Details of the Target
General Information of Target
| Target ID | LDTP12533 | |||||
|---|---|---|---|---|---|---|
| Target Name | Ubiquilin-4 (UBQLN4) | |||||
| Gene Name | UBQLN4 | |||||
| Gene ID | 56893 | |||||
| Synonyms |
C1orf6; CIP75; UBIN; Ubiquilin-4; Ataxin-1 interacting ubiquitin-like protein; A1Up; Ataxin-1 ubiquitin-like-interacting protein A1U; Connexin43-interacting protein of 75 kDa; CIP75 |
|||||
| 3D Structure | ||||||
| Sequence |
MMQGNTCHRMSFHPGRGCPRGRGGHGARPSAPSFRPQNLRLLHPQQPPVQYQYEPPSAPS
TTFSNSPAPNFLPPRPDFVPFPPPMPPSAQGPLPPCPIRPPFPNHQMRHPFPVPPCFPPM PPPMPCPNNPPVPGAPPGQGTFPFMMPPPSMPHPPPPPVMPQQVNYQYPPGYSHHNFPPP SFNSFQNNPSSFLPSANNSSSPHFRHLPPYPLPKAPSERRSPERLKHYDDHRHRDHSHGR GERHRSLDRRERGRSPDRRRQDSRYRSDYDRGRTPSRHRSYERSRERERERHRHRDNRRS PSLERSYKKEYKRSGRSYGLSVVPEPAGCTPELPGEIIKNTDSWAPPLEIVNHRSPSREK KRARWEEEKDRWSDNQSSGKDKNYTSIKEKEPEETMPDKNEEEEEELLKPVWIRCTHSEN YYSSDPMDQVGDSTVVGTSRLRDLYDKFEEELGSRQEKAKAARPPWEPPKTKLDEDLESS SESECESDEDSTCSSSSDSEVFDVIAEIKRKKAHPDRLHDELWYNDPGQMNDGPLCKCSA KARRTGIRHSIYPGEEAIKPCRPMTNNAGRLFHYRITVSPPTNFLTDRPTVIEYDDHEYI FEGFSMFAHAPLTNIPLCKVIRFNIDYTIHFIEEMMPENFCVKGLELFSLFLFRDILELY DWNLKGPLFEDSPPCCPRFHFMPRFVRFLPDGGKEVLSMHQILLYLLRCSKALVPEEEIA NMLQWEELEWQKYAEECKGMIVTNPGTKPSSVRIDQLDREQFNPDVITFPIIVHFGIRPA QLSYAGDPQYQKLWKSYVKLRHLLANSPKVKQTDKQKLAQREEALQKIRQKNTMRREVTV ELSSQGFWKTGIRSDVCQHAMMLPVLTHHIRYHQCLMHLDKLIGYTFQDRCLLQLAMTHP SHHLNFGMNPDHARNSLSNCGIRQPKYGDRKVHHMHMRKKGINTLINIMSRLGQDDPTPS RINHNERLEFLGDAVVEFLTSVHLYYLFPSLEEGGLATYRTAIVQNQHLAMLAKKLELDR FMLYAHGPDLCRESDLRHAMANCFEALIGAVYLEGSLEEAKQLFGRLLFNDPDLREVWLN YPLHPLQLQEPNTDRQLIETSPVLQKLTEFEEAIGVIFTHVRLLARAFTLRTVGFNHLTL GHNQRMEFLGDSIMQLVATEYLFIHFPDHHEGHLTLLRSSLVNNRTQAKVAEELGMQEYA ITNDKTKRPVALRTKTLADLLESFIAALYIDKDLEYVHTFMNVCFFPRLKEFILNQDWND PKSQLQQCCLTLRTEGKEPDIPLYKTLQTVGPSHARTYTVAVYFKGERIGCGKGPSIQQA EMGAAMDALEKYNFPQMAHQKRFIERKYRQELKEMRWEREHQEREPDETEDIKK |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
Regulator of protein degradation that mediates the proteasomal targeting of misfolded, mislocalized or accumulated proteins. Acts by binding polyubiquitin chains of target proteins via its UBA domain and by interacting with subunits of the proteasome via its ubiquitin-like domain. Key regulator of DNA repair that represses homologous recombination repair: in response to DNA damage, recruited to sites of DNA damage following phosphorylation by ATM and acts by binding and removing ubiquitinated MRE11 from damaged chromatin, leading to MRE11 degradation by the proteasome. MRE11 degradation prevents homologous recombination repair, redirecting double-strand break repair toward non-homologous end joining (NHEJ). Specifically recognizes and binds mislocalized transmembrane-containing proteins and targets them to proteasomal degradation. Collaborates with DESI1/POST in the export of ubiquitinated proteins from the nucleus to the cytoplasm. Also plays a role in the regulation of the proteasomal degradation of non-ubiquitinated GJA1. Acts as an adapter protein that recruits UBQLN1 to the autophagy machinery. Mediates the association of UBQLN1 with autophagosomes and the autophagy-related protein LC3 (MAP1LC3A/B/C) and may assist in the maturation of autophagosomes to autolysosomes by mediating autophagosome-lysosome fusion.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
14.14 | LDD0402 | [1] | |
|
TH211 Probe Info |
 |
Y279(9.19) | LDD0260 | [2] | |
