Details of the Target
General Information of Target
| Target ID | LDTP03618 | |||||
|---|---|---|---|---|---|---|
| Target Name | 14-3-3 protein sigma (SFN) | |||||
| Gene Name | SFN | |||||
| Gene ID | 2810 | |||||
| Synonyms |
HME1; 14-3-3 protein sigma; Epithelial cell marker protein 1; Stratifin |
|||||
| 3D Structure | ||||||
| Sequence |
MERASLIQKAKLAEQAERYEDMAAFMKGAVEKGEELSCEERNLLSVAYKNVVGGQRAAWR
VLSSIEQKSNEEGSEEKGPEVREYREKVETELQGVCDTVLGLLDSHLIKEAGDAESRVFY LKMKGDYYRYLAEVATGDDKKRIIDSARSAYQEAMDISKKEMPPTNPIRLGLALNFSVFH YEIANSPEEAISLAKTTFDEAMADLHTLSEDSYKDSTLIMQLLRDNLTLWTADNAGEEGG EAPQEPQS |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
14-3-3 family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Adapter protein implicated in the regulation of a large spectrum of both general and specialized signaling pathways. Binds to a large number of partners, usually by recognition of a phosphoserine or phosphothreonine motif. Binding generally results in the modulation of the activity of the binding partner. Promotes cytosolic retention of GBP1 GTPase by binding to phosphorylated GBP1, thereby inhibiting the innate immune response. Also acts as a TP53/p53-regulated inhibitor of G2/M progression. When bound to KRT17, regulates protein synthesis and epithelial cell growth by stimulating Akt/mTOR pathway. May also regulate MDM2 autoubiquitination and degradation and thereby activate p53/TP53.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| CHL1 | SNV: p.A135T; p.G137D | . | |||
| Ishikawa (Heraklio) 02 ER | SNV: p.Y181H | DBIA Probe Info | |||
| MOLT4 | SNV: p.R82C; p.D126N | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
P1 Probe Info |
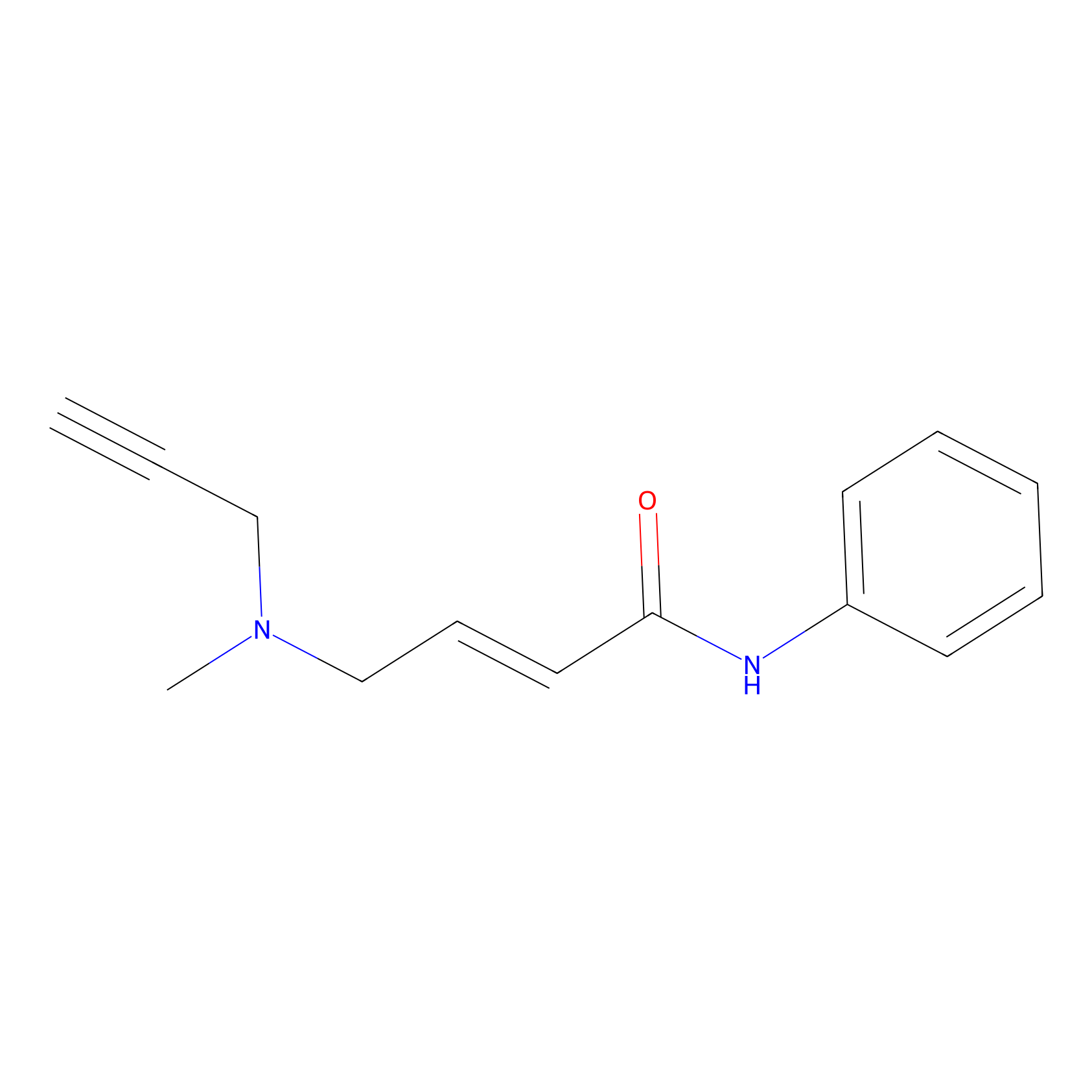 |
4.62 | LDD0448 | [1] | |
|
P2 Probe Info |
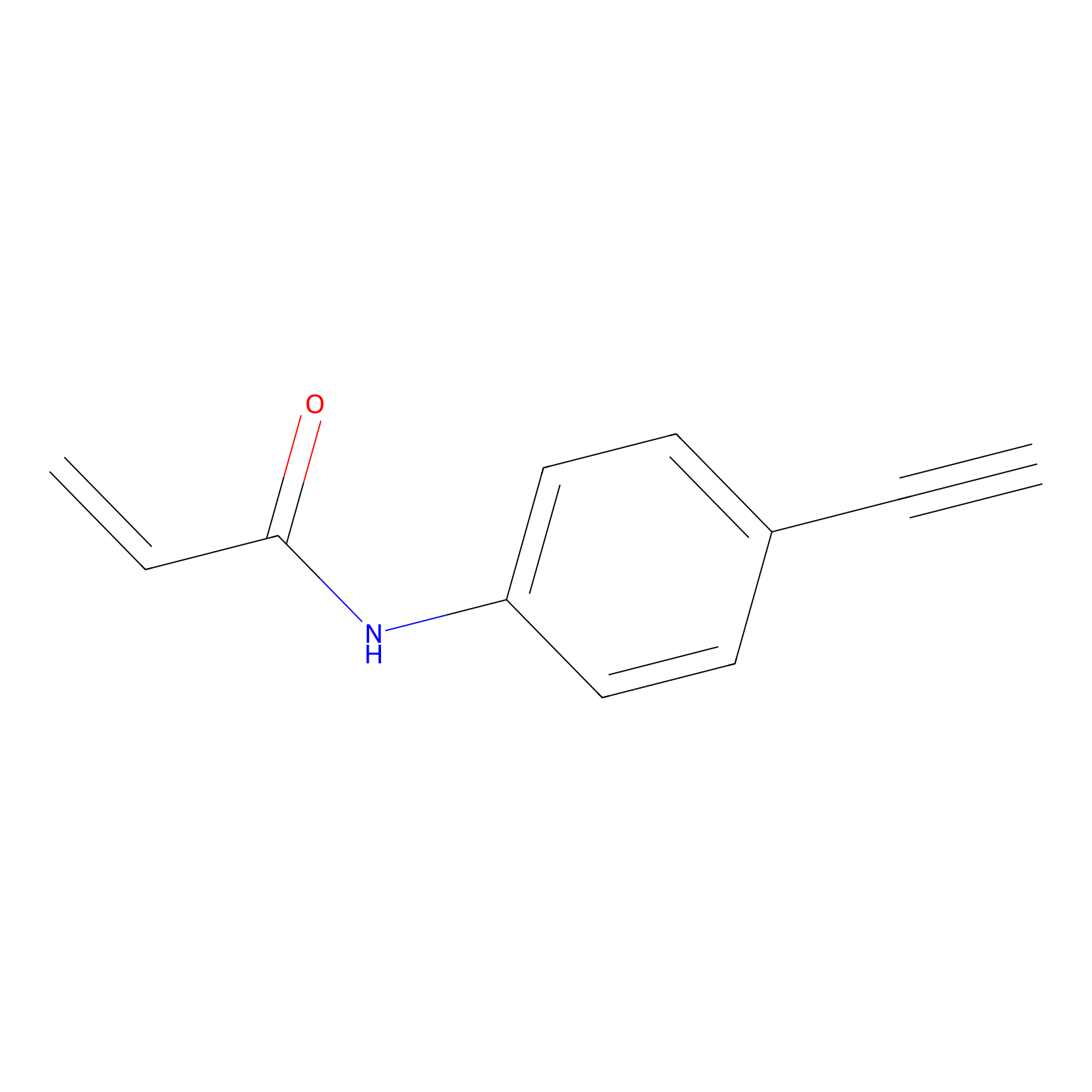 |
8.09 | LDD0449 | [1] | |
|
P3 Probe Info |
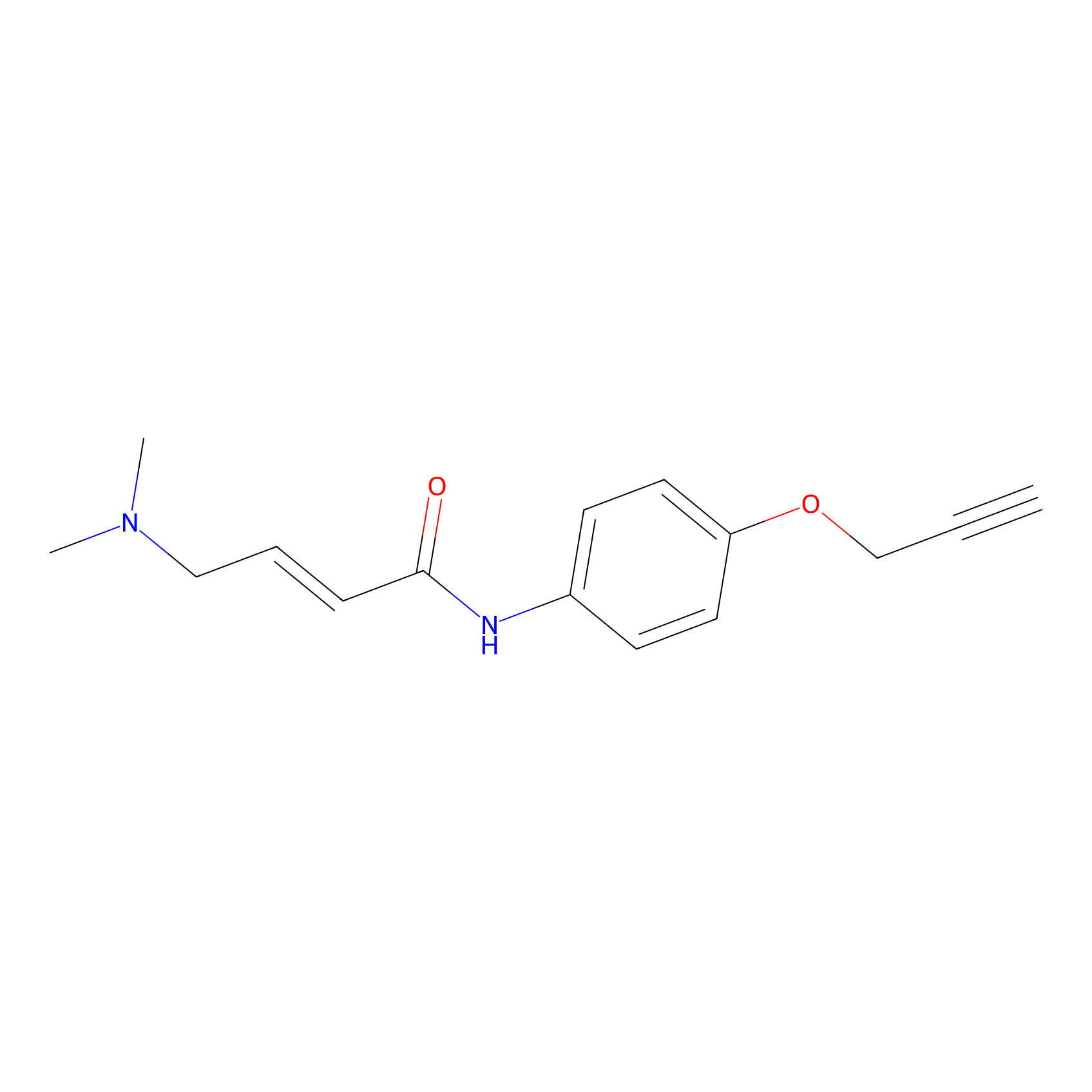 |
10.00 | LDD0450 | [1] | |
|
P8 Probe Info |
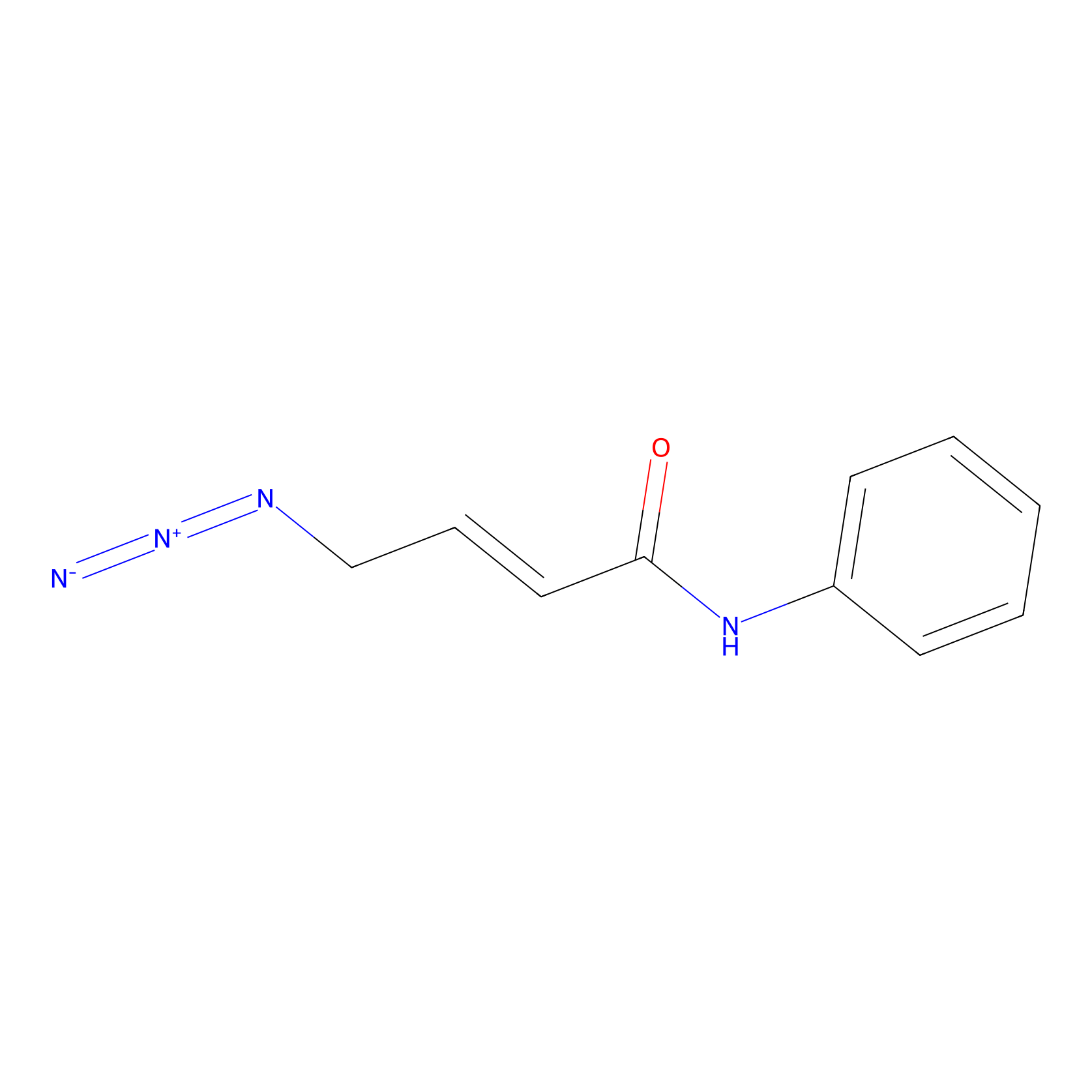 |
5.48 | LDD0451 | [1] | |
|
AZ-9 Probe Info |
 |
2.30 | LDD0393 | [2] | |
|
CY-1 Probe Info |
 |
4.50 | LDD0243 | [3] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [4] | |
|
ONAyne Probe Info |
 |
K32(5.00); K68(8.06); K9(1.31) | LDD0274 | [5] | |
|
STPyne Probe Info |
 |
K140(8.75); K141(0.34); K159(1.28); K49(5.61) | LDD0277 | [5] | |
|
BTD Probe Info |
 |
C38(8.91) | LDD1699 | [6] | |
|
OPA-S-S-alkyne Probe Info |
 |
K77(3.35); K124(4.42); K49(5.33) | LDD3494 | [7] | |
|
Probe 1 Probe Info |
 |
Y19(66.73); Y130(109.22); Y213(29.67) | LDD3495 | [8] | |
|
JZ128-DTB Probe Info |
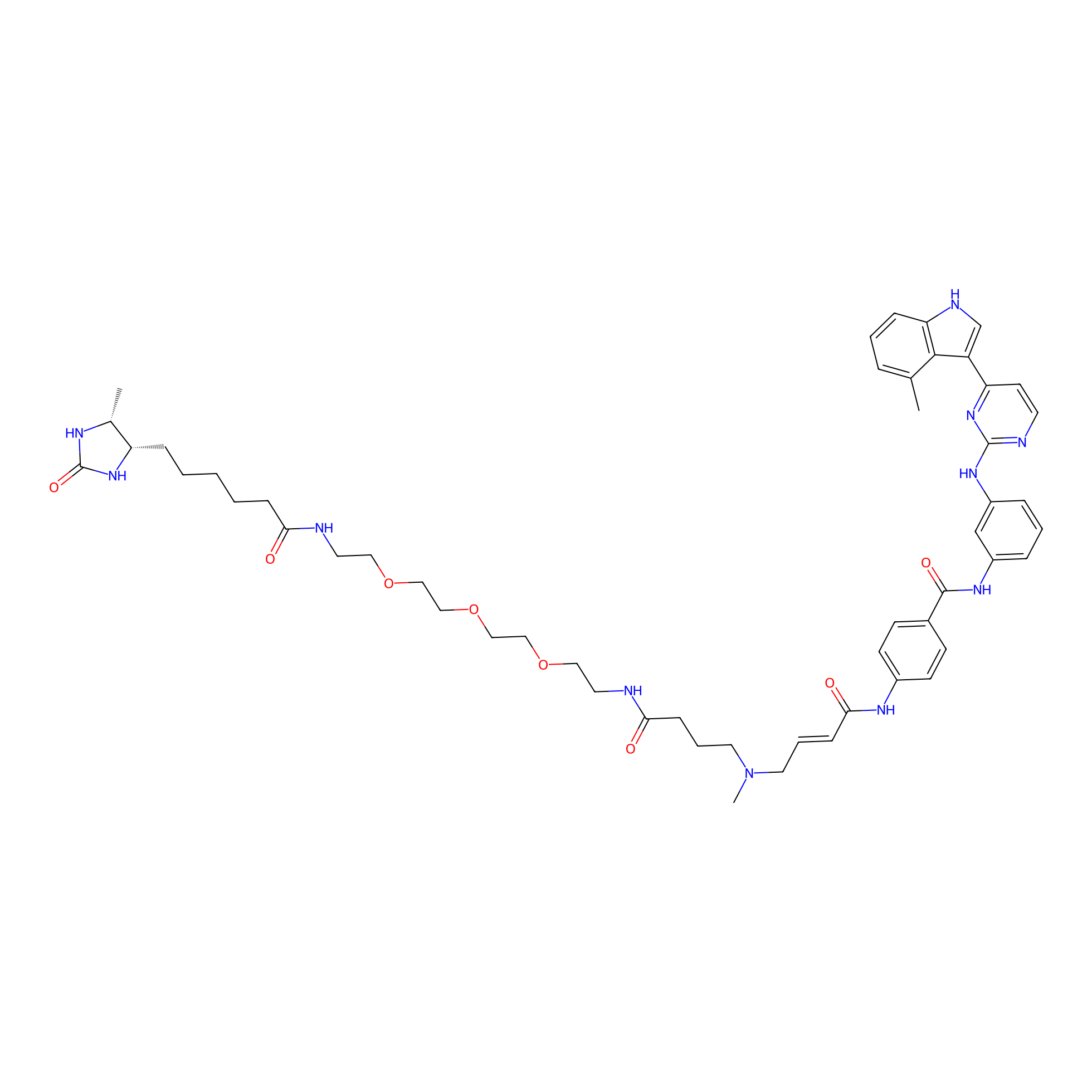 |
N.A. | LDD0462 | [9] | |
|
THZ1-DTB Probe Info |
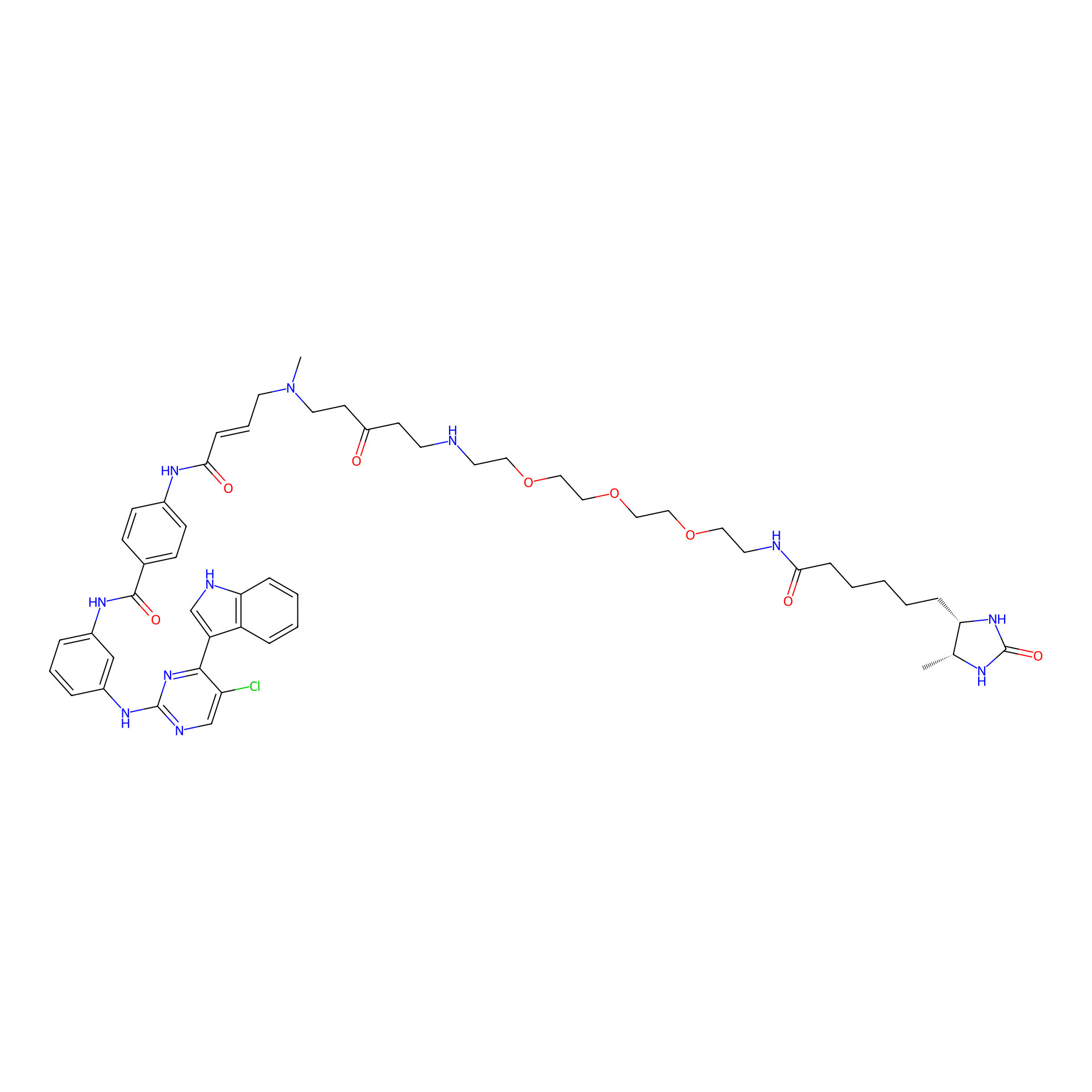 |
C38(1.04) | LDD0460 | [9] | |
|
AHL-Pu-1 Probe Info |
 |
C38(2.89) | LDD0169 | [10] | |
|
HHS-465 Probe Info |
 |
Y127(2.82); Y128(1.47); Y48(10.00) | LDD2237 | [11] | |
|
DBIA Probe Info |
 |
C38(0.98) | LDD0078 | [12] | |
|
m-APA Probe Info |
 |
H106(0.00); H206(0.00) | LDD2231 | [13] | |
|
ATP probe Probe Info |
 |
K11(0.00); K49(0.00); K124(0.00); K140(0.00) | LDD0035 | [14] | |
|
IPM Probe Info |
 |
N.A. | LDD0025 | [15] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [15] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [16] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [15] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0149 | [17] | |
