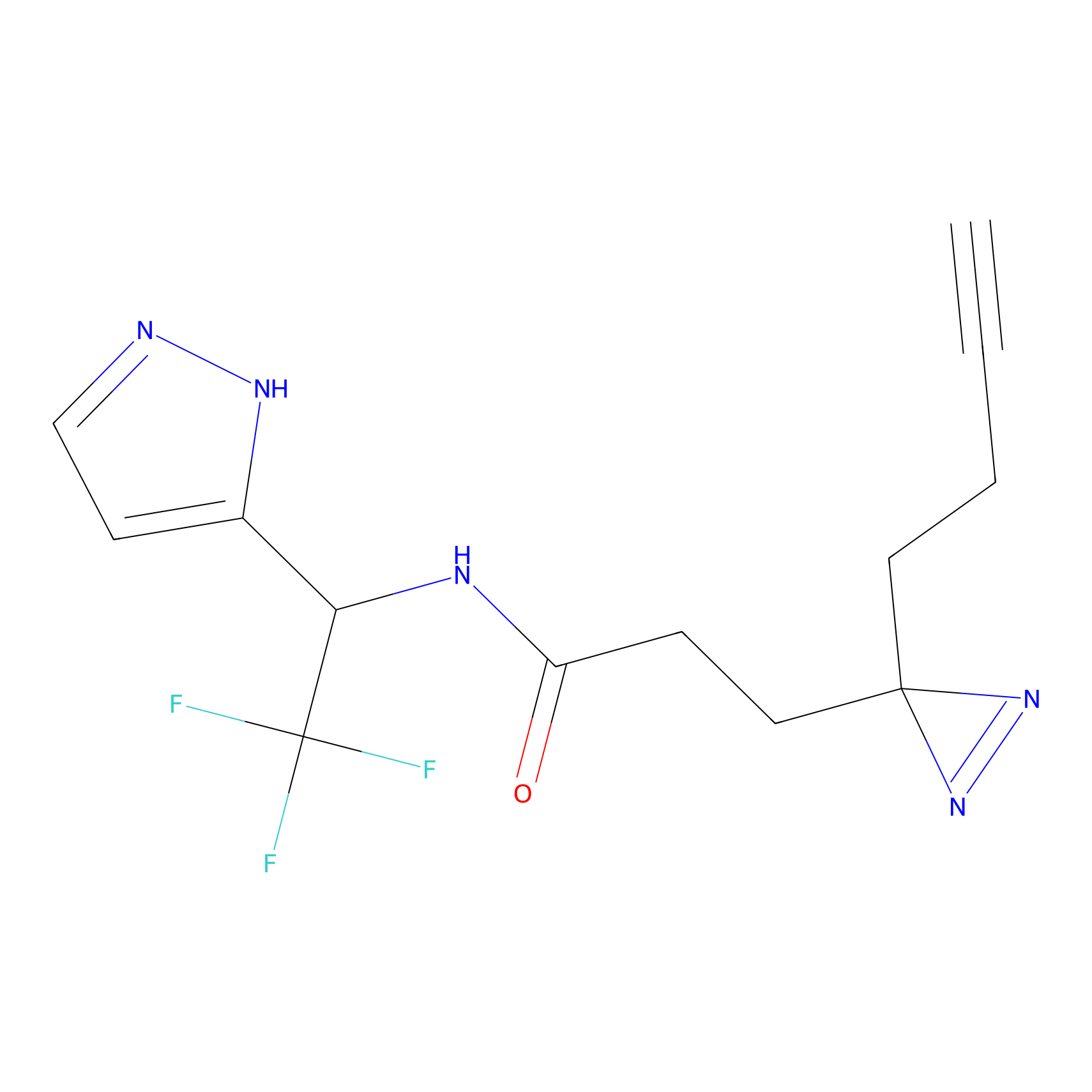 |
5.10 | LDD1970 | [35] | |
|
C389 Probe Info |
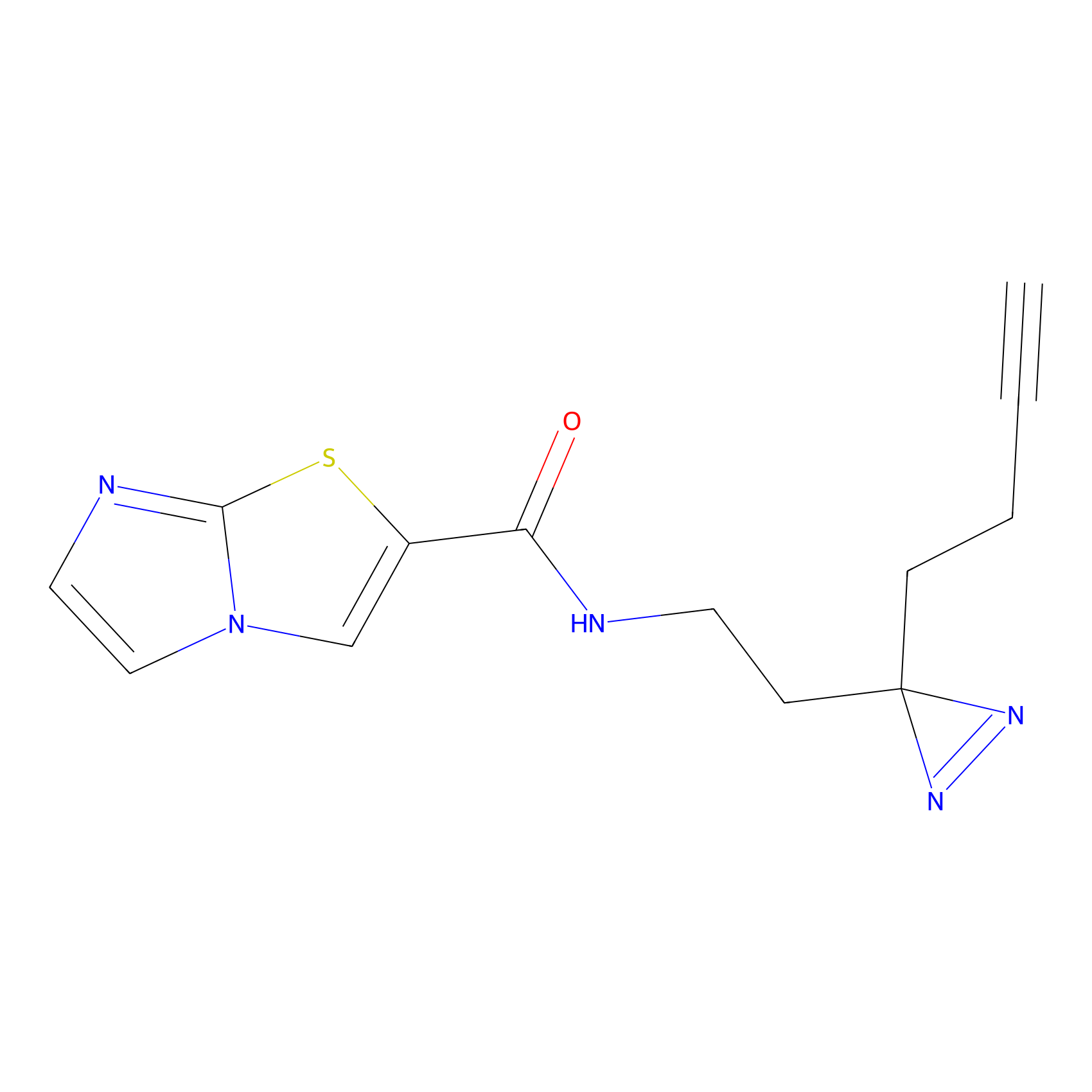 |
12.64 | LDD2048 | [35] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0138 | [36] | |
|
Probe 3 Probe Info |
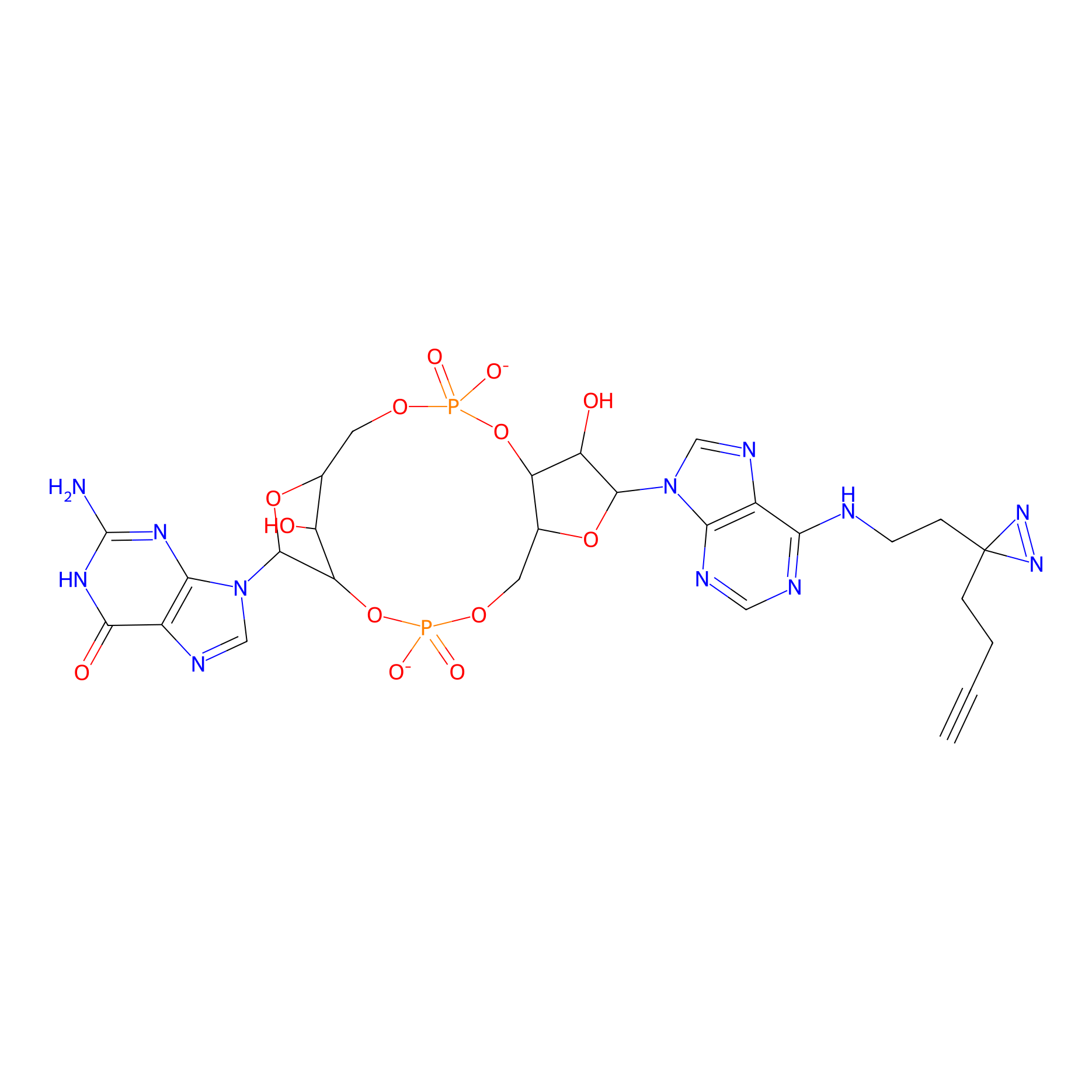 |
N.A. | LDD0195 | [37] | |
|
Staurosporine capture compound Probe Info |
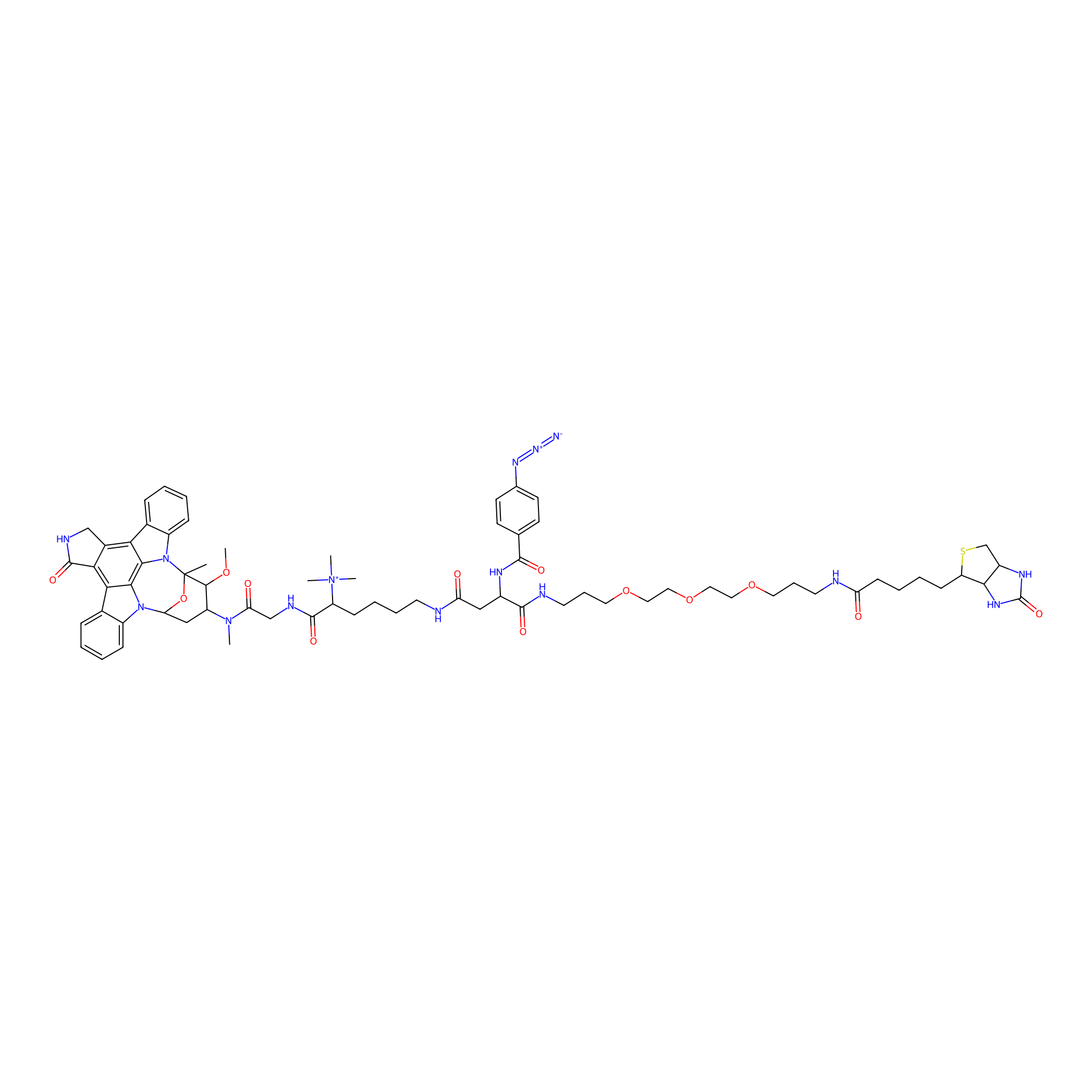 |
N.A. | LDD0083 | [38] | |
|
DA-2 Probe Info |
 |
N.A. | LDD0070 | [39] | |
|
OEA-DA Probe Info |
 |
7.38 | LDD0046 | [40] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0068 | [41] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0101 | 23-cGAMP | HeLa | N.A. | LDD0195 | [37] |