|
TH214 Probe Info |
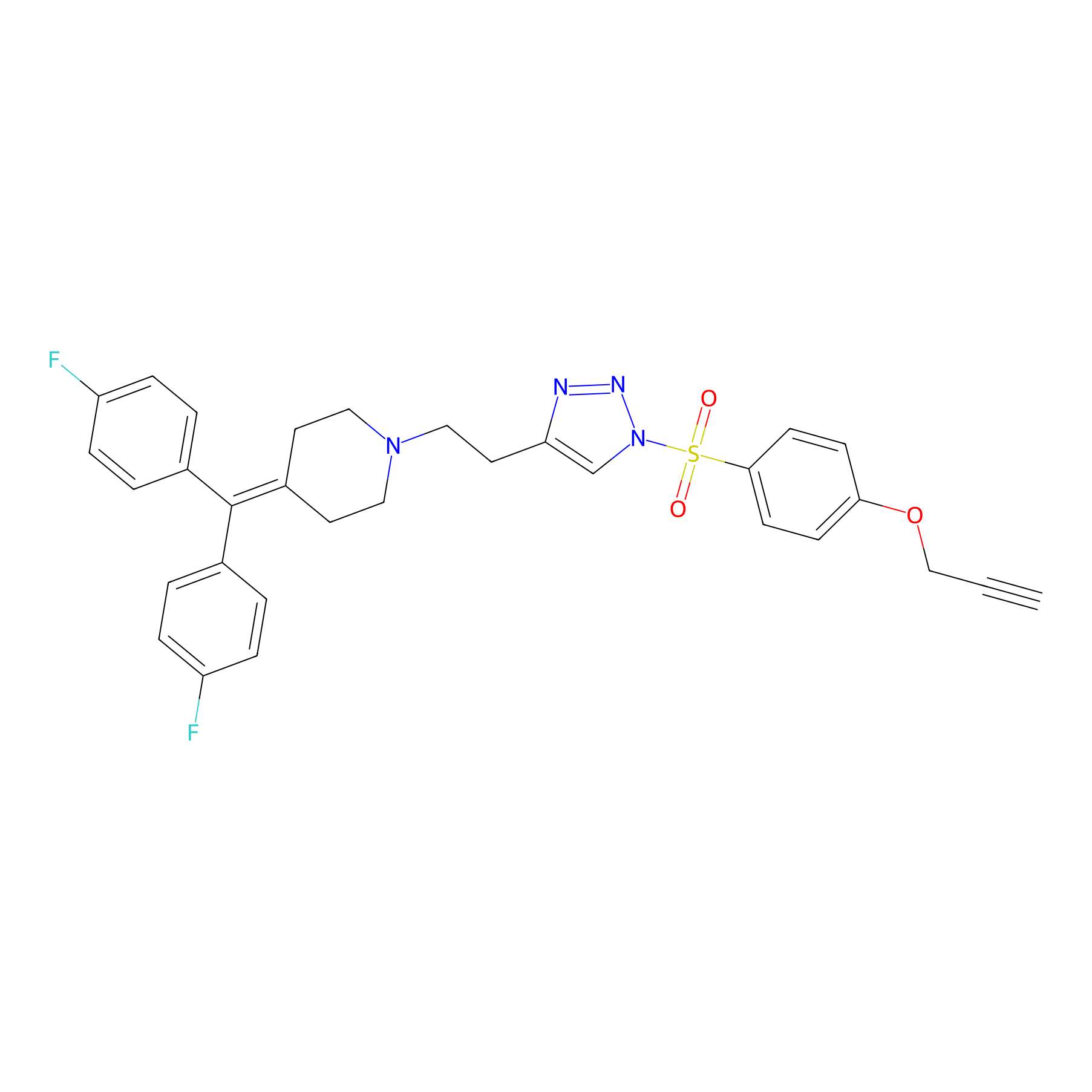 |
Y279(11.72) | LDD0258 | [2] | |
|
TH216 Probe Info |
 |
Y286(20.00); Y279(15.22) | LDD0259 | [2] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [3] | |
|
AZ-9 Probe Info |
 |
E260(10.00) | LDD2208 | [4] | |
|
Probe 1 Probe Info |
 |
Y279(19.34); Y286(19.62) | LDD3495 | [5] | |
|
BTD Probe Info |
 |
C29(0.99) | LDD2091 | [6] | |
|
DA-P3 Probe Info |
 |
5.78 | LDD0180 | [7] | |
|
AHL-Pu-1 Probe Info |
 |
C29(2.88) | LDD0169 | [8] | |
|
HHS-482 Probe Info |
 |
Y279(0.84) | LDD0285 | [9] | |
|
HHS-475 Probe Info |
 |
Y286(0.43); Y279(0.93) | LDD0264 | [10] | |
|
HHS-465 Probe Info |
 |
Y279(9.16); Y286(10.00) | LDD2237 | [11] | |
|
DBIA Probe Info |
 |
C29(0.96) | LDD0078 | [12] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [13] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [14] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [14] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [14] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [15] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2224 | [16] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [17] | |
|
WYneO Probe Info |
 |
N.A. | LDD0022 | [17] | |
|
ENE Probe Info |
 |
N.A. | LDD0006 | [17] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [17] | |
|
PPMS Probe Info |
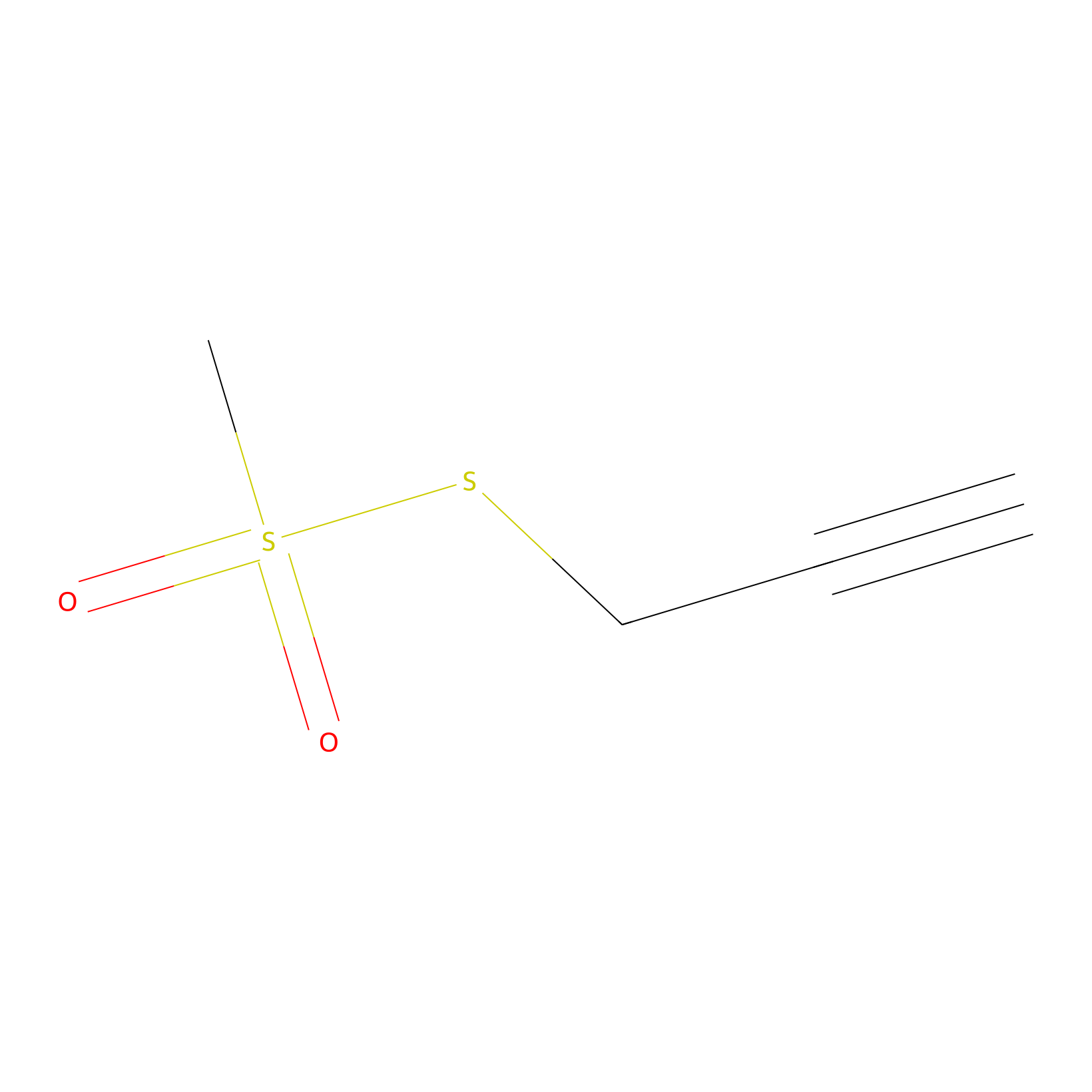 |
N.A. | LDD0008 | [17] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [17] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [18] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [19] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [19] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [19] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C235 Probe Info |
 |
22.47 | LDD1908 | [20] | |
|
FFF probe11 Probe Info |
 |
18.62 | LDD0471 | [21] | |
|
FFF probe12 Probe Info |
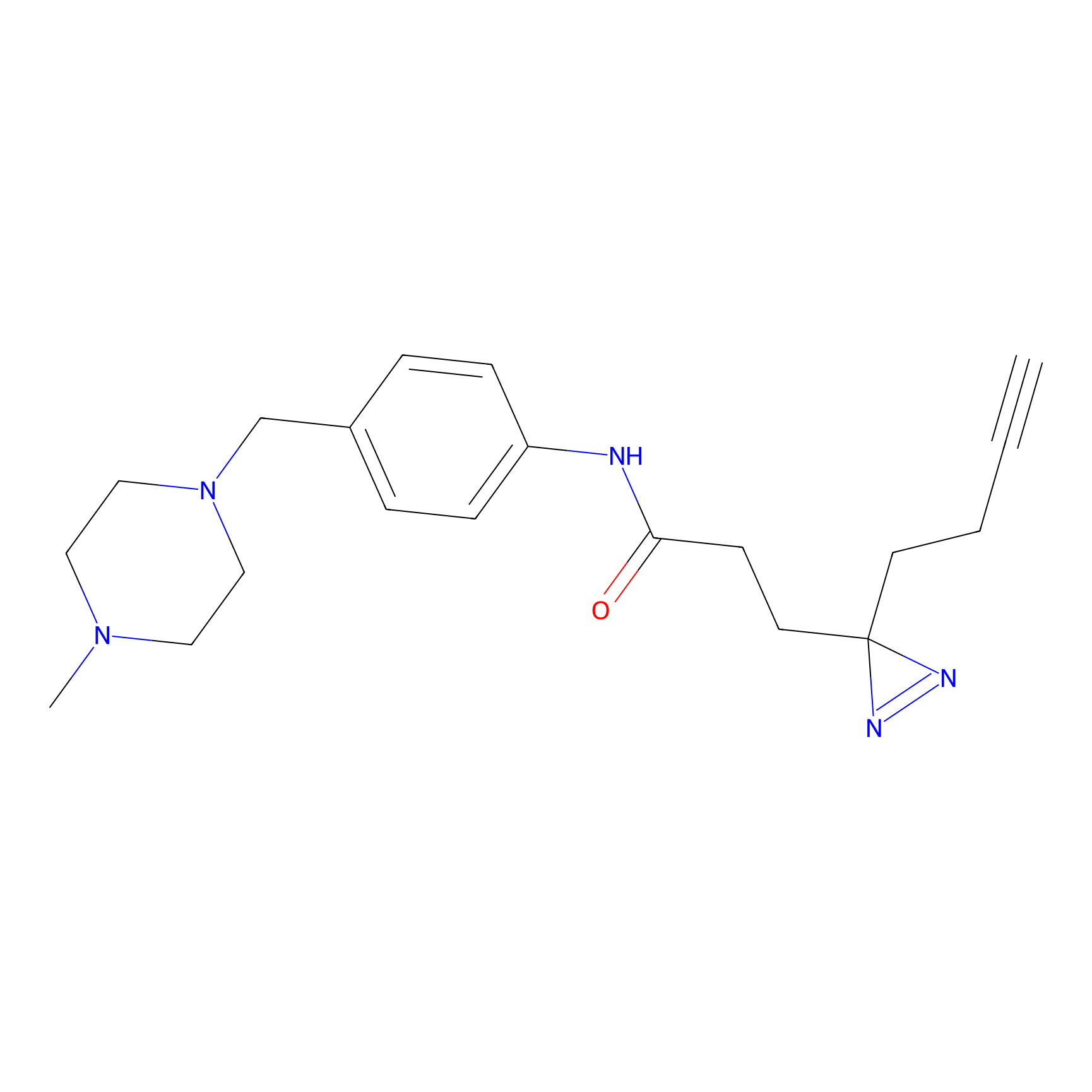 |
12.22 | LDD0473 | [21] | |
|
FFF probe13 Probe Info |
 |
20.00 | LDD0475 | [21] | |
|
FFF probe14 Probe Info |
 |
20.00 | LDD0477 | [21] | |
|
FFF probe2 Probe Info |
 |
20.00 | LDD0463 | [21] | |
|
FFF probe3 Probe Info |
 |
20.00 | LDD0464 | [21] | |
|
JN0003 Probe Info |
 |
20.00 | LDD0469 | [21] | |
|
DFG-out-3 Probe Info |
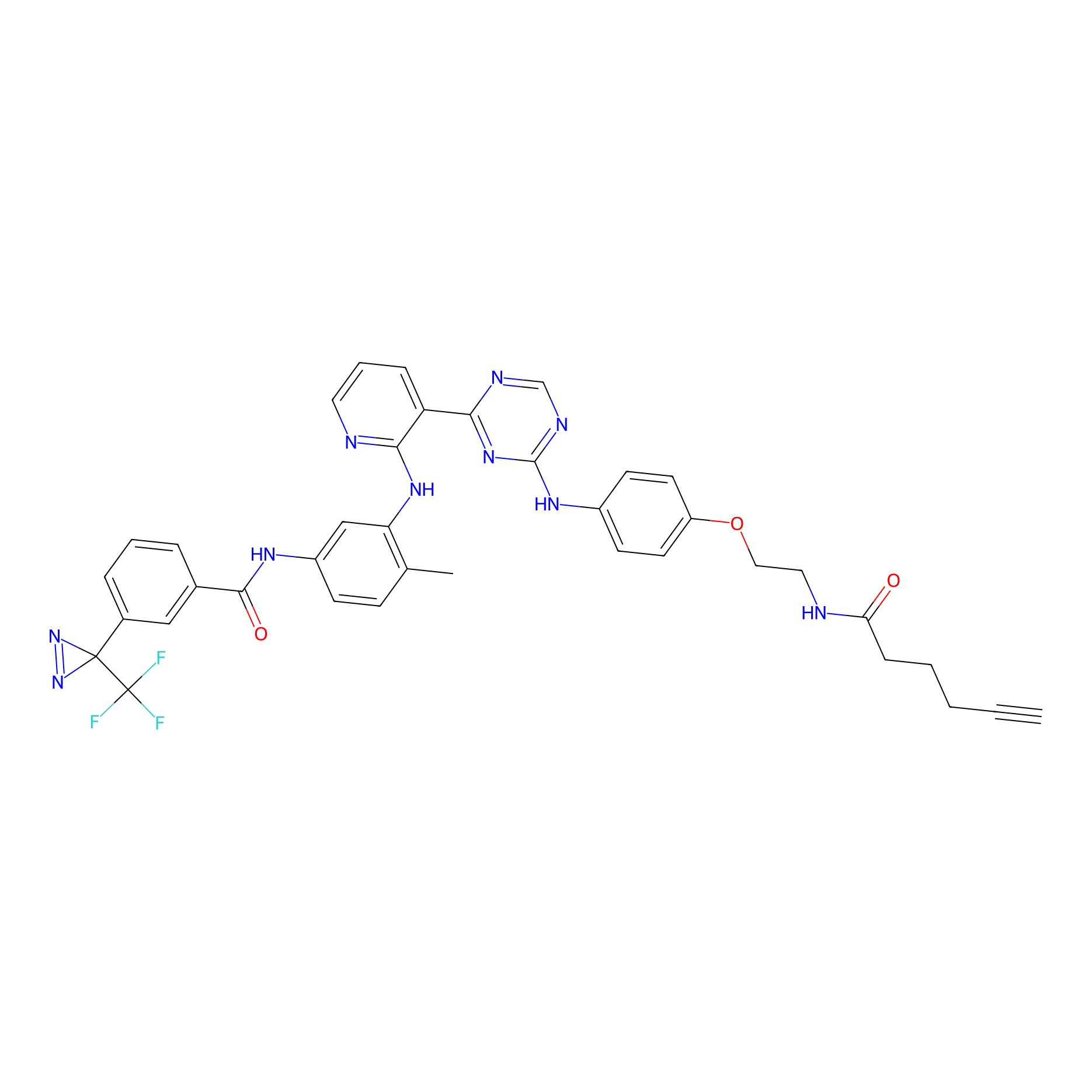 |
2.10 | LDD0074 | [22] | |
|
Kambe_3 Probe Info |
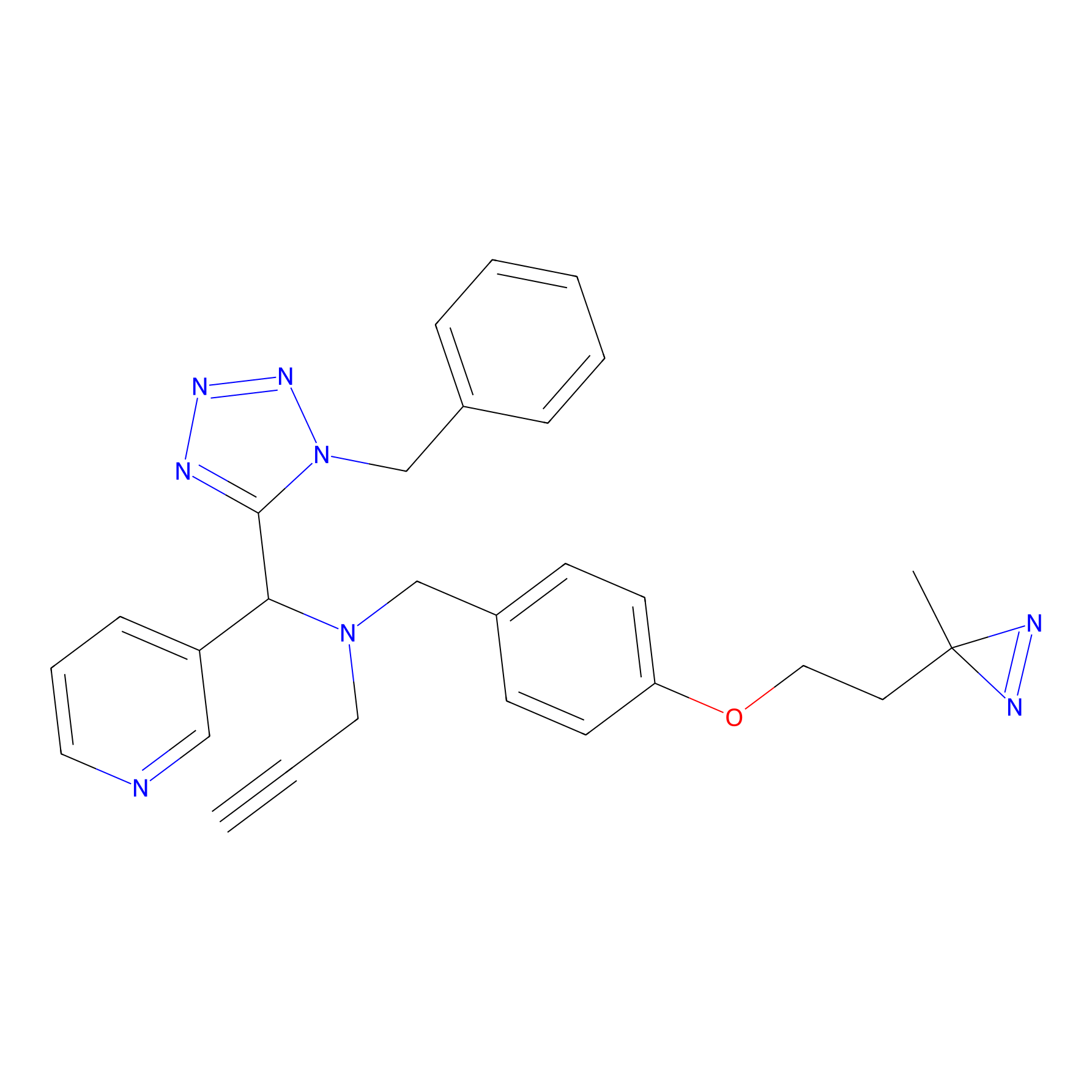 |
2.00 | LDD0130 | [23] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C29(1.12) | LDD2117 | [6] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C29(0.96) | LDD2152 | [6] |
| LDCM0510 | 3-(4-(Hydroxydiphenylmethyl)piperidin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C29(0.66) | LDD2103 | [6] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C29(2.88) | LDD0169 | [8] |
| LDCM0214 | AC1 | HCT 116 | C29(1.06) | LDD0531 | [12] |
| LDCM0215 | AC10 | HCT 116 | C29(0.87) | LDD0532 | [12] |
| LDCM0216 | AC100 | HCT 116 | C29(1.01) | LDD0533 | [12] |
| LDCM0217 | AC101 | HCT 116 | C29(1.02) | LDD0534 | [12] |
| LDCM0218 | AC102 | HCT 116 | C29(0.93) | LDD0535 | [12] |
| LDCM0219 | AC103 | HCT 116 | C29(1.21) | LDD0536 | [12] |
| LDCM0220 | AC104 | HCT 116 | C29(0.99) | LDD0537 | [12] |
| LDCM0221 | AC105 | HCT 116 | C29(1.13) | LDD0538 | [12] |
| LDCM0222 | AC106 | HCT 116 | C29(1.14) | LDD0539 | [12] |
| LDCM0223 | AC107 | HCT 116 | C29(0.97) | LDD0540 | [12] |