|
PF-06672131 Probe Info |
 |
N.A. | LDD0017 | [18] | |
|
Acrolein Probe Info |
 |
C38(0.00); H206(0.00) | LDD0217 | [19] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [19] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [19] | |
|
W1 Probe Info |
 |
N.A. | LDD0236 | [20] | |
|
MPP-AC Probe Info |
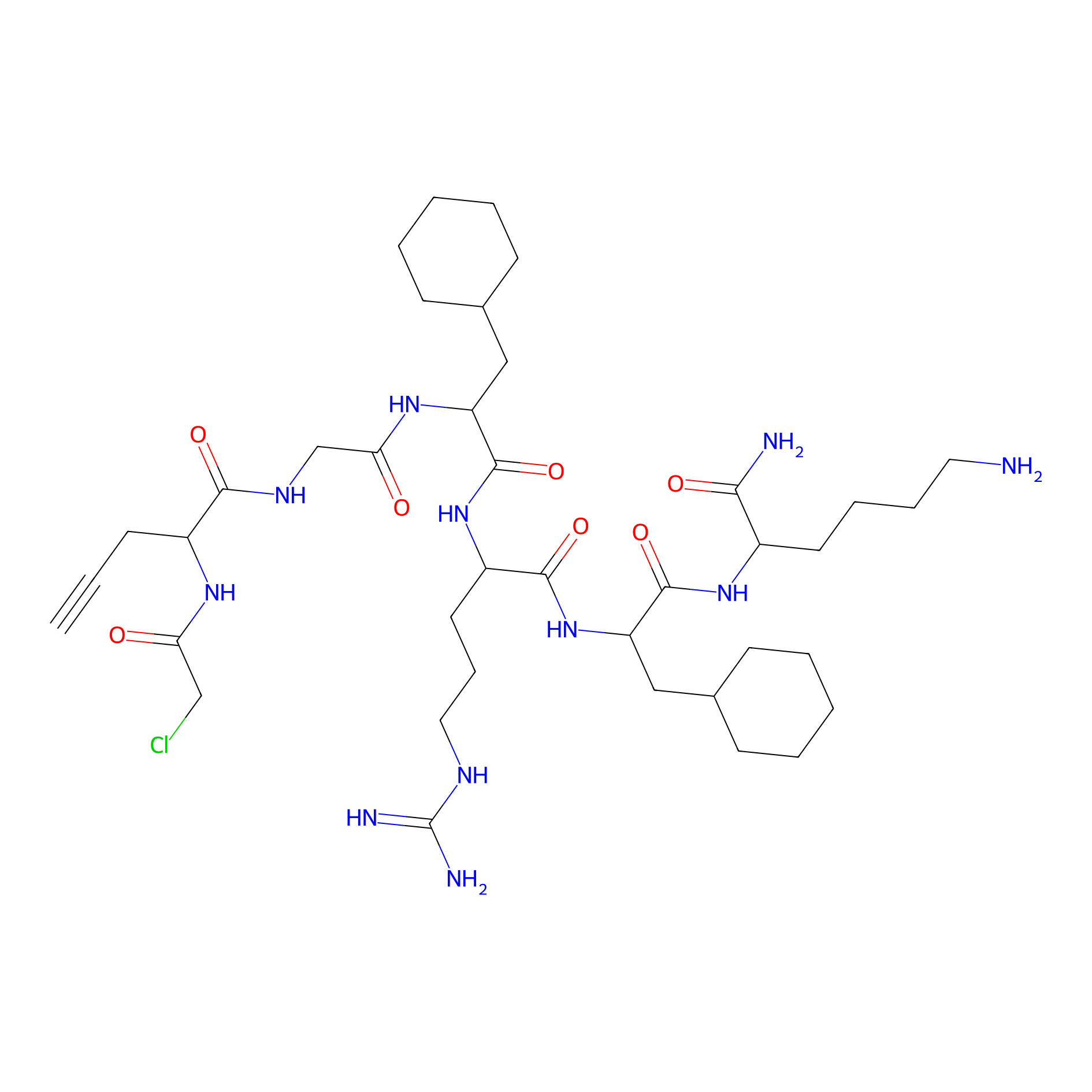 |
N.A. | LDD0428 | [21] | |
|
NAIA_5 Probe Info |
 |
C96(0.00); C38(0.00) | LDD2223 | [16] | |
|
HHS-475 Probe Info |
 |
Y213(1.54) | LDD2238 | [11] | |
|
HHS-482 Probe Info |
 |
Y213(1.11) | LDD2239 | [11] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
DA-2 Probe Info |
 |
N.A. | LDD0070 | [22] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C38(0.89) | LDD2142 | [6] |
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C38(1.20) | LDD2112 | [6] |
| LDCM0502 | 1-(Cyanoacetyl)piperidine | MDA-MB-231 | C38(1.09) | LDD2095 | [6] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C38(1.11) | LDD2117 | [6] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C38(1.08) | LDD2152 | [6] |
| LDCM0510 | 3-(4-(Hydroxydiphenylmethyl)piperidin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C38(0.98) | LDD2103 | [6] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C38(2.89) | LDD0169 | [10] |
| LDCM0214 | AC1 | HCT 116 | C38(1.27) | LDD0531 | [12] |
| LDCM0215 | AC10 | HCT 116 | C38(1.34) | LDD0532 | [12] |
| LDCM0216 | AC100 | HCT 116 | C38(1.23) | LDD0533 | [12] |
| LDCM0217 | AC101 | HCT 116 | C38(1.14) | LDD0534 | [12] |
| LDCM0218 | AC102 | HCT 116 | C38(1.49) | LDD0535 | [12] |
| LDCM0219 | AC103 | HCT 116 | C38(1.28) | LDD0536 | [12] |
| LDCM0220 | AC104 | HCT 116 | C38(1.46) | LDD0537 | [12] |
| LDCM0221 | AC105 | HCT 116 | C38(1.41) | LDD0538 | [12] |
| LDCM0222 | AC106 | HCT 116 | C38(1.24) | LDD0539 | [12] |
| LDCM0223 | AC107 | HCT 116 | C38(1.48) | LDD0540 | [12] |
| LDCM0224 | AC108 | HCT 116 | C38(1.18) | LDD0541 | [12] |
| LDCM0225 | AC109 | HCT 116 | C38(0.88) | LDD0542 | [12] |
| LDCM0226 | AC11 | HCT 116 | C38(1.37) | LDD0543 | [12] |
| LDCM0227 | AC110 | HCT 116 | C38(1.20) | LDD0544 | [12] |
| LDCM0228 | AC111 | HCT 116 | C38(1.12) | LDD0545 | [12] |
| LDCM0229 | AC112 | HCT 116 | C38(1.33) | LDD0546 | [12] |
| LDCM0230 | AC113 | HCT 116 | C38(1.03) | LDD0547 | [12] |
| LDCM0231 | AC114 | HCT 116 | C38(1.12) | LDD0548 | [12] |
| LDCM0232 | AC115 | HCT 116 | C38(1.22) | LDD0549 | [12] |
| LDCM0233 | AC116 | HCT 116 | C38(1.29) | LDD0550 | [12] |
| LDCM0234 | AC117 | HCT 116 | C38(1.26) | LDD0551 | [12] |
| LDCM0235 | AC118 | HCT 116 | C38(1.19) | LDD0552 | [12] |
| LDCM0236 | AC119 | HCT 116 | C38(1.43) | LDD0553 | [12] |
| LDCM0237 | AC12 | HCT 116 | C38(1.23) | LDD0554 | [12] |
| LDCM0238 | AC120 | HCT 116 | C38(1.31) | LDD0555 | [12] |
| LDCM0239 | AC121 | HCT 116 | C38(1.12) | LDD0556 | [12] |
| LDCM0240 | AC122 | HCT 116 | C38(1.25) | LDD0557 | [12] |
| LDCM0241 | AC123 | HCT 116 | C38(1.02) | LDD0558 | [12] |
| LDCM0242 | AC124 | HCT 116 | C38(1.00) | LDD0559 | [12] |
| LDCM0243 | AC125 | HCT 116 | C38(1.16) | LDD0560 | [12] |
| LDCM0244 | AC126 | HCT 116 | C38(1.16) | LDD0561 | [12] |
| LDCM0245 | AC127 | HCT 116 | C38(1.16) | LDD0562 | [12] |
| LDCM0246 | AC128 | HCT 116 | C38(1.22) | LDD0563 | [12] |
