Details of the Target
General Information of Target
| Target ID | LDTP07439 | |||||
|---|---|---|---|---|---|---|
| Target Name | Mitochondrial adenyl nucleotide antiporter SLC25A25 (SLC25A25) | |||||
| Gene Name | SLC25A25 | |||||
| Gene ID | 114789 | |||||
| Synonyms |
APC3; KIAA1896; MCSC3; SCAMC2; Mitochondrial adenyl nucleotide antiporter SLC25A25; Mitochondrial ATP-Mg/Pi carrier protein 3; Mitochondrial Ca(2+)-dependent solute carrier protein 3; Short calcium-binding mitochondrial carrier protein 2; SCaMC-2; Solute carrier family 25 member 25
|
|||||
| 3D Structure | ||||||
| Sequence |
MLCLCLYVPVIGEAQTEFQYFESKGLPAELKSIFKLSVFIPSQEFSTYRQWKQKIVQAGD
KDLDGQLDFEEFVHYLQDHEKKLRLVFKSLDKKNDGRIDAQEIMQSLRDLGVKISEQQAE KILKSMDKNGTMTIDWNEWRDYHLLHPVENIPEIILYWKHSTIFDVGENLTVPDEFTVEE RQTGMWWRHLVAGGGAGAVSRTCTAPLDRLKVLMQVHASRSNNMGIVGGFTQMIREGGAR SLWRGNGINVLKIAPESAIKFMAYEQIKRLVGSDQETLRIHERLVAGSLAGAIAQSSIYP