| LDCM0224 | AC108 | HCT 116 | C29(1.06) | LDD0541 | [12] |
| LDCM0225 | AC109 | HCT 116 | C29(0.88) | LDD0542 | [12] |
| LDCM0226 | AC11 | HCT 116 | C29(1.02) | LDD0543 | [12] |
| LDCM0227 | AC110 | HCT 116 | C29(0.96) | LDD0544 | [12] |
| LDCM0228 | AC111 | HCT 116 | C29(0.94) | LDD0545 | [12] |
| LDCM0229 | AC112 | HCT 116 | C29(1.06) | LDD0546 | [12] |
| LDCM0230 | AC113 | HCT 116 | C29(1.02) | LDD0547 | [12] |
| LDCM0231 | AC114 | HCT 116 | C29(1.30) | LDD0548 | [12] |
| LDCM0232 | AC115 | HCT 116 | C29(1.22) | LDD0549 | [12] |
| LDCM0233 | AC116 | HCT 116 | C29(1.31) | LDD0550 | [12] |
| LDCM0234 | AC117 | HCT 116 | C29(1.15) | LDD0551 | [12] |
| LDCM0235 | AC118 | HCT 116 | C29(1.15) | LDD0552 | [12] |
| LDCM0236 | AC119 | HCT 116 | C29(1.17) | LDD0553 | [12] |
| LDCM0237 | AC12 | HCT 116 | C29(0.85) | LDD0554 | [12] |
| LDCM0238 | AC120 | HCT 116 | C29(1.04) | LDD0555 | [12] |
| LDCM0239 | AC121 | HCT 116 | C29(1.16) | LDD0556 | [12] |
| LDCM0240 | AC122 | HCT 116 | C29(1.19) | LDD0557 | [12] |
| LDCM0241 | AC123 | HCT 116 | C29(1.04) | LDD0558 | [12] |
| LDCM0242 | AC124 | HCT 116 | C29(1.08) | LDD0559 | [12] |
| LDCM0243 | AC125 | HCT 116 | C29(1.27) | LDD0560 | [12] |
| LDCM0244 | AC126 | HCT 116 | C29(1.29) | LDD0561 | [12] |
| LDCM0245 | AC127 | HCT 116 | C29(1.50) | LDD0562 | [12] |
| LDCM0246 | AC128 | HCT 116 | C29(0.82) | LDD0563 | [12] |
| LDCM0247 | AC129 | HCT 116 | C29(1.02) | LDD0564 | [12] |
| LDCM0249 | AC130 | HCT 116 | C29(1.03) | LDD0566 | [12] |
| LDCM0250 | AC131 | HCT 116 | C29(1.04) | LDD0567 | [12] |
| LDCM0251 | AC132 | HCT 116 | C29(1.13) | LDD0568 | [12] |
| LDCM0252 | AC133 | HCT 116 | C29(1.07) | LDD0569 | [12] |
| LDCM0253 | AC134 | HCT 116 | C29(1.17) | LDD0570 | [12] |
| LDCM0254 | AC135 | HCT 116 | C29(0.96) | LDD0571 | [12] |
| LDCM0255 | AC136 | HCT 116 | C29(1.10) | LDD0572 | [12] |
| LDCM0256 | AC137 | HCT 116 | C29(1.09) | LDD0573 | [12] |
| LDCM0257 | AC138 | HCT 116 | C29(1.22) | LDD0574 | [12] |
| LDCM0258 | AC139 | HCT 116 | C29(1.23) | LDD0575 | [12] |
| LDCM0259 | AC14 | HCT 116 | C29(0.86) | LDD0576 | [12] |
| LDCM0260 | AC140 | HCT 116 | C29(1.33) | LDD0577 | [12] |
| LDCM0261 | AC141 | HCT 116 | C29(1.32) | LDD0578 | [12] |
| LDCM0262 | AC142 | HCT 116 | C29(1.00) | LDD0579 | [12] |
| LDCM0263 | AC143 | HCT 116 | C29(1.20) | LDD0580 | [12] |
| LDCM0264 | AC144 | HCT 116 | C29(1.43) | LDD0581 | [12] |
| LDCM0265 | AC145 | HCT 116 | C29(1.26) | LDD0582 | [12] |
| LDCM0266 | AC146 | HCT 116 | C29(1.57) | LDD0583 | [12] |
| LDCM0267 | AC147 | HCT 116 | C29(1.46) | LDD0584 | [12] |
| LDCM0268 | AC148 | HCT 116 | C29(2.00) | LDD0585 | [12] |
| LDCM0269 | AC149 | HCT 116 | C29(1.70) | LDD0586 | [12] |
| LDCM0270 | AC15 | HCT 116 | C29(0.91) | LDD0587 | [12] |
| LDCM0271 | AC150 | HCT 116 | C29(1.04) | LDD0588 | [12] |
| LDCM0272 | AC151 | HCT 116 | C29(1.15) | LDD0589 | [12] |
| LDCM0273 | AC152 | HCT 116 | C29(1.41) | LDD0590 | [12] |
| LDCM0274 | AC153 | HCT 116 | C29(2.06) | LDD0591 | [12] |
| LDCM0621 | AC154 | HCT 116 | C29(1.43) | LDD2158 | [12] |
| LDCM0622 | AC155 | HCT 116 | C29(1.39) | LDD2159 | [12] |
| LDCM0623 | AC156 | HCT 116 | C29(1.04) | LDD2160 | [12] |
| LDCM0624 | AC157 | HCT 116 | C29(1.00) | LDD2161 | [12] |
| LDCM0276 | AC17 | HCT 116 | C29(1.20) | LDD0593 | [12] |
| LDCM0277 | AC18 | HCT 116 | C29(1.26) | LDD0594 | [12] |