| LDCM0247 | AC129 | HCT 116 | C38(1.15) | LDD0564 | [12] |
| LDCM0249 | AC130 | HCT 116 | C38(1.25) | LDD0566 | [12] |
| LDCM0250 | AC131 | HCT 116 | C38(0.92) | LDD0567 | [12] |
| LDCM0251 | AC132 | HCT 116 | C38(1.14) | LDD0568 | [12] |
| LDCM0252 | AC133 | HCT 116 | C38(1.22) | LDD0569 | [12] |
| LDCM0253 | AC134 | HCT 116 | C38(1.25) | LDD0570 | [12] |
| LDCM0254 | AC135 | HCT 116 | C38(1.15) | LDD0571 | [12] |
| LDCM0255 | AC136 | HCT 116 | C38(1.19) | LDD0572 | [12] |
| LDCM0256 | AC137 | HCT 116 | C38(1.40) | LDD0573 | [12] |
| LDCM0257 | AC138 | HCT 116 | C38(1.57) | LDD0574 | [12] |
| LDCM0258 | AC139 | HCT 116 | C38(1.36) | LDD0575 | [12] |
| LDCM0259 | AC14 | HCT 116 | C38(1.15) | LDD0576 | [12] |
| LDCM0260 | AC140 | HCT 116 | C38(1.30) | LDD0577 | [12] |
| LDCM0261 | AC141 | HCT 116 | C38(1.46) | LDD0578 | [12] |
| LDCM0262 | AC142 | HCT 116 | C38(1.13) | LDD0579 | [12] |
| LDCM0263 | AC143 | HCT 116 | C38(1.06) | LDD0580 | [12] |
| LDCM0264 | AC144 | HCT 116 | C38(1.02) | LDD0581 | [12] |
| LDCM0265 | AC145 | HCT 116 | C38(0.95) | LDD0582 | [12] |
| LDCM0266 | AC146 | HCT 116 | C38(1.38) | LDD0583 | [12] |
| LDCM0267 | AC147 | HCT 116 | C38(1.11) | LDD0584 | [12] |
| LDCM0268 | AC148 | HCT 116 | C38(1.82) | LDD0585 | [12] |
| LDCM0269 | AC149 | HCT 116 | C38(1.39) | LDD0586 | [12] |
| LDCM0270 | AC15 | HCT 116 | C38(1.16) | LDD0587 | [12] |
| LDCM0271 | AC150 | HCT 116 | C38(1.17) | LDD0588 | [12] |
| LDCM0272 | AC151 | HCT 116 | C38(1.06) | LDD0589 | [12] |
| LDCM0273 | AC152 | HCT 116 | C38(1.12) | LDD0590 | [12] |
| LDCM0274 | AC153 | HCT 116 | C38(1.77) | LDD0591 | [12] |
| LDCM0621 | AC154 | HCT 116 | C38(1.04) | LDD2158 | [12] |
| LDCM0622 | AC155 | HCT 116 | C38(1.03) | LDD2159 | [12] |
| LDCM0623 | AC156 | HCT 116 | C38(1.17) | LDD2160 | [12] |
| LDCM0624 | AC157 | HCT 116 | C38(0.90) | LDD2161 | [12] |
| LDCM0276 | AC17 | HCT 116 | C38(1.24) | LDD0593 | [12] |
| LDCM0277 | AC18 | HCT 116 | C38(1.28) | LDD0594 | [12] |
| LDCM0278 | AC19 | HCT 116 | C38(1.21) | LDD0595 | [12] |
| LDCM0279 | AC2 | HCT 116 | C38(1.06) | LDD0596 | [12] |
| LDCM0280 | AC20 | HCT 116 | C38(1.39) | LDD0597 | [12] |
| LDCM0281 | AC21 | HCT 116 | C38(1.49) | LDD0598 | [12] |
| LDCM0282 | AC22 | HCT 116 | C38(1.23) | LDD0599 | [12] |
| LDCM0283 | AC23 | HCT 116 | C38(1.36) | LDD0600 | [12] |
| LDCM0284 | AC24 | HCT 116 | C38(1.20) | LDD0601 | [12] |
| LDCM0285 | AC25 | HCT 116 | C38(1.08) | LDD0602 | [12] |
| LDCM0286 | AC26 | HCT 116 | C38(1.29) | LDD0603 | [12] |
| LDCM0287 | AC27 | HCT 116 | C38(1.21) | LDD0604 | [12] |
| LDCM0288 | AC28 | HCT 116 | C38(1.28) | LDD0605 | [12] |
| LDCM0289 | AC29 | HCT 116 | C38(1.18) | LDD0606 | [12] |
| LDCM0290 | AC3 | HCT 116 | C38(1.10) | LDD0607 | [12] |
| LDCM0291 | AC30 | HCT 116 | C38(1.27) | LDD0608 | [12] |
| LDCM0292 | AC31 | HCT 116 | C38(1.17) | LDD0609 | [12] |
| LDCM0293 | AC32 | HCT 116 | C38(1.57) | LDD0610 | [12] |
| LDCM0294 | AC33 | HCT 116 | C38(1.32) | LDD0611 | [12] |
| LDCM0295 | AC34 | HCT 116 | C38(1.52) | LDD0612 | [12] |
| LDCM0296 | AC35 | HCT 116 | C38(0.97) | LDD0613 | [12] |
| LDCM0297 | AC36 | HCT 116 | C38(1.01) | LDD0614 | [12] |
| LDCM0298 | AC37 | HCT 116 | C38(1.04) | LDD0615 | [12] |
| LDCM0299 | AC38 | HCT 116 | C38(1.07) | LDD0616 | [12] |
| LDCM0300 | AC39 | HCT 116 | C38(1.29) | LDD0617 | [12] |