MEVLKTRMALRKTGQYSGMLDCARRILAREGVAAFYKGYVPNMLGIIPYAGIDLAVYETL KNAWLQHYAVNSADPGVFVLLACGTMSSTCGQLASYPLALVRTRMQAQASIEGAPEVTMS SLFKHILRTEGAFGLYRGLAPNFMKVIPAVSISYVVYENLKITLGVQSR |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
Mitochondrial carrier (TC 2.A.29) family
|
|||||
| Subcellular location |
Mitochondrion inner membrane
|
|||||
| Function |
Electroneutral antiporter that most probably mediates the transport of adenyl nucleotides through the inner mitochondrial membrane. Originally identified as an ATP-magnesium/inorganic phosphate antiporter, it could have a broader specificity for adenyl nucleotides. By regulating the mitochondrial matrix adenyl nucleotide pool could adapt to changing cellular energetic demands and indirectly regulate adenyl nucleotide-dependent metabolic pathways.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
DBIA Probe Info |
 |
C249(3.63) | LDD3311 | [1] | |
|
Curcusone 37 Probe Info |
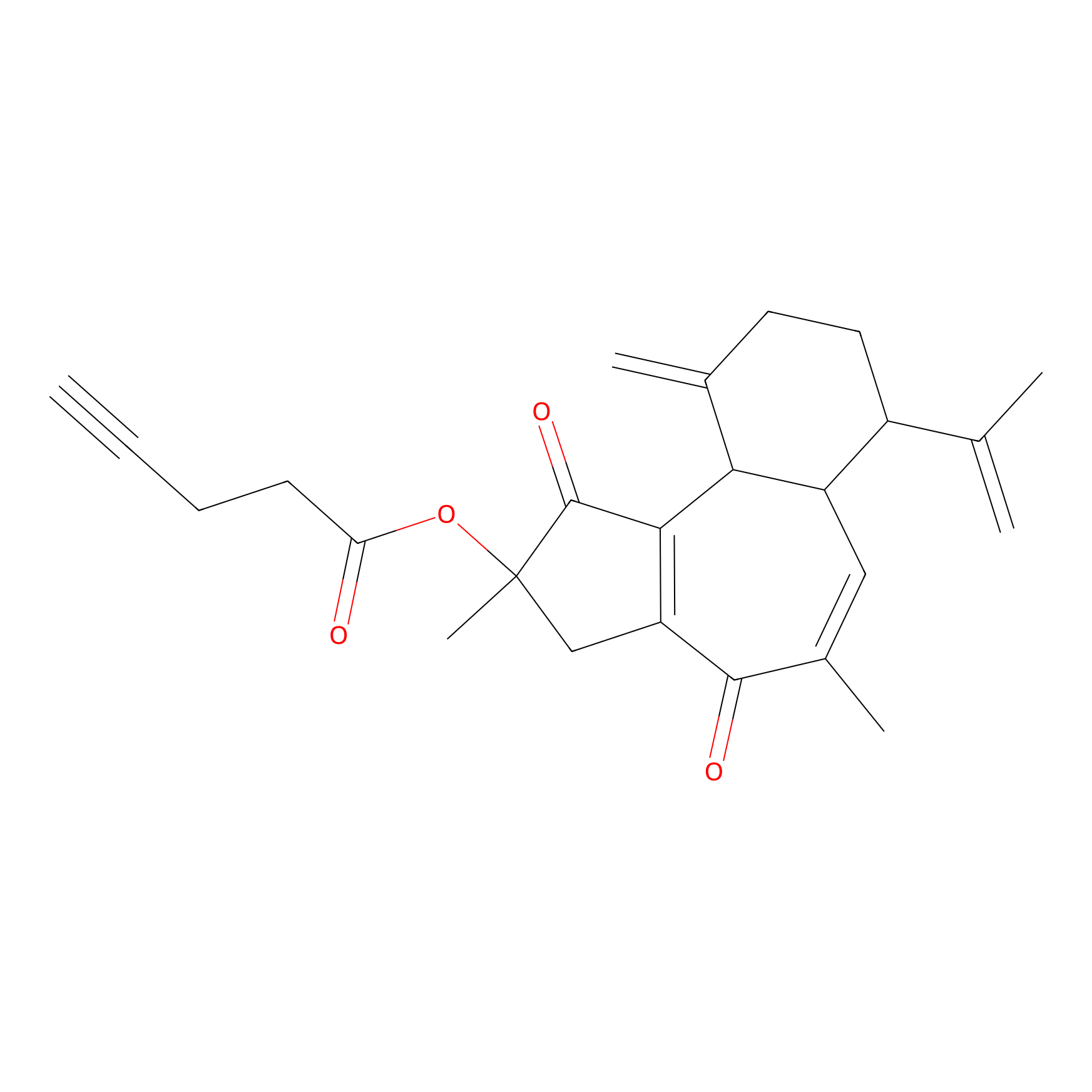 |
2.02 | LDD0188 | [2] | |
|
IA-alkyne Probe Info |
 |
C203(5.08) | LDD1706 | [3] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [4] | |
|
IPM Probe Info |
 |
C203(0.00); C322(0.00) | LDD2156 | [5] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C040 Probe Info |
 |
9.32 | LDD1740 | [6] | |
|
C055 Probe Info |