| LDCM0025 | 4SU-RNA | HEK-293T | C217(2.33) | LDD0168 | [15] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C217(2.44) | LDD0169 | [15] |
| LDCM0156 | Aniline | NCI-H1299 | 7.32 | LDD0403 | [11] |
| LDCM0108 | Chloroacetamide | HeLa | C217(0.00); H77(0.00); H131(0.00); C247(0.00) | LDD0222 | [31] |
| LDCM0116 | HHS-0101 | DM93 | Y18(0.81); Y26(0.82); Y182(0.86); Y238(3.92) | LDD0264 | [17] |
| LDCM0117 | HHS-0201 | DM93 | Y18(0.68); Y26(0.86); Y182(1.22); Y238(4.04) | LDD0265 | [17] |
| LDCM0118 | HHS-0301 | DM93 | Y18(0.69); Y26(1.48); Y182(2.70); Y238(2.73) | LDD0266 | [17] |
| LDCM0119 | HHS-0401 | DM93 | Y26(0.72); Y18(0.74); Y182(1.72); Y238(2.03) | LDD0267 | [17] |
| LDCM0120 | HHS-0701 | DM93 | Y18(0.64); Y26(0.68); Y182(1.93); Y238(2.08) | LDD0268 | [17] |
| LDCM0107 | IAA | HeLa | H77(0.00); H131(0.00); C247(0.00); C217(0.00) | LDD0221 | [31] |
| LDCM0123 | JWB131 | DM93 | Y18(0.95); Y182(1.01); Y238(0.44); Y26(1.01) | LDD0285 | [16] |
| LDCM0124 | JWB142 | DM93 | Y18(0.57); Y182(0.90); Y238(0.92); Y26(0.68) | LDD0286 | [16] |
| LDCM0125 | JWB146 | DM93 | Y18(0.93); Y182(0.91); Y238(1.14); Y26(1.09) | LDD0287 | [16] |