| LDCM0301 | AC4 | HCT 116 | C38(1.19) | LDD0618 | [12] |
| LDCM0302 | AC40 | HCT 116 | C38(1.39) | LDD0619 | [12] |
| LDCM0303 | AC41 | HCT 116 | C38(1.25) | LDD0620 | [12] |
| LDCM0304 | AC42 | HCT 116 | C38(1.15) | LDD0621 | [12] |
| LDCM0305 | AC43 | HCT 116 | C38(1.26) | LDD0622 | [12] |
| LDCM0306 | AC44 | HCT 116 | C38(1.23) | LDD0623 | [12] |
| LDCM0307 | AC45 | HCT 116 | C38(1.34) | LDD0624 | [12] |
| LDCM0308 | AC46 | HCT 116 | C38(1.18) | LDD0625 | [12] |
| LDCM0309 | AC47 | HCT 116 | C38(1.11) | LDD0626 | [12] |
| LDCM0310 | AC48 | HCT 116 | C38(1.38) | LDD0627 | [12] |
| LDCM0311 | AC49 | HCT 116 | C38(1.67) | LDD0628 | [12] |
| LDCM0312 | AC5 | HCT 116 | C38(1.30) | LDD0629 | [12] |
| LDCM0313 | AC50 | HCT 116 | C38(1.44) | LDD0630 | [12] |
| LDCM0314 | AC51 | HCT 116 | C38(1.09) | LDD0631 | [12] |
| LDCM0315 | AC52 | HCT 116 | C38(1.18) | LDD0632 | [12] |
| LDCM0316 | AC53 | HCT 116 | C38(1.20) | LDD0633 | [12] |
| LDCM0317 | AC54 | HCT 116 | C38(1.34) | LDD0634 | [12] |
| LDCM0318 | AC55 | HCT 116 | C38(1.50) | LDD0635 | [12] |
| LDCM0319 | AC56 | HCT 116 | C38(2.40) | LDD0636 | [12] |
| LDCM0320 | AC57 | HCT 116 | C38(1.51) | LDD0637 | [12] |
| LDCM0321 | AC58 | HCT 116 | C38(1.68) | LDD0638 | [12] |
| LDCM0322 | AC59 | HCT 116 | C38(1.47) | LDD0639 | [12] |
| LDCM0323 | AC6 | HCT 116 | C38(1.12) | LDD0640 | [12] |
| LDCM0324 | AC60 | HCT 116 | C38(1.55) | LDD0641 | [12] |
| LDCM0325 | AC61 | HCT 116 | C38(1.40) | LDD0642 | [12] |
| LDCM0326 | AC62 | HCT 116 | C38(1.63) | LDD0643 | [12] |
| LDCM0327 | AC63 | HCT 116 | C38(1.51) | LDD0644 | [12] |
| LDCM0328 | AC64 | HCT 116 | C38(1.66) | LDD0645 | [12] |
| LDCM0329 | AC65 | HCT 116 | C38(1.68) | LDD0646 | [12] |
| LDCM0330 | AC66 | HCT 116 | C38(1.86) | LDD0647 | [12] |
| LDCM0331 | AC67 | HCT 116 | C38(2.18) | LDD0648 | [12] |
| LDCM0332 | AC68 | HCT 116 | C38(1.10) | LDD0649 | [12] |
| LDCM0333 | AC69 | HCT 116 | C38(1.38) | LDD0650 | [12] |
| LDCM0334 | AC7 | HCT 116 | C38(1.20) | LDD0651 | [12] |
| LDCM0335 | AC70 | HCT 116 | C38(1.31) | LDD0652 | [12] |
| LDCM0336 | AC71 | HCT 116 | C38(1.12) | LDD0653 | [12] |
| LDCM0337 | AC72 | HCT 116 | C38(1.14) | LDD0654 | [12] |
| LDCM0338 | AC73 | HCT 116 | C38(1.64) | LDD0655 | [12] |
| LDCM0339 | AC74 | HCT 116 | C38(1.55) | LDD0656 | [12] |
| LDCM0340 | AC75 | HCT 116 | C38(1.94) | LDD0657 | [12] |
| LDCM0341 | AC76 | HCT 116 | C38(1.35) | LDD0658 | [12] |
| LDCM0342 | AC77 | HCT 116 | C38(1.25) | LDD0659 | [12] |
| LDCM0343 | AC78 | HCT 116 | C38(1.22) | LDD0660 | [12] |
| LDCM0344 | AC79 | HCT 116 | C38(1.06) | LDD0661 | [12] |
| LDCM0345 | AC8 | HCT 116 | C38(1.33) | LDD0662 | [12] |
| LDCM0346 | AC80 | HCT 116 | C38(1.02) | LDD0663 | [12] |
| LDCM0347 | AC81 | HCT 116 | C38(1.30) | LDD0664 | [12] |
| LDCM0348 | AC82 | HCT 116 | C38(2.07) | LDD0665 | [12] |
| LDCM0349 | AC83 | HCT 116 | C38(1.61) | LDD0666 | [12] |
| LDCM0350 | AC84 | HCT 116 | C38(1.97) | LDD0667 | [12] |
| LDCM0351 | AC85 | HCT 116 | C38(1.68) | LDD0668 | [12] |
| LDCM0352 | AC86 | HCT 116 | C38(1.10) | LDD0669 | [12] |
| LDCM0353 | AC87 | HCT 116 | C38(1.09) | LDD0670 | [12] |
| LDCM0354 | AC88 | HCT 116 | C38(1.16) | LDD0671 | [12] |
| LDCM0355 | AC89 | HCT 116 | C38(1.38) | LDD0672 | [12] |
| LDCM0357 | AC90 | HCT 116 | C38(1.06) | LDD0674 | [12] |