| LDCM0278 | AC19 | HCT 116 | C29(1.24) | LDD0595 | [12] |
| LDCM0279 | AC2 | HCT 116 | C29(1.24) | LDD0596 | [12] |
| LDCM0280 | AC20 | HCT 116 | C29(0.99) | LDD0597 | [12] |
| LDCM0281 | AC21 | HCT 116 | C29(1.08) | LDD0598 | [12] |
| LDCM0282 | AC22 | HCT 116 | C29(1.15) | LDD0599 | [12] |
| LDCM0283 | AC23 | HCT 116 | C29(1.17) | LDD0600 | [12] |
| LDCM0284 | AC24 | HCT 116 | C29(1.13) | LDD0601 | [12] |
| LDCM0285 | AC25 | HEK-293T | C29(0.92) | LDD0883 | [12] |
| LDCM0286 | AC26 | HEK-293T | C29(0.91) | LDD0884 | [12] |
| LDCM0287 | AC27 | HEK-293T | C29(0.94) | LDD0885 | [12] |
| LDCM0288 | AC28 | HEK-293T | C29(0.79) | LDD0886 | [12] |
| LDCM0289 | AC29 | HEK-293T | C29(0.83) | LDD0887 | [12] |
| LDCM0290 | AC3 | HCT 116 | C29(1.02) | LDD0607 | [12] |
| LDCM0291 | AC30 | HEK-293T | C29(0.85) | LDD0889 | [12] |
| LDCM0292 | AC31 | HEK-293T | C29(0.87) | LDD0890 | [12] |
| LDCM0293 | AC32 | HEK-293T | C29(0.90) | LDD0891 | [12] |
| LDCM0294 | AC33 | HEK-293T | C29(0.93) | LDD0892 | [12] |
| LDCM0295 | AC34 | HEK-293T | C29(0.94) | LDD0893 | [12] |
| LDCM0296 | AC35 | HCT 116 | C29(1.01) | LDD0613 | [12] |
| LDCM0297 | AC36 | HCT 116 | C29(1.06) | LDD0614 | [12] |
| LDCM0298 | AC37 | HCT 116 | C29(0.87) | LDD0615 | [12] |
| LDCM0299 | AC38 | HCT 116 | C29(1.09) | LDD0616 | [12] |
| LDCM0300 | AC39 | HCT 116 | C29(1.09) | LDD0617 | [12] |
| LDCM0301 | AC4 | HCT 116 | C29(1.23) | LDD0618 | [12] |
| LDCM0302 | AC40 | HCT 116 | C29(1.21) | LDD0619 | [12] |
| LDCM0303 | AC41 | HCT 116 | C29(1.17) | LDD0620 | [12] |
| LDCM0304 | AC42 | HCT 116 | C29(1.09) | LDD0621 | [12] |
| LDCM0305 | AC43 | HCT 116 | C29(1.08) | LDD0622 | [12] |
| LDCM0306 | AC44 | HCT 116 | C29(1.06) | LDD0623 | [12] |
| LDCM0307 | AC45 | HCT 116 | C29(1.31) | LDD0624 | [12] |
| LDCM0308 | AC46 | HCT 116 | C29(1.21) | LDD0625 | [12] |
| LDCM0309 | AC47 | HCT 116 | C29(1.07) | LDD0626 | [12] |
| LDCM0310 | AC48 | HCT 116 | C29(1.12) | LDD0627 | [12] |
| LDCM0311 | AC49 | HCT 116 | C29(1.15) | LDD0628 | [12] |
| LDCM0312 | AC5 | HCT 116 | C29(1.25) | LDD0629 | [12] |
| LDCM0313 | AC50 | HCT 116 | C29(1.14) | LDD0630 | [12] |
| LDCM0314 | AC51 | HCT 116 | C29(1.02) | LDD0631 | [12] |
| LDCM0315 | AC52 | HCT 116 | C29(1.02) | LDD0632 | [12] |
| LDCM0316 | AC53 | HCT 116 | C29(1.03) | LDD0633 | [12] |
| LDCM0317 | AC54 | HCT 116 | C29(0.97) | LDD0634 | [12] |
| LDCM0318 | AC55 | HCT 116 | C29(0.95) | LDD0635 | [12] |
| LDCM0319 | AC56 | HCT 116 | C29(1.05) | LDD0636 | [12] |
| LDCM0320 | AC57 | HCT 116 | C29(1.56) | LDD0637 | [12] |
| LDCM0321 | AC58 | HCT 116 | C29(1.65) | LDD0638 | [12] |
| LDCM0322 | AC59 | HCT 116 | C29(1.83) | LDD0639 | [12] |
| LDCM0323 | AC6 | HCT 116 | C29(1.08) | LDD0640 | [12] |
| LDCM0324 | AC60 | HCT 116 | C29(1.60) | LDD0641 | [12] |
| LDCM0325 | AC61 | HCT 116 | C29(1.41) | LDD0642 | [12] |
| LDCM0326 | AC62 | HCT 116 | C29(1.66) | LDD0643 | [12] |
| LDCM0327 | AC63 | HCT 116 | C29(1.58) | LDD0644 | [12] |
| LDCM0328 | AC64 | HCT 116 | C29(1.69) | LDD0645 | [12] |
| LDCM0329 | AC65 | HCT 116 | C29(1.64) | LDD0646 | [12] |
| LDCM0330 | AC66 | HCT 116 | C29(1.63) | LDD0647 | [12] |
| LDCM0331 | AC67 | HCT 116 | C29(1.81) | LDD0648 | [12] |
| LDCM0332 | AC68 | HCT 116 | C29(0.99) | LDD0649 | [12] |
| LDCM0333 | AC69 | HCT 116 | C29(1.05) | LDD0650 | [12] |
| LDCM0334 | AC7 | HCT 116 | C29(0.96) | LDD0651 | [12] |