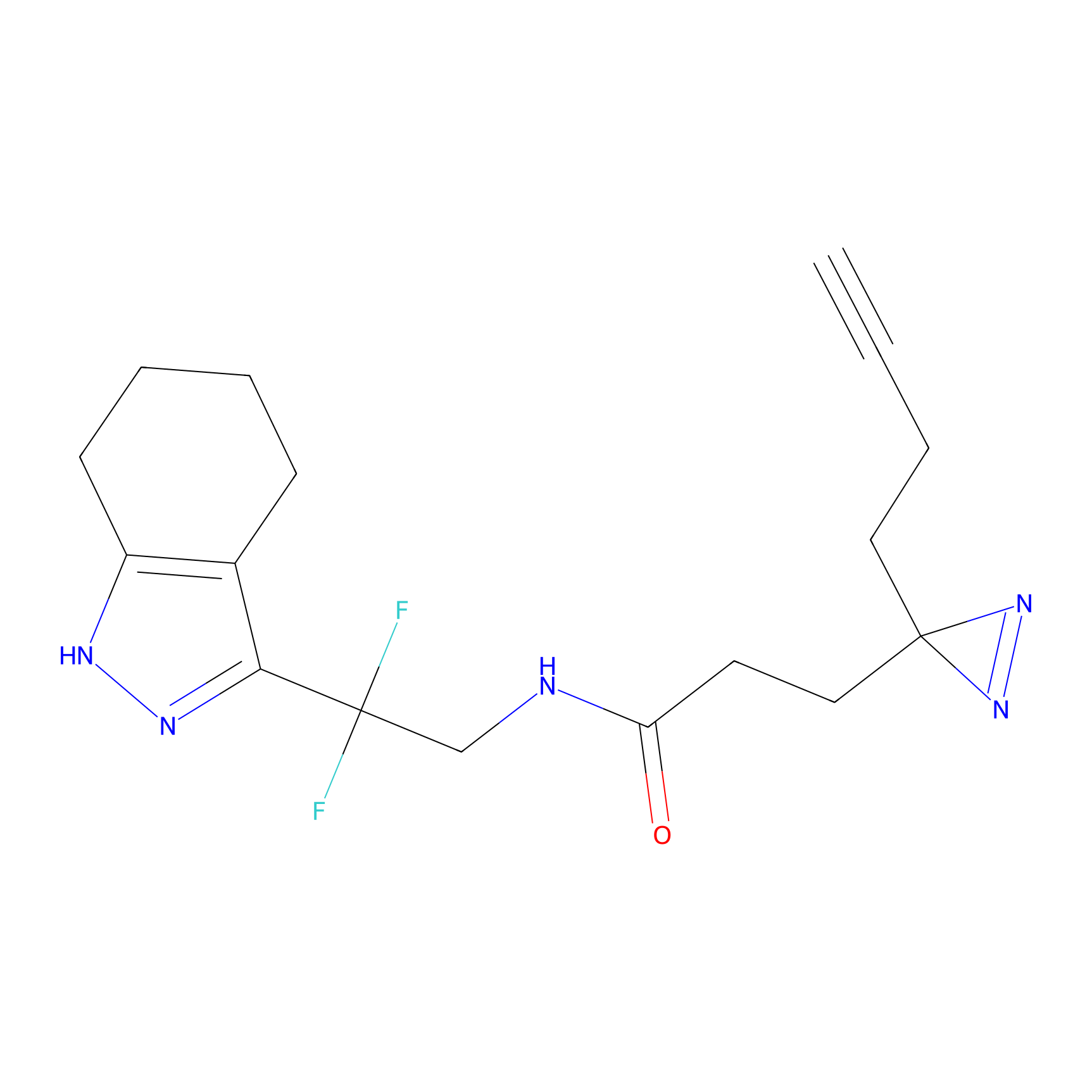 |
12.21 | LDD1752 | [6] | |
|
C056 Probe Info |
 |
30.27 | LDD1753 | [6] | |
|
C063 Probe Info |
 |
14.32 | LDD1760 | [6] | |
|
C070 Probe Info |
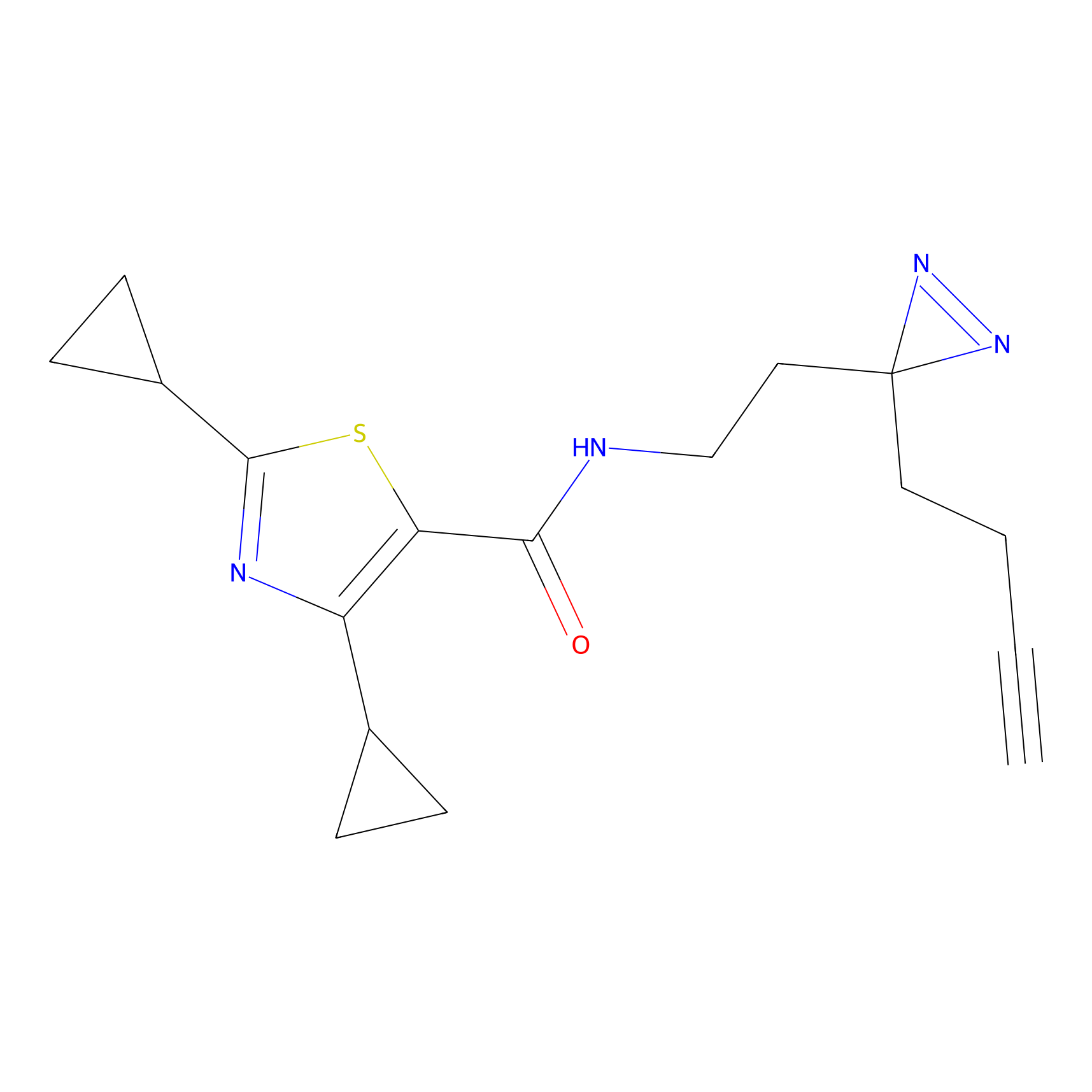 |
10.48 | LDD1766 | [6] | |
|
C072 Probe Info |
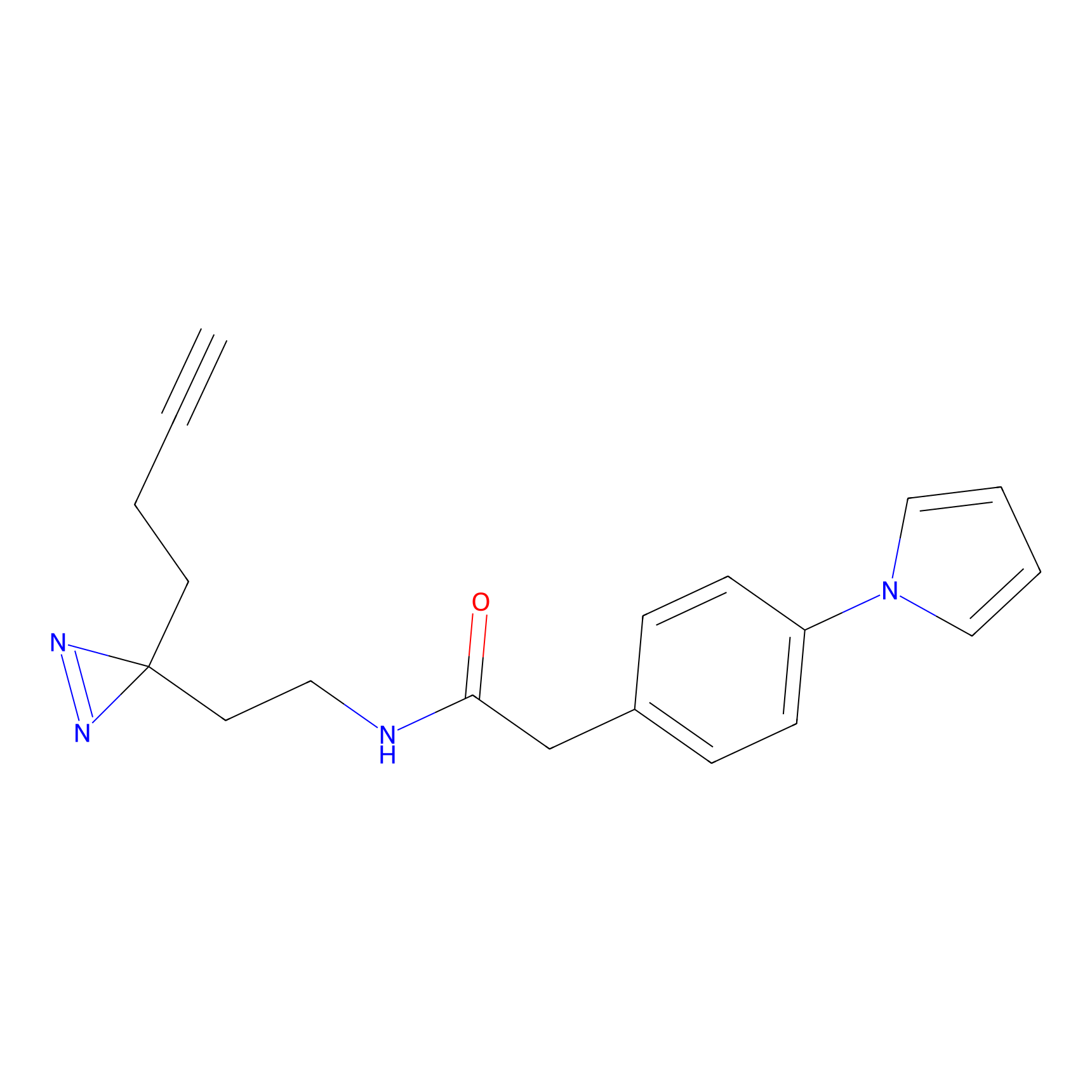 |
7.36 | LDD1768 | [6] | |
|
C087 Probe Info |
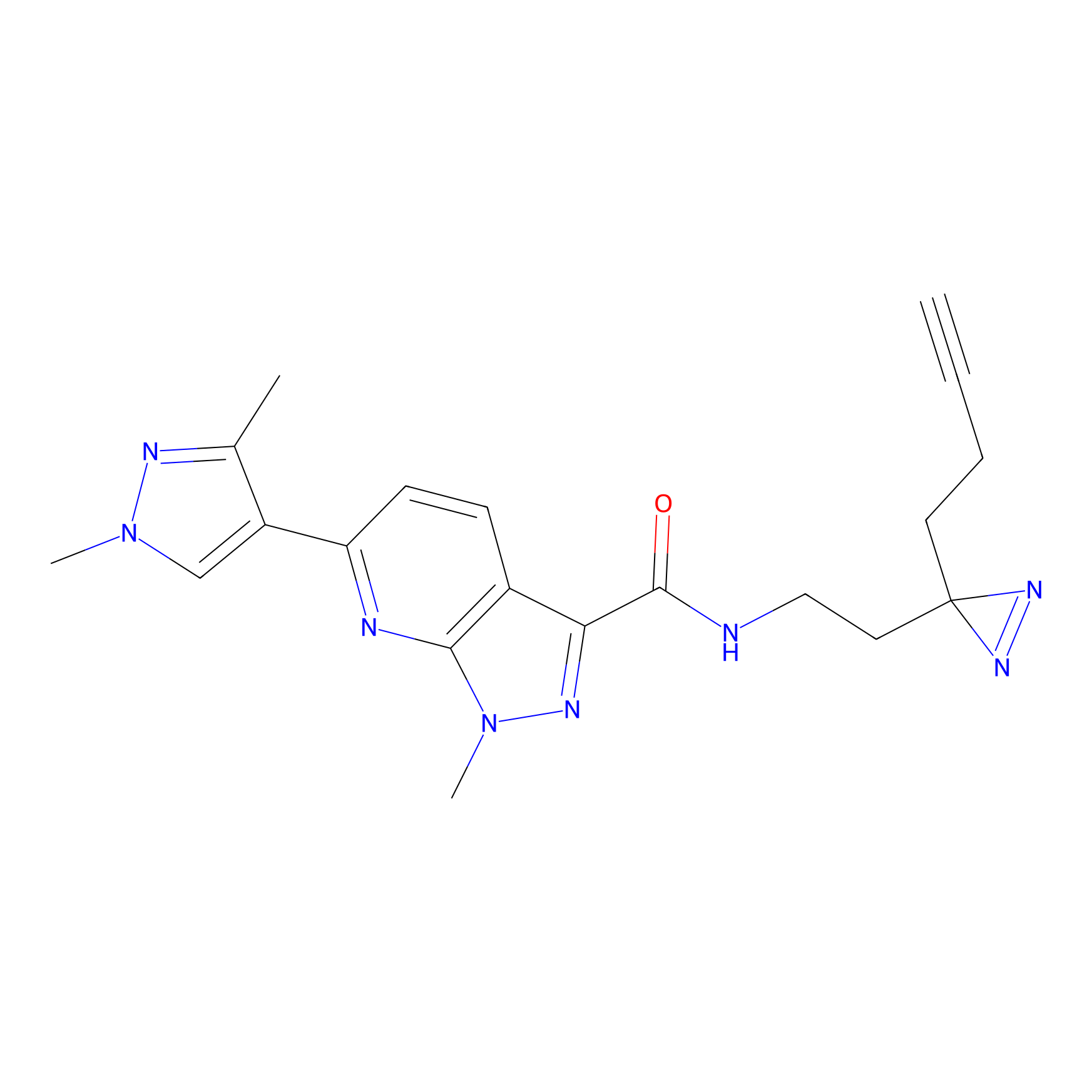 |
8.11 | LDD1779 | [6] | |
|
C112 Probe Info |