| LDCM0358 | AC91 | HCT 116 | C38(2.24) | LDD0675 | [12] |
| LDCM0359 | AC92 | HCT 116 | C38(1.44) | LDD0676 | [12] |
| LDCM0360 | AC93 | HCT 116 | C38(1.37) | LDD0677 | [12] |
| LDCM0361 | AC94 | HCT 116 | C38(1.09) | LDD0678 | [12] |
| LDCM0362 | AC95 | HCT 116 | C38(1.18) | LDD0679 | [12] |
| LDCM0363 | AC96 | HCT 116 | C38(1.42) | LDD0680 | [12] |
| LDCM0364 | AC97 | HCT 116 | C38(1.44) | LDD0681 | [12] |
| LDCM0365 | AC98 | HCT 116 | C38(2.24) | LDD0682 | [12] |
| LDCM0366 | AC99 | HCT 116 | C38(1.25) | LDD0683 | [12] |
| LDCM0248 | AKOS034007472 | HCT 116 | C38(0.97) | LDD0565 | [12] |
| LDCM0356 | AKOS034007680 | HCT 116 | C38(1.08) | LDD0673 | [12] |
| LDCM0275 | AKOS034007705 | HCT 116 | C38(1.74) | LDD0592 | [12] |
| LDCM0020 | ARS-1620 | HCC44 | C38(0.98) | LDD0078 | [12] |
| LDCM0630 | CCW28-3 | 231MFP | C38(1.16); C96(1.12) | LDD2214 | [23] |
| LDCM0108 | Chloroacetamide | HeLa | C38(0.00); H206(0.00) | LDD0222 | [19] |
| LDCM0632 | CL-Sc | Hep-G2 | C38(1.07); C38(1.05); C38(0.36) | LDD2227 | [16] |
| LDCM0367 | CL1 | HCT 116 | C38(1.01) | LDD0684 | [12] |
| LDCM0368 | CL10 | HCT 116 | C38(1.18) | LDD0685 | [12] |
| LDCM0369 | CL100 | HCT 116 | C38(1.24) | LDD0686 | [12] |
| LDCM0370 | CL101 | HCT 116 | C38(1.22) | LDD0687 | [12] |
| LDCM0371 | CL102 | HCT 116 | C38(1.13) | LDD0688 | [12] |
| LDCM0372 | CL103 | HCT 116 | C38(1.14) | LDD0689 | [12] |
| LDCM0373 | CL104 | HCT 116 | C38(1.05) | LDD0690 | [12] |
| LDCM0374 | CL105 | HCT 116 | C38(1.37) | LDD0691 | [12] |
| LDCM0375 | CL106 | HCT 116 | C38(1.33) | LDD0692 | [12] |
| LDCM0376 | CL107 | HCT 116 | C38(1.27) | LDD0693 | [12] |
| LDCM0377 | CL108 | HCT 116 | C38(1.10) | LDD0694 | [12] |
| LDCM0378 | CL109 | HCT 116 | C38(1.38) | LDD0695 | [12] |
| LDCM0379 | CL11 | HCT 116 | C38(1.37) | LDD0696 | [12] |
| LDCM0380 | CL110 | HCT 116 | C38(1.10) | LDD0697 | [12] |
| LDCM0381 | CL111 | HCT 116 | C38(1.35) | LDD0698 | [12] |
| LDCM0382 | CL112 | HCT 116 | C38(1.11) | LDD0699 | [12] |
| LDCM0383 | CL113 | HCT 116 | C38(1.42) | LDD0700 | [12] |
| LDCM0384 | CL114 | HCT 116 | C38(1.77) | LDD0701 | [12] |
| LDCM0385 | CL115 | HCT 116 | C38(1.23) | LDD0702 | [12] |
| LDCM0386 | CL116 | HCT 116 | C38(1.24) | LDD0703 | [12] |
| LDCM0387 | CL117 | HCT 116 | C38(1.83) | LDD0704 | [12] |
| LDCM0388 | CL118 | HCT 116 | C38(1.22) | LDD0705 | [12] |
| LDCM0389 | CL119 | HCT 116 | C38(1.29) | LDD0706 | [12] |
| LDCM0390 | CL12 | HCT 116 | C38(1.33) | LDD0707 | [12] |
| LDCM0391 | CL120 | HCT 116 | C38(1.24) | LDD0708 | [12] |
| LDCM0392 | CL121 | HCT 116 | C38(1.17) | LDD0709 | [12] |
| LDCM0393 | CL122 | HCT 116 | C38(1.23) | LDD0710 | [12] |
| LDCM0394 | CL123 | HCT 116 | C38(1.20) | LDD0711 | [12] |
| LDCM0395 | CL124 | HCT 116 | C38(1.66) | LDD0712 | [12] |
| LDCM0396 | CL125 | HCT 116 | C38(1.19) | LDD0713 | [12] |
| LDCM0397 | CL126 | HCT 116 | C38(1.37) | LDD0714 | [12] |
| LDCM0398 | CL127 | HCT 116 | C38(1.35) | LDD0715 | [12] |
| LDCM0399 | CL128 | HCT 116 | C38(1.41) | LDD0716 | [12] |
| LDCM0400 | CL13 | HCT 116 | C38(1.13) | LDD0717 | [12] |
| LDCM0401 | CL14 | HCT 116 | C38(0.97) | LDD0718 | [12] |
| LDCM0402 | CL15 | HCT 116 | C38(1.10) | LDD0719 | [12] |
| LDCM0403 | CL16 | HCT 116 | C38(1.09) | LDD0720 | [12] |
| LDCM0404 | CL17 | HCT 116 | C38(1.18) | LDD0721 | [12] |