| LDCM0335 | AC70 | HCT 116 | C29(1.27) | LDD0652 | [12] |
| LDCM0336 | AC71 | HCT 116 | C29(1.06) | LDD0653 | [12] |
| LDCM0337 | AC72 | HCT 116 | C29(1.09) | LDD0654 | [12] |
| LDCM0338 | AC73 | HCT 116 | C29(1.37) | LDD0655 | [12] |
| LDCM0339 | AC74 | HCT 116 | C29(1.37) | LDD0656 | [12] |
| LDCM0340 | AC75 | HCT 116 | C29(1.50) | LDD0657 | [12] |
| LDCM0341 | AC76 | HCT 116 | C29(1.23) | LDD0658 | [12] |
| LDCM0342 | AC77 | HCT 116 | C29(1.24) | LDD0659 | [12] |
| LDCM0343 | AC78 | HCT 116 | C29(0.96) | LDD0660 | [12] |
| LDCM0344 | AC79 | HCT 116 | C29(1.10) | LDD0661 | [12] |
| LDCM0345 | AC8 | HCT 116 | C29(1.02) | LDD0662 | [12] |
| LDCM0346 | AC80 | HCT 116 | C29(1.03) | LDD0663 | [12] |
| LDCM0347 | AC81 | HCT 116 | C29(1.12) | LDD0664 | [12] |
| LDCM0348 | AC82 | HCT 116 | C29(1.68) | LDD0665 | [12] |
| LDCM0349 | AC83 | HCT 116 | C29(1.59) | LDD0666 | [12] |
| LDCM0350 | AC84 | HCT 116 | C29(1.43) | LDD0667 | [12] |
| LDCM0351 | AC85 | HCT 116 | C29(1.48) | LDD0668 | [12] |
| LDCM0352 | AC86 | HCT 116 | C29(1.36) | LDD0669 | [12] |
| LDCM0353 | AC87 | HCT 116 | C29(1.02) | LDD0670 | [12] |
| LDCM0354 | AC88 | HCT 116 | C29(1.23) | LDD0671 | [12] |
| LDCM0355 | AC89 | HCT 116 | C29(1.12) | LDD0672 | [12] |
| LDCM0357 | AC90 | HCT 116 | C29(0.93) | LDD0674 | [12] |
| LDCM0358 | AC91 | HCT 116 | C29(1.27) | LDD0675 | [12] |
| LDCM0359 | AC92 | HCT 116 | C29(1.11) | LDD0676 | [12] |
| LDCM0360 | AC93 | HCT 116 | C29(0.92) | LDD0677 | [12] |
| LDCM0361 | AC94 | HCT 116 | C29(0.80) | LDD0678 | [12] |
| LDCM0362 | AC95 | HCT 116 | C29(0.89) | LDD0679 | [12] |
| LDCM0363 | AC96 | HCT 116 | C29(0.94) | LDD0680 | [12] |
| LDCM0364 | AC97 | HCT 116 | C29(1.33) | LDD0681 | [12] |
| LDCM0365 | AC98 | HCT 116 | C29(1.30) | LDD0682 | [12] |
| LDCM0366 | AC99 | HCT 116 | C29(1.02) | LDD0683 | [12] |
| LDCM0248 | AKOS034007472 | HCT 116 | C29(0.81) | LDD0565 | [12] |
| LDCM0356 | AKOS034007680 | HCT 116 | C29(1.02) | LDD0673 | [12] |
| LDCM0275 | AKOS034007705 | HCT 116 | C29(1.19) | LDD0592 | [12] |
| LDCM0156 | Aniline | NCI-H1299 | 13.56 | LDD0403 | [1] |
| LDCM0020 | ARS-1620 | HCC44 | C29(0.96) | LDD0078 | [12] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C29(0.99) | LDD2091 | [6] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [19] |
| LDCM0632 | CL-Sc | Hep-G2 | C29(0.89) | LDD2227 | [16] |
| LDCM0367 | CL1 | HCT 116 | C29(0.98) | LDD0684 | [12] |
| LDCM0368 | CL10 | HCT 116 | C29(1.26) | LDD0685 | [12] |
| LDCM0369 | CL100 | HCT 116 | C29(1.21) | LDD0686 | [12] |
| LDCM0370 | CL101 | HCT 116 | C29(1.03) | LDD0687 | [12] |
| LDCM0371 | CL102 | HCT 116 | C29(0.94) | LDD0688 | [12] |
| LDCM0372 | CL103 | HCT 116 | C29(1.06) | LDD0689 | [12] |
| LDCM0373 | CL104 | HCT 116 | C29(1.04) | LDD0690 | [12] |
| LDCM0374 | CL105 | HCT 116 | C29(1.08) | LDD0691 | [12] |
| LDCM0375 | CL106 | HCT 116 | C29(0.99) | LDD0692 | [12] |
| LDCM0376 | CL107 | HCT 116 | C29(1.14) | LDD0693 | [12] |
| LDCM0377 | CL108 | HCT 116 | C29(1.14) | LDD0694 | [12] |
| LDCM0378 | CL109 | HCT 116 | C29(1.20) | LDD0695 | [12] |
| LDCM0379 | CL11 | HCT 116 | C29(1.36) | LDD0696 | [12] |
| LDCM0380 | CL110 | HCT 116 | C29(1.20) | LDD0697 | [12] |
| LDCM0381 | CL111 | HCT 116 | C29(1.28) | LDD0698 | [12] |
| LDCM0382 | CL112 | HEK-293T | C29(1.04) | LDD0980 | [12] |
| LDCM0383 | CL113 | HEK-293T | C29(0.84) | LDD0981 | [12] |