 |
24.93 | LDD1799 | [6] | |
|
C134 Probe Info |
 |
26.72 | LDD1816 | [6] | |
|
C201 Probe Info |
 |
36.25 | LDD1877 | [6] | |
|
C206 Probe Info |
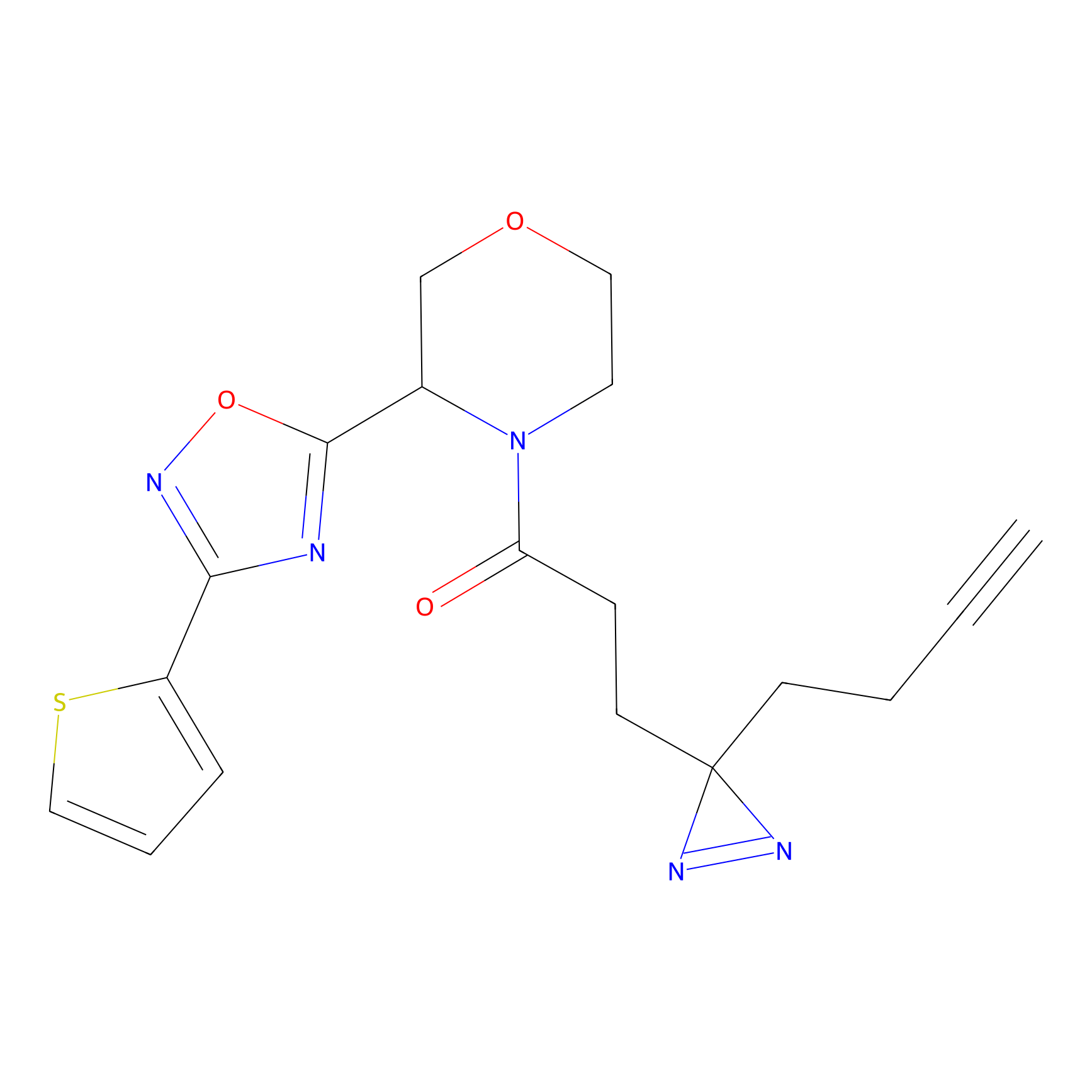 |
17.88 | LDD1881 | [6] | |
|
C210 Probe Info |
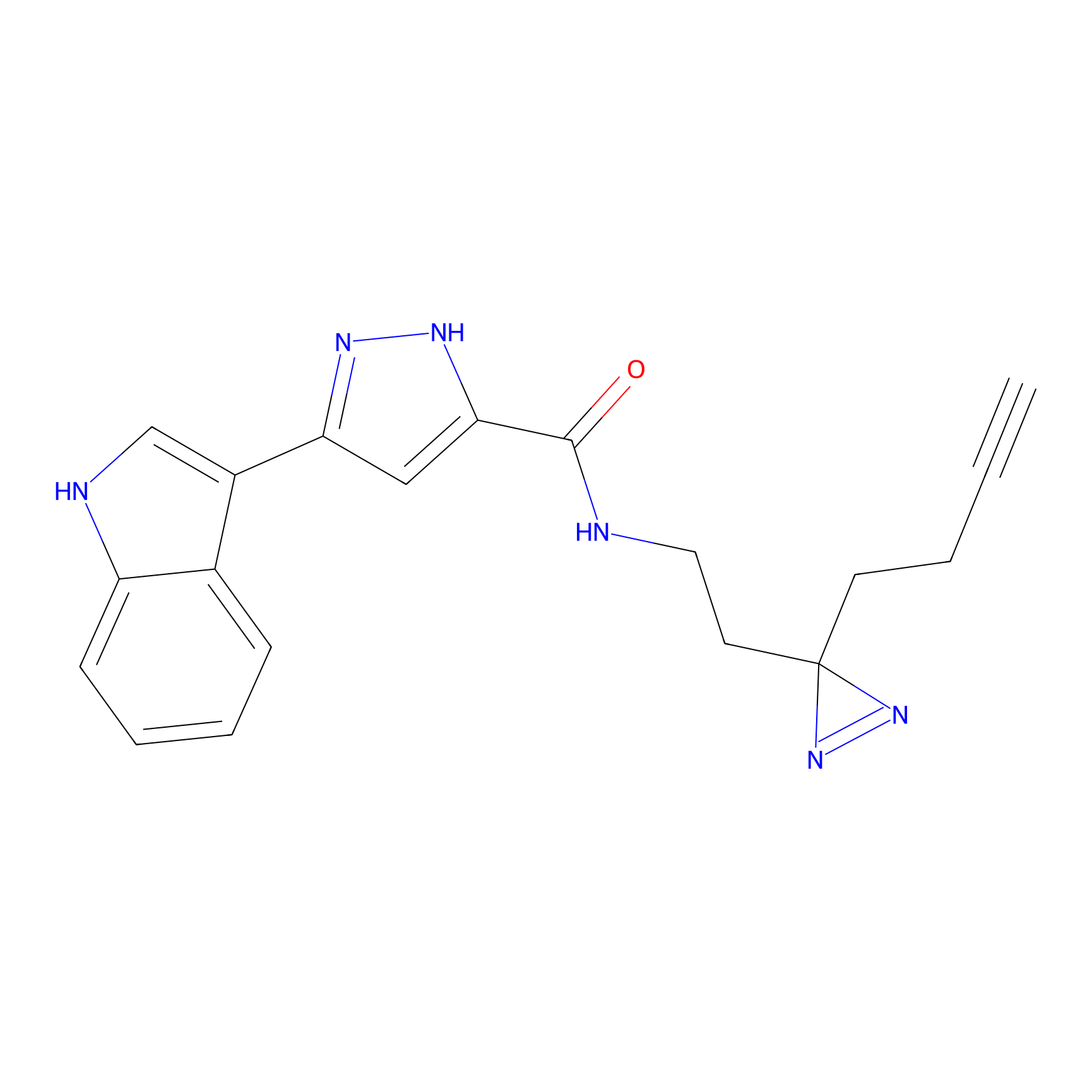 |
62.68 | LDD1884 | [6] | |
|
C225 Probe Info |
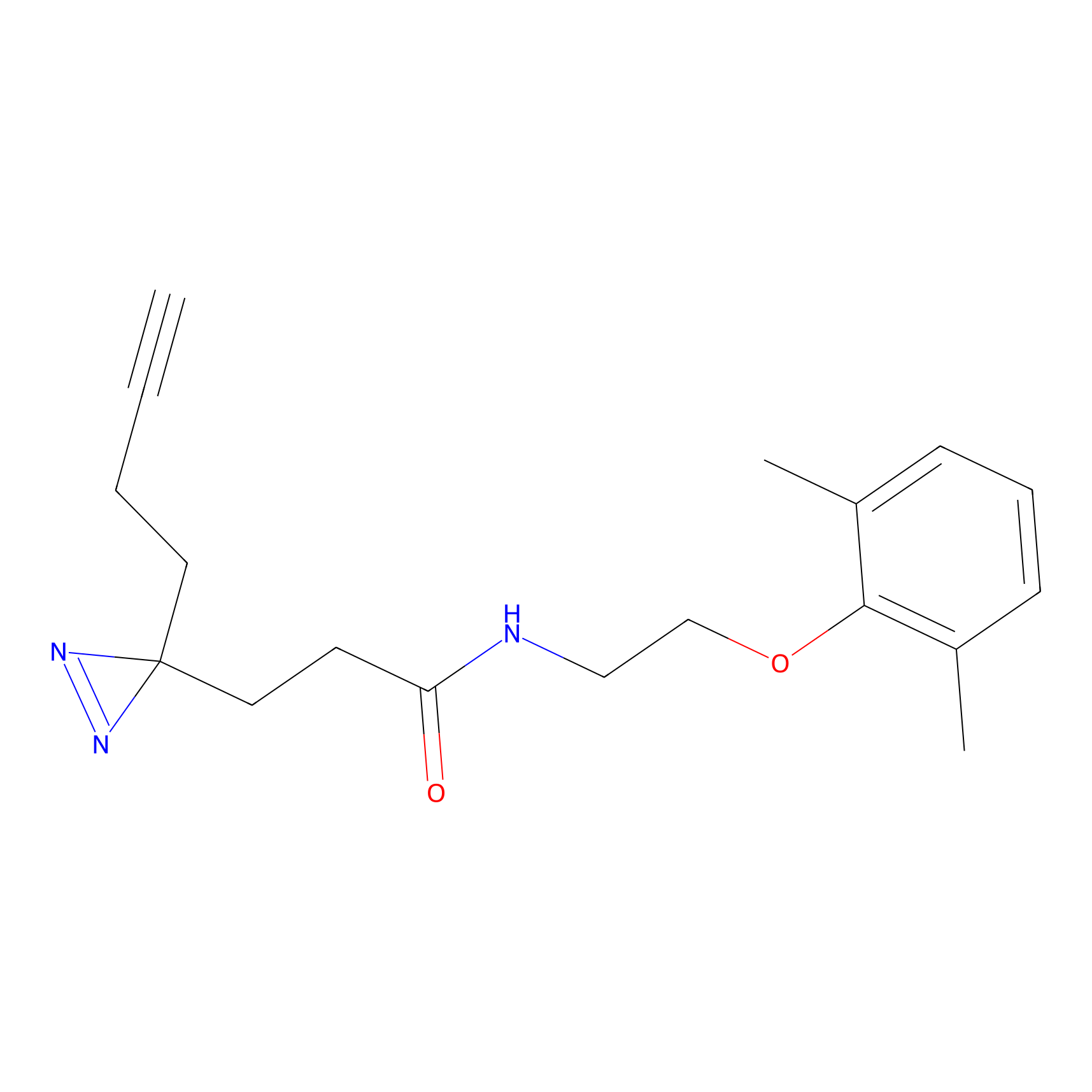 |
5.62 | LDD1898 | [6] | |
|
C226 Probe Info |