| LDCM0405 | CL18 | HCT 116 | C38(1.16) | LDD0722 | [12] |
| LDCM0406 | CL19 | HCT 116 | C38(1.07) | LDD0723 | [12] |
| LDCM0407 | CL2 | HCT 116 | C38(0.95) | LDD0724 | [12] |
| LDCM0408 | CL20 | HCT 116 | C38(1.13) | LDD0725 | [12] |
| LDCM0409 | CL21 | HCT 116 | C38(1.37) | LDD0726 | [12] |
| LDCM0410 | CL22 | HCT 116 | C38(2.26) | LDD0727 | [12] |
| LDCM0411 | CL23 | HCT 116 | C38(1.18) | LDD0728 | [12] |
| LDCM0412 | CL24 | HCT 116 | C38(1.27) | LDD0729 | [12] |
| LDCM0413 | CL25 | HCT 116 | C38(1.52) | LDD0730 | [12] |
| LDCM0414 | CL26 | HCT 116 | C38(1.18) | LDD0731 | [12] |
| LDCM0415 | CL27 | HCT 116 | C38(1.07) | LDD0732 | [12] |
| LDCM0416 | CL28 | HCT 116 | C38(1.29) | LDD0733 | [12] |
| LDCM0417 | CL29 | HCT 116 | C38(1.23) | LDD0734 | [12] |
| LDCM0418 | CL3 | HCT 116 | C38(1.07) | LDD0735 | [12] |
| LDCM0419 | CL30 | HCT 116 | C38(1.05) | LDD0736 | [12] |
| LDCM0420 | CL31 | HCT 116 | C38(1.10) | LDD0737 | [12] |
| LDCM0421 | CL32 | HCT 116 | C38(1.37) | LDD0738 | [12] |
| LDCM0422 | CL33 | HCT 116 | C38(1.63) | LDD0739 | [12] |
| LDCM0423 | CL34 | HCT 116 | C38(0.78) | LDD0740 | [12] |
| LDCM0424 | CL35 | HCT 116 | C38(0.84) | LDD0741 | [12] |
| LDCM0425 | CL36 | HCT 116 | C38(0.75) | LDD0742 | [12] |
| LDCM0426 | CL37 | HCT 116 | C38(0.72) | LDD0743 | [12] |
| LDCM0428 | CL39 | HCT 116 | C38(0.79) | LDD0745 | [12] |
| LDCM0429 | CL4 | HCT 116 | C38(1.38) | LDD0746 | [12] |
| LDCM0430 | CL40 | HCT 116 | C38(0.80) | LDD0747 | [12] |
| LDCM0431 | CL41 | HCT 116 | C38(0.87) | LDD0748 | [12] |
| LDCM0432 | CL42 | HCT 116 | C38(0.90) | LDD0749 | [12] |
| LDCM0433 | CL43 | HCT 116 | C38(0.99) | LDD0750 | [12] |
| LDCM0434 | CL44 | HCT 116 | C38(0.83) | LDD0751 | [12] |
| LDCM0435 | CL45 | HCT 116 | C38(0.85) | LDD0752 | [12] |
| LDCM0436 | CL46 | HCT 116 | C38(1.20) | LDD0753 | [12] |
| LDCM0437 | CL47 | HCT 116 | C38(1.09) | LDD0754 | [12] |
| LDCM0438 | CL48 | HCT 116 | C38(1.21) | LDD0755 | [12] |
| LDCM0439 | CL49 | HCT 116 | C38(1.08) | LDD0756 | [12] |
| LDCM0440 | CL5 | HCT 116 | C38(0.96) | LDD0757 | [12] |
| LDCM0441 | CL50 | HCT 116 | C38(1.15) | LDD0758 | [12] |
| LDCM0442 | CL51 | HCT 116 | C38(1.24) | LDD0759 | [12] |
| LDCM0443 | CL52 | HCT 116 | C38(1.19) | LDD0760 | [12] |
| LDCM0444 | CL53 | HCT 116 | C38(1.04) | LDD0761 | [12] |
| LDCM0445 | CL54 | HCT 116 | C38(1.27) | LDD0762 | [12] |
| LDCM0446 | CL55 | HCT 116 | C38(1.08) | LDD0763 | [12] |
| LDCM0447 | CL56 | HCT 116 | C38(1.21) | LDD0764 | [12] |
| LDCM0448 | CL57 | HCT 116 | C38(0.95) | LDD0765 | [12] |
| LDCM0449 | CL58 | HCT 116 | C38(1.13) | LDD0766 | [12] |
| LDCM0450 | CL59 | HCT 116 | C38(1.06) | LDD0767 | [12] |
| LDCM0451 | CL6 | HCT 116 | C38(1.14) | LDD0768 | [12] |
| LDCM0452 | CL60 | HCT 116 | C38(1.09) | LDD0769 | [12] |
| LDCM0453 | CL61 | HCT 116 | C38(1.17) | LDD0770 | [12] |
| LDCM0454 | CL62 | HCT 116 | C38(1.30) | LDD0771 | [12] |
| LDCM0455 | CL63 | HCT 116 | C38(1.48) | LDD0772 | [12] |
| LDCM0456 | CL64 | HCT 116 | C38(1.38) | LDD0773 | [12] |
| LDCM0457 | CL65 | HCT 116 | C38(1.36) | LDD0774 | [12] |
| LDCM0458 | CL66 | HCT 116 | C38(1.79) | LDD0775 | [12] |
| LDCM0459 | CL67 | HCT 116 | C38(1.36) | LDD0776 | [12] |
| LDCM0460 | CL68 | HCT 116 | C38(1.26) | LDD0777 | [12] |
| LDCM0461 | CL69 | HCT 116 | C38(1.38) | LDD0778 | [12] |