| LDCM0384 | CL114 | HEK-293T | C29(0.87) | LDD0982 | [12] |
| LDCM0385 | CL115 | HEK-293T | C29(1.00) | LDD0983 | [12] |
| LDCM0386 | CL116 | HEK-293T | C29(0.85) | LDD0984 | [12] |
| LDCM0387 | CL117 | HCT 116 | C29(1.49) | LDD0704 | [12] |
| LDCM0388 | CL118 | HCT 116 | C29(0.98) | LDD0705 | [12] |
| LDCM0389 | CL119 | HCT 116 | C29(1.13) | LDD0706 | [12] |
| LDCM0390 | CL12 | HCT 116 | C29(1.23) | LDD0707 | [12] |
| LDCM0391 | CL120 | HCT 116 | C29(1.05) | LDD0708 | [12] |
| LDCM0392 | CL121 | HCT 116 | C29(1.01) | LDD0709 | [12] |
| LDCM0393 | CL122 | HCT 116 | C29(0.95) | LDD0710 | [12] |
| LDCM0394 | CL123 | HCT 116 | C29(0.96) | LDD0711 | [12] |
| LDCM0395 | CL124 | HCT 116 | C29(1.05) | LDD0712 | [12] |
| LDCM0396 | CL125 | HCT 116 | C29(1.23) | LDD0713 | [12] |
| LDCM0397 | CL126 | HCT 116 | C29(1.24) | LDD0714 | [12] |
| LDCM0398 | CL127 | HCT 116 | C29(1.23) | LDD0715 | [12] |
| LDCM0399 | CL128 | HCT 116 | C29(1.56) | LDD0716 | [12] |
| LDCM0400 | CL13 | HCT 116 | C29(1.16) | LDD0717 | [12] |
| LDCM0401 | CL14 | HCT 116 | C29(0.89) | LDD0718 | [12] |
| LDCM0402 | CL15 | HCT 116 | C29(1.34) | LDD0719 | [12] |
| LDCM0403 | CL16 | HCT 116 | C29(1.26) | LDD0720 | [12] |
| LDCM0404 | CL17 | HCT 116 | C29(1.44) | LDD0721 | [12] |
| LDCM0405 | CL18 | HCT 116 | C29(1.41) | LDD0722 | [12] |
| LDCM0406 | CL19 | HCT 116 | C29(1.06) | LDD0723 | [12] |
| LDCM0407 | CL2 | HCT 116 | C29(1.11) | LDD0724 | [12] |
| LDCM0408 | CL20 | HCT 116 | C29(1.39) | LDD0725 | [12] |
| LDCM0409 | CL21 | HCT 116 | C29(1.61) | LDD0726 | [12] |
| LDCM0410 | CL22 | HCT 116 | C29(2.07) | LDD0727 | [12] |
| LDCM0411 | CL23 | HCT 116 | C29(1.20) | LDD0728 | [12] |
| LDCM0412 | CL24 | HCT 116 | C29(1.38) | LDD0729 | [12] |
| LDCM0413 | CL25 | HCT 116 | C29(1.30) | LDD0730 | [12] |
| LDCM0414 | CL26 | HCT 116 | C29(1.18) | LDD0731 | [12] |
| LDCM0415 | CL27 | HCT 116 | C29(1.32) | LDD0732 | [12] |
| LDCM0416 | CL28 | HCT 116 | C29(1.27) | LDD0733 | [12] |
| LDCM0417 | CL29 | HCT 116 | C29(1.58) | LDD0734 | [12] |
| LDCM0418 | CL3 | HCT 116 | C29(0.93) | LDD0735 | [12] |
| LDCM0419 | CL30 | HCT 116 | C29(1.14) | LDD0736 | [12] |
| LDCM0420 | CL31 | HCT 116 | C29(1.21) | LDD0737 | [12] |
| LDCM0421 | CL32 | HCT 116 | C29(1.13) | LDD0738 | [12] |
| LDCM0422 | CL33 | HCT 116 | C29(1.08) | LDD0739 | [12] |
| LDCM0423 | CL34 | HCT 116 | C29(0.93) | LDD0740 | [12] |
| LDCM0424 | CL35 | HCT 116 | C29(1.07) | LDD0741 | [12] |
| LDCM0425 | CL36 | HCT 116 | C29(1.01) | LDD0742 | [12] |
| LDCM0426 | CL37 | HCT 116 | C29(1.00) | LDD0743 | [12] |
| LDCM0428 | CL39 | HCT 116 | C29(0.97) | LDD0745 | [12] |
| LDCM0429 | CL4 | HCT 116 | C29(1.39) | LDD0746 | [12] |
| LDCM0430 | CL40 | HCT 116 | C29(1.21) | LDD0747 | [12] |
| LDCM0431 | CL41 | HCT 116 | C29(0.97) | LDD0748 | [12] |
| LDCM0432 | CL42 | HCT 116 | C29(1.05) | LDD0749 | [12] |
| LDCM0433 | CL43 | HCT 116 | C29(1.01) | LDD0750 | [12] |
| LDCM0434 | CL44 | HCT 116 | C29(0.93) | LDD0751 | [12] |
| LDCM0435 | CL45 | HCT 116 | C29(0.96) | LDD0752 | [12] |
| LDCM0436 | CL46 | HCT 116 | C29(0.90) | LDD0753 | [12] |
| LDCM0437 | CL47 | HCT 116 | C29(0.89) | LDD0754 | [12] |
| LDCM0438 | CL48 | HCT 116 | C29(0.86) | LDD0755 | [12] |
| LDCM0439 | CL49 | HCT 116 | C29(0.85) | LDD0756 | [12] |
| LDCM0440 | CL5 | HCT 116 | C29(1.06) | LDD0757 | [12] |
| LDCM0441 | CL50 | HCT 116 | C29(0.97) | LDD0758 | [12] |