| LDCM0126 | JWB150 | DM93 | Y18(3.49); Y182(3.53); Y238(1.96); Y26(3.99) | LDD0288 | [16] |
| LDCM0127 | JWB152 | DM93 | Y18(1.96); Y182(2.15); Y238(1.22); Y26(2.21) | LDD0289 | [16] |
| LDCM0128 | JWB198 | DM93 | Y18(1.09); Y182(1.64); Y238(0.43); Y26(1.19) | LDD0290 | [16] |
| LDCM0129 | JWB202 | DM93 | Y18(0.50); Y182(0.80); Y26(0.55) | LDD0291 | [16] |
| LDCM0130 | JWB211 | DM93 | Y18(0.93); Y182(1.19); Y238(0.85); Y26(1.02) | LDD0292 | [16] |
| LDCM0179 | JZ128 | PC-3 | N.A. | LDD0462 | [13] |
| LDCM0022 | KB02 | 22RV1 | C613(1.19) | LDD2243 | [12] |
| LDCM0023 | KB03 | 22RV1 | C613(1.02) | LDD2660 | [12] |
| LDCM0024 | KB05 | COLO792 | C613(0.96) | LDD3310 | [12] |
| LDCM0109 | NEM | HeLa | H131(0.00); H77(0.00) | LDD0223 | [31] |
| LDCM0551 | Nucleophilic fragment 5b | MDA-MB-231 | C247(0.63) | LDD2145 | [14] |
| LDCM0131 | RA190 | MM1.R | C247(1.26) | LDD0304 | [42] |
| LDCM0019 | Staurosporine | Hep-G2 | N.A. | LDD0083 | [38] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Protein max (MAX) | MAX family | P61244 | |||
| ALX homeobox protein 1 (ALX1) | Paired homeobox family | Q15699 | |||
| PHD finger protein 19 (PHF19) | Polycomblike family | Q5T6S3 | |||
Other
References