| LDCM0462 | CL7 | HCT 116 | C38(1.27) | LDD0779 | [12] |
| LDCM0463 | CL70 | HCT 116 | C38(1.18) | LDD0780 | [12] |
| LDCM0464 | CL71 | HCT 116 | C38(1.31) | LDD0781 | [12] |
| LDCM0465 | CL72 | HCT 116 | C38(1.17) | LDD0782 | [12] |
| LDCM0466 | CL73 | HCT 116 | C38(1.18) | LDD0783 | [12] |
| LDCM0467 | CL74 | HCT 116 | C38(1.21) | LDD0784 | [12] |
| LDCM0469 | CL76 | HCT 116 | C38(1.22) | LDD0786 | [12] |
| LDCM0470 | CL77 | HCT 116 | C38(1.16) | LDD0787 | [12] |
| LDCM0471 | CL78 | HCT 116 | C38(1.28) | LDD0788 | [12] |
| LDCM0472 | CL79 | HCT 116 | C38(1.33) | LDD0789 | [12] |
| LDCM0473 | CL8 | HCT 116 | C38(1.65) | LDD0790 | [12] |
| LDCM0474 | CL80 | HCT 116 | C38(1.18) | LDD0791 | [12] |
| LDCM0475 | CL81 | HCT 116 | C38(1.23) | LDD0792 | [12] |
| LDCM0476 | CL82 | HCT 116 | C38(1.64) | LDD0793 | [12] |
| LDCM0477 | CL83 | HCT 116 | C38(1.63) | LDD0794 | [12] |
| LDCM0478 | CL84 | HCT 116 | C38(2.15) | LDD0795 | [12] |
| LDCM0479 | CL85 | HCT 116 | C38(1.33) | LDD0796 | [12] |
| LDCM0480 | CL86 | HCT 116 | C38(1.23) | LDD0797 | [12] |
| LDCM0481 | CL87 | HCT 116 | C38(1.31) | LDD0798 | [12] |
| LDCM0482 | CL88 | HCT 116 | C38(1.42) | LDD0799 | [12] |
| LDCM0483 | CL89 | HCT 116 | C38(2.64) | LDD0800 | [12] |
| LDCM0484 | CL9 | HCT 116 | C38(1.15) | LDD0801 | [12] |
| LDCM0485 | CL90 | HCT 116 | C38(1.38) | LDD0802 | [12] |
| LDCM0486 | CL91 | HCT 116 | C38(1.31) | LDD0803 | [12] |
| LDCM0487 | CL92 | HCT 116 | C38(1.24) | LDD0804 | [12] |
| LDCM0488 | CL93 | HCT 116 | C38(1.21) | LDD0805 | [12] |
| LDCM0489 | CL94 | HCT 116 | C38(1.30) | LDD0806 | [12] |
| LDCM0490 | CL95 | HCT 116 | C38(1.52) | LDD0807 | [12] |
| LDCM0491 | CL96 | HCT 116 | C38(1.20) | LDD0808 | [12] |
| LDCM0492 | CL97 | HCT 116 | C38(1.19) | LDD0809 | [12] |
| LDCM0493 | CL98 | HCT 116 | C38(1.25) | LDD0810 | [12] |
| LDCM0494 | CL99 | HCT 116 | C38(1.32) | LDD0811 | [12] |
| LDCM0634 | CY-0357 | Hep-G2 | C38(1.18); C38(0.95) | LDD2228 | [16] |
| LDCM0625 | F8 | Ramos | C38(1.32) | LDD2187 | [24] |
| LDCM0572 | Fragment10 | Ramos | C38(1.20) | LDD2189 | [24] |
| LDCM0574 | Fragment12 | Ramos | C38(0.63) | LDD2191 | [24] |
| LDCM0575 | Fragment13 | Ramos | C38(0.80) | LDD2192 | [24] |
| LDCM0576 | Fragment14 | Ramos | C38(0.81) | LDD2193 | [24] |
| LDCM0579 | Fragment20 | Ramos | C38(0.97) | LDD2194 | [24] |
| LDCM0580 | Fragment21 | Ramos | C38(0.78) | LDD2195 | [24] |
| LDCM0582 | Fragment23 | Ramos | C38(0.61) | LDD2196 | [24] |
| LDCM0578 | Fragment27 | Ramos | C38(0.66) | LDD2197 | [24] |
| LDCM0588 | Fragment30 | Ramos | C38(0.98) | LDD2199 | [24] |
| LDCM0589 | Fragment31 | Ramos | C38(0.54) | LDD2200 | [24] |
| LDCM0590 | Fragment32 | Ramos | C38(1.29) | LDD2201 | [24] |
| LDCM0468 | Fragment33 | HCT 116 | C38(1.27) | LDD0785 | [12] |
| LDCM0596 | Fragment38 | Ramos | C38(0.41) | LDD2203 | [24] |
| LDCM0566 | Fragment4 | Ramos | C38(0.59) | LDD2184 | [24] |
| LDCM0427 | Fragment51 | HCT 116 | C38(0.70) | LDD0744 | [12] |
| LDCM0610 | Fragment52 | Ramos | C38(0.84) | LDD2204 | [24] |
| LDCM0614 | Fragment56 | Ramos | C38(0.74) | LDD2205 | [24] |
| LDCM0569 | Fragment7 | Ramos | C38(0.80) | LDD2186 | [24] |
| LDCM0571 | Fragment9 | Ramos | C38(1.05) | LDD2188 | [24] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [19] |
| LDCM0179 | JZ128 | PC-3 | N.A. | LDD0462 | [9] |