| LDCM0442 | CL51 | HCT 116 | C29(1.01) | LDD0759 | [12] |
| LDCM0443 | CL52 | HCT 116 | C29(0.97) | LDD0760 | [12] |
| LDCM0444 | CL53 | HCT 116 | C29(1.03) | LDD0761 | [12] |
| LDCM0445 | CL54 | HCT 116 | C29(0.91) | LDD0762 | [12] |
| LDCM0446 | CL55 | HCT 116 | C29(0.88) | LDD0763 | [12] |
| LDCM0447 | CL56 | HCT 116 | C29(1.10) | LDD0764 | [12] |
| LDCM0448 | CL57 | HCT 116 | C29(0.96) | LDD0765 | [12] |
| LDCM0449 | CL58 | HCT 116 | C29(0.86) | LDD0766 | [12] |
| LDCM0450 | CL59 | HCT 116 | C29(0.97) | LDD0767 | [12] |
| LDCM0451 | CL6 | HCT 116 | C29(1.16) | LDD0768 | [12] |
| LDCM0452 | CL60 | HCT 116 | C29(1.01) | LDD0769 | [12] |
| LDCM0453 | CL61 | HCT 116 | C29(1.06) | LDD0770 | [12] |
| LDCM0454 | CL62 | HCT 116 | C29(1.09) | LDD0771 | [12] |
| LDCM0455 | CL63 | HCT 116 | C29(1.03) | LDD0772 | [12] |
| LDCM0456 | CL64 | HCT 116 | C29(1.10) | LDD0773 | [12] |
| LDCM0457 | CL65 | HCT 116 | C29(0.96) | LDD0774 | [12] |
| LDCM0458 | CL66 | HCT 116 | C29(1.29) | LDD0775 | [12] |
| LDCM0459 | CL67 | HCT 116 | C29(1.15) | LDD0776 | [12] |
| LDCM0460 | CL68 | HCT 116 | C29(1.14) | LDD0777 | [12] |
| LDCM0461 | CL69 | HCT 116 | C29(1.07) | LDD0778 | [12] |
| LDCM0462 | CL7 | HCT 116 | C29(1.25) | LDD0779 | [12] |
| LDCM0463 | CL70 | HCT 116 | C29(0.92) | LDD0780 | [12] |
| LDCM0464 | CL71 | HCT 116 | C29(1.10) | LDD0781 | [12] |
| LDCM0465 | CL72 | HCT 116 | C29(1.01) | LDD0782 | [12] |
| LDCM0466 | CL73 | HCT 116 | C29(1.10) | LDD0783 | [12] |
| LDCM0467 | CL74 | HCT 116 | C29(1.23) | LDD0784 | [12] |
| LDCM0469 | CL76 | HCT 116 | C29(1.12) | LDD0786 | [12] |
| LDCM0470 | CL77 | HCT 116 | C29(1.11) | LDD0787 | [12] |
| LDCM0471 | CL78 | HCT 116 | C29(1.32) | LDD0788 | [12] |
| LDCM0472 | CL79 | HCT 116 | C29(1.56) | LDD0789 | [12] |
| LDCM0473 | CL8 | HCT 116 | C29(1.16) | LDD0790 | [12] |
| LDCM0474 | CL80 | HCT 116 | C29(1.09) | LDD0791 | [12] |
| LDCM0475 | CL81 | HCT 116 | C29(1.27) | LDD0792 | [12] |
| LDCM0476 | CL82 | HCT 116 | C29(1.52) | LDD0793 | [12] |
| LDCM0477 | CL83 | HCT 116 | C29(1.55) | LDD0794 | [12] |
| LDCM0478 | CL84 | HCT 116 | C29(1.90) | LDD0795 | [12] |
| LDCM0479 | CL85 | HCT 116 | C29(1.29) | LDD0796 | [12] |
| LDCM0480 | CL86 | HCT 116 | C29(1.19) | LDD0797 | [12] |
| LDCM0481 | CL87 | HCT 116 | C29(1.44) | LDD0798 | [12] |
| LDCM0482 | CL88 | HCT 116 | C29(1.45) | LDD0799 | [12] |
| LDCM0483 | CL89 | HCT 116 | C29(1.70) | LDD0800 | [12] |
| LDCM0484 | CL9 | HCT 116 | C29(1.37) | LDD0801 | [12] |
| LDCM0485 | CL90 | HCT 116 | C29(1.49) | LDD0802 | [12] |
| LDCM0486 | CL91 | HCT 116 | C29(1.30) | LDD0803 | [12] |
| LDCM0487 | CL92 | HCT 116 | C29(1.32) | LDD0804 | [12] |
| LDCM0488 | CL93 | HCT 116 | C29(1.08) | LDD0805 | [12] |
| LDCM0489 | CL94 | HCT 116 | C29(1.69) | LDD0806 | [12] |
| LDCM0490 | CL95 | HCT 116 | C29(1.59) | LDD0807 | [12] |
| LDCM0491 | CL96 | HCT 116 | C29(1.19) | LDD0808 | [12] |
| LDCM0492 | CL97 | HCT 116 | C29(1.19) | LDD0809 | [12] |
| LDCM0493 | CL98 | HCT 116 | C29(1.43) | LDD0810 | [12] |
| LDCM0494 | CL99 | HCT 116 | C29(1.23) | LDD0811 | [12] |
| LDCM0634 | CY-0357 | Hep-G2 | C29(0.80) | LDD2228 | [16] |
| LDCM0028 | Dobutamine | HEK-293T | 5.78 | LDD0180 | [7] |
| LDCM0495 | E2913 | HEK-293T | C29(1.63) | LDD1698 | [24] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C29(1.34) | LDD1702 | [6] |
| LDCM0625 | F8 | Ramos | C29(1.78) | LDD2187 | [25] |
| LDCM0573 | Fragment11 | Ramos | C29(0.13) | LDD2190 | [25] |
| LDCM0576 | Fragment14 | Ramos | C29(0.93) | LDD2193 | [25] |
| LDCM0468 | Fragment33 | HCT 116 | C29(1.08) | LDD0785 | [12] |
| LDCM0566 | Fragment4 | Ramos | C29(0.75) | LDD2184 | [25] |
| LDCM0427 | Fragment51 | HCT 116 | C29(1.12) | LDD0744 | [12] |
| LDCM0569 | Fragment7 | Ramos | C29(1.66) | LDD2186 | [25] |
| LDCM0116 | HHS-0101 | DM93 | Y286(0.43); Y279(0.93) | LDD0264 | [10] |
| LDCM0117 | HHS-0201 | DM93 | Y279(0.85) | LDD0265 | [10] |
| LDCM0118 | HHS-0301 | DM93 | Y279(0.84) | LDD0266 | [10] |
| LDCM0119 | HHS-0401 | DM93 | Y279(0.94) | LDD0267 | [10] |
| LDCM0120 | HHS-0701 | DM93 | Y279(1.09); Y286(4.94) | LDD0268 | [10] |
| LDCM0123 | JWB131 | DM93 | Y279(0.84) | LDD0285 | [9] |
| LDCM0124 | JWB142 | DM93 | Y279(0.65) | LDD0286 | [9] |
| LDCM0125 | JWB146 | DM93 | Y279(1.10) | LDD0287 | [9] |
| LDCM0126 | JWB150 | DM93 | Y279(2.42) | LDD0288 | [9] |
| LDCM0127 | JWB152 | DM93 | Y279(2.04) | LDD0289 | [9] |
| LDCM0128 | JWB198 | DM93 | Y279(1.02) | LDD0290 | [9] |
| LDCM0129 | JWB202 | DM93 | Y279(0.54) | LDD0291 | [9] |
| LDCM0130 | JWB211 | DM93 | Y279(0.94) | LDD0292 | [9] |
| LDCM0074 | Kambe_cp66 | PC-3 | 2.00 | LDD0130 | [23] |
| LDCM0022 | KB02 | HCT 116 | C29(3.44) | LDD0080 | [12] |
| LDCM0023 | KB03 | HCT 116 | C29(1.29) | LDD0081 | [12] |
| LDCM0024 | KB05 | HCT 116 | C29(1.42) | LDD0082 | [12] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C29(1.03) | LDD2101 | [6] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C29(0.88) | LDD2104 | [6] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C29(0.97) | LDD2107 | [6] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C29(0.94) | LDD2114 | [6] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C29(0.39) | LDD2115 | [6] |
| LDCM0525 | Nucleophilic fragment 25b | MDA-MB-231 | C29(1.92) | LDD2118 | [6] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C29(1.43) | LDD2119 | [6] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C29(0.97) | LDD2123 | [6] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C29(0.72) | LDD2129 | [6] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C29(0.62) | LDD2134 | [6] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C29(0.86) | LDD2135 | [6] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C29(0.88) | LDD2137 | [6] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C29(0.87) | LDD2141 | [6] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C29(1.30) | LDD2143 | [6] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C29(1.64) | LDD2144 | [6] |
| LDCM0029 | Quercetin | HEK-293T | 9.46 | LDD0181 | [7] |
| LDCM0131 | RA190 | MM1.R | C29(1.21) | LDD0304 | [26] |
| LDCM0016 | Ranjitkar_cp1 | A431 | 2.10 | LDD0074 | [22] |
| LDCM0021 | THZ1 | HCT 116 | C29(1.17) | LDD2173 | [12] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Peptidyl-prolyl cis-trans isomerase FKBP2 (FKBP2) | FKBP-type PPIase family | P26885 | |||
| E3 ubiquitin-protein ligase TRIM32 (TRIM32) | TRIM/RBCC family | Q13049 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Ubiquilin-4 (UBQLN4) | . | Q9NRR5 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Transcriptional repressor RHIT (ZNF205) | Krueppel C2H2-type zinc-finger protein family | O95201 | |||
| Protein AF-17 (MLLT6) | . | P55198 | |||
Other
References