| LDCM0022 | KB02 | Ramos | C38(1.82) | LDD2182 | [24] |
| LDCM0023 | KB03 | Jurkat | C38(4.16) | LDD0209 | [25] |
| LDCM0024 | KB05 | HMCB | C96(1.55) | LDD3312 | [26] |
| LDCM0509 | N-(4-bromo-3,5-dimethylphenyl)-2-nitroacetamide | MDA-MB-231 | C38(0.96) | LDD2102 | [6] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C38(0.89) | LDD2121 | [6] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [19] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C38(1.02) | LDD2089 | [6] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C38(1.23) | LDD2090 | [6] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C38(1.06) | LDD2092 | [6] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C38(1.04) | LDD2094 | [6] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C38(0.95) | LDD2098 | [6] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C38(1.03) | LDD2099 | [6] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C38(1.16) | LDD2100 | [6] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C38(0.97) | LDD2104 | [6] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C38(1.06) | LDD2105 | [6] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C38(0.87) | LDD2106 | [6] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C38(0.99) | LDD2108 | [6] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C38(1.08) | LDD2109 | [6] |
| LDCM0517 | Nucleophilic fragment 21b | MDA-MB-231 | C38(1.90) | LDD2110 | [6] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C38(0.65) | LDD2114 | [6] |
| LDCM0523 | Nucleophilic fragment 24b | MDA-MB-231 | C38(0.88) | LDD2116 | [6] |
| LDCM0525 | Nucleophilic fragment 25b | MDA-MB-231 | C38(0.19) | LDD2118 | [6] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C38(0.83) | LDD2120 | [6] |
| LDCM0531 | Nucleophilic fragment 28b | MDA-MB-231 | C38(1.20) | LDD2124 | [6] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C38(1.07) | LDD2125 | [6] |
| LDCM0533 | Nucleophilic fragment 29b | MDA-MB-231 | C38(0.89) | LDD2126 | [6] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C38(1.40) | LDD2128 | [6] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C38(0.89) | LDD2136 | [6] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C38(0.92) | LDD2137 | [6] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C38(0.87) | LDD2140 | [6] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C38(1.75) | LDD2141 | [6] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C38(1.04) | LDD2150 | [6] |
| LDCM0557 | Nucleophilic fragment 8b | MDA-MB-231 | C38(0.22) | LDD2151 | [6] |
| LDCM0559 | Nucleophilic fragment 9b | MDA-MB-231 | C38(1.72) | LDD2153 | [6] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C38(1.04) | LDD2206 | [27] |
| LDCM0131 | RA190 | MM1.R | C38(1.03) | LDD0304 | [28] |
| LDCM0021 | THZ1 | HeLa S3 | C38(1.04) | LDD0460 | [9] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| 14-3-3 protein gamma (YWHAG) | 14-3-3 family | P61981 | |||
| 14-3-3 protein zeta/delta (YWHAZ) | 14-3-3 family | P63104 | |||
| Cellular tumor antigen p53 (TP53) | P53 family | P04637 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Forkhead box protein O4 (FOXO4) | . | P98177 | |||
| Paired box protein Pax-9 (PAX9) | . | P55771 | |||
| Transcription factor HIVEP3 (HIVEP3) | . | Q5T1R4 | |||
Other
References