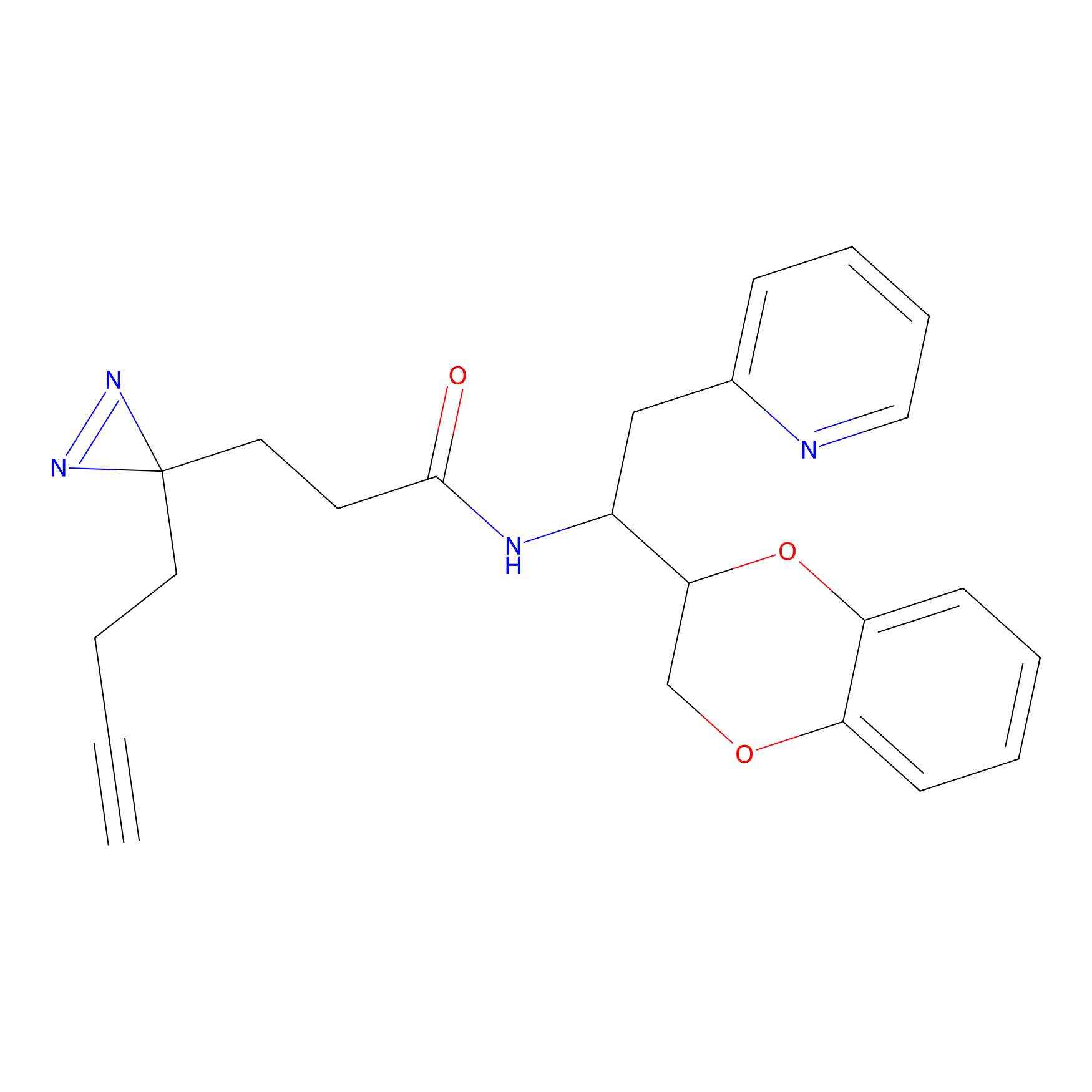 |
6.15 | LDD1899 | [6] | |
|
C228 Probe Info |
 |
34.30 | LDD1901 | [6] | |
|
C278 Probe Info |
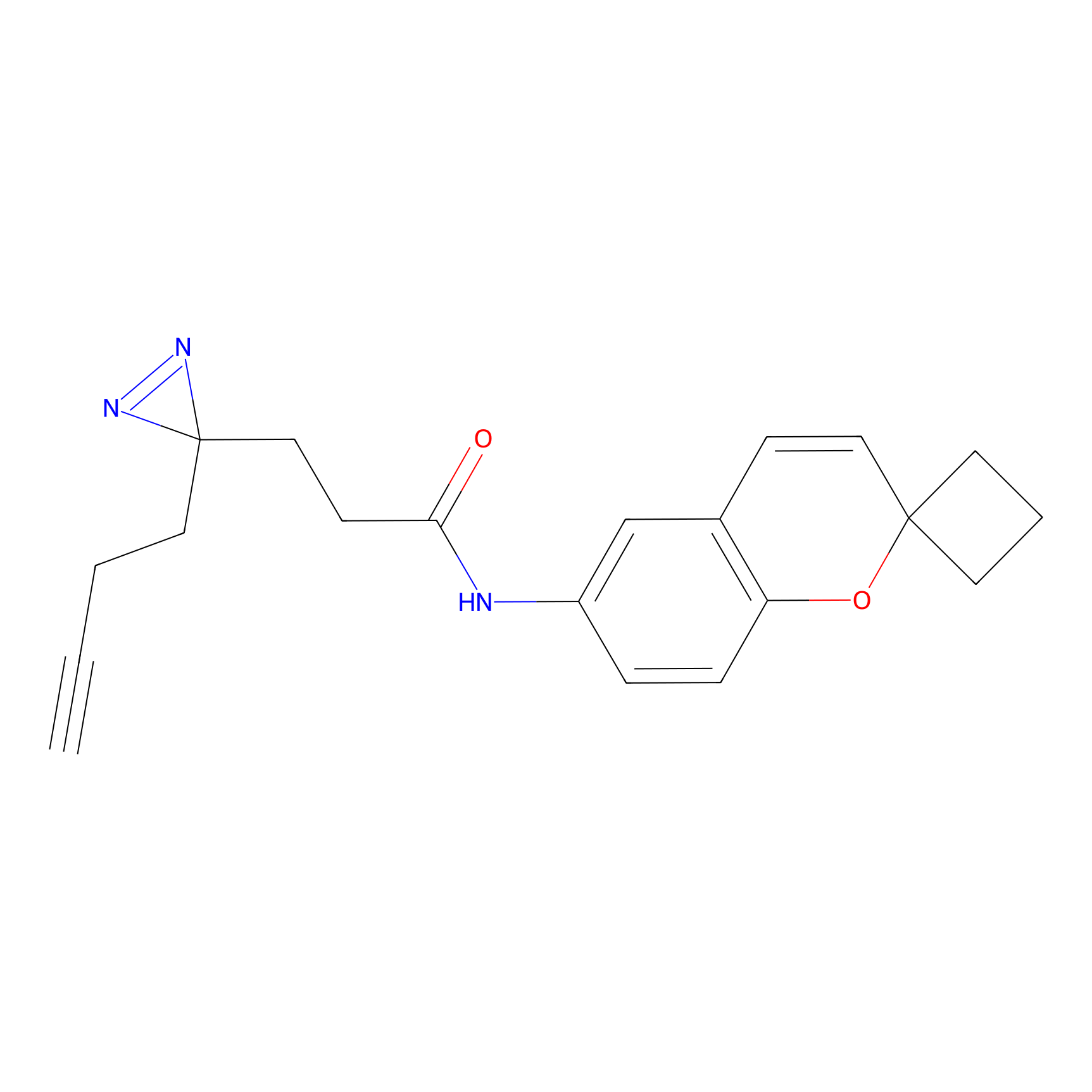 |
61.39 | LDD1948 | [6] | |
|
C362 Probe Info |
 |
51.98 | LDD2023 | [6] | |
|
C363 Probe Info |
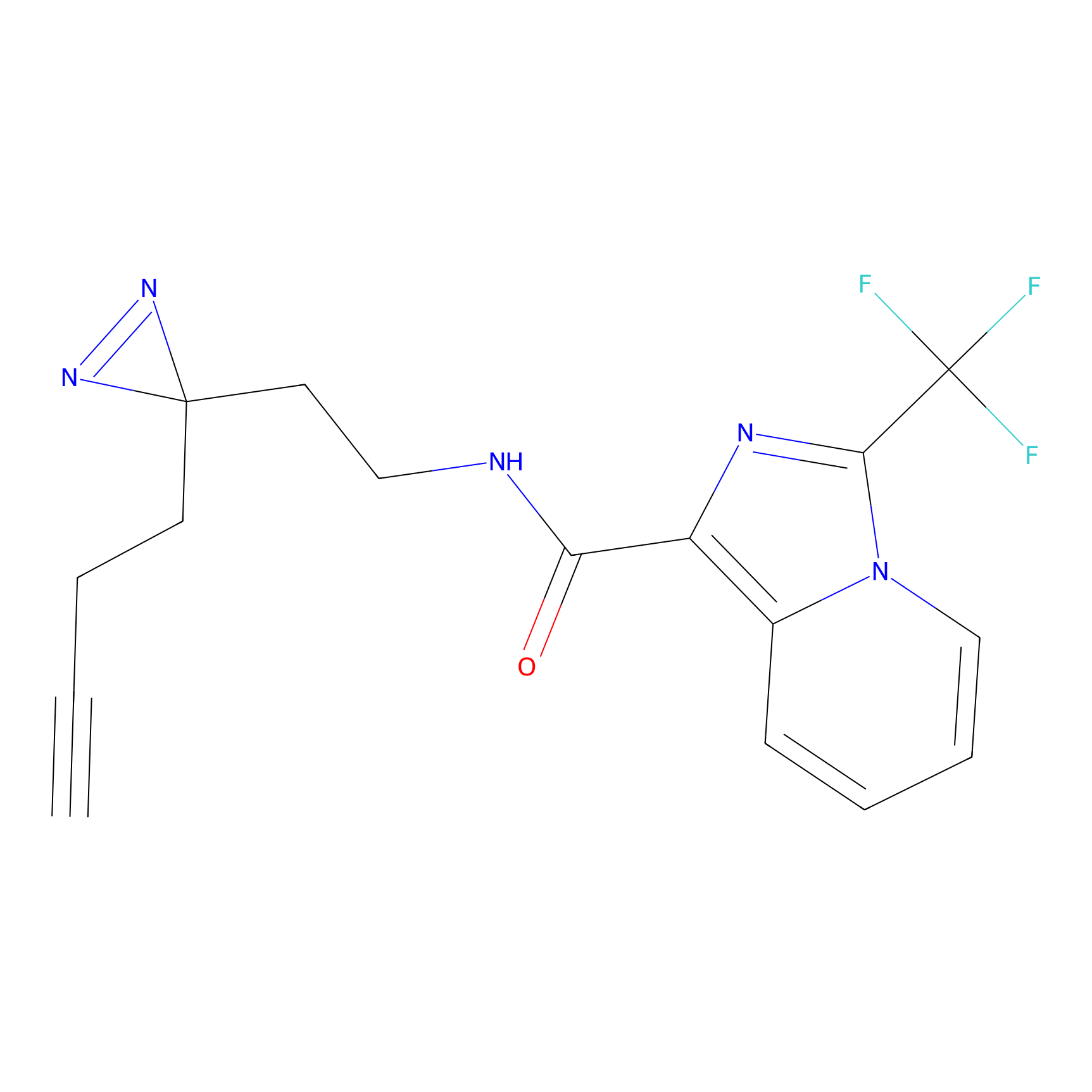 |
16.34 | LDD2024 | [6] | |
|
C364 Probe Info |
 |
17.88 | LDD2025 | [6] | |
|
C366 Probe Info |
 |
5.74 | LDD2027 | [6] | |
|
C399 Probe Info |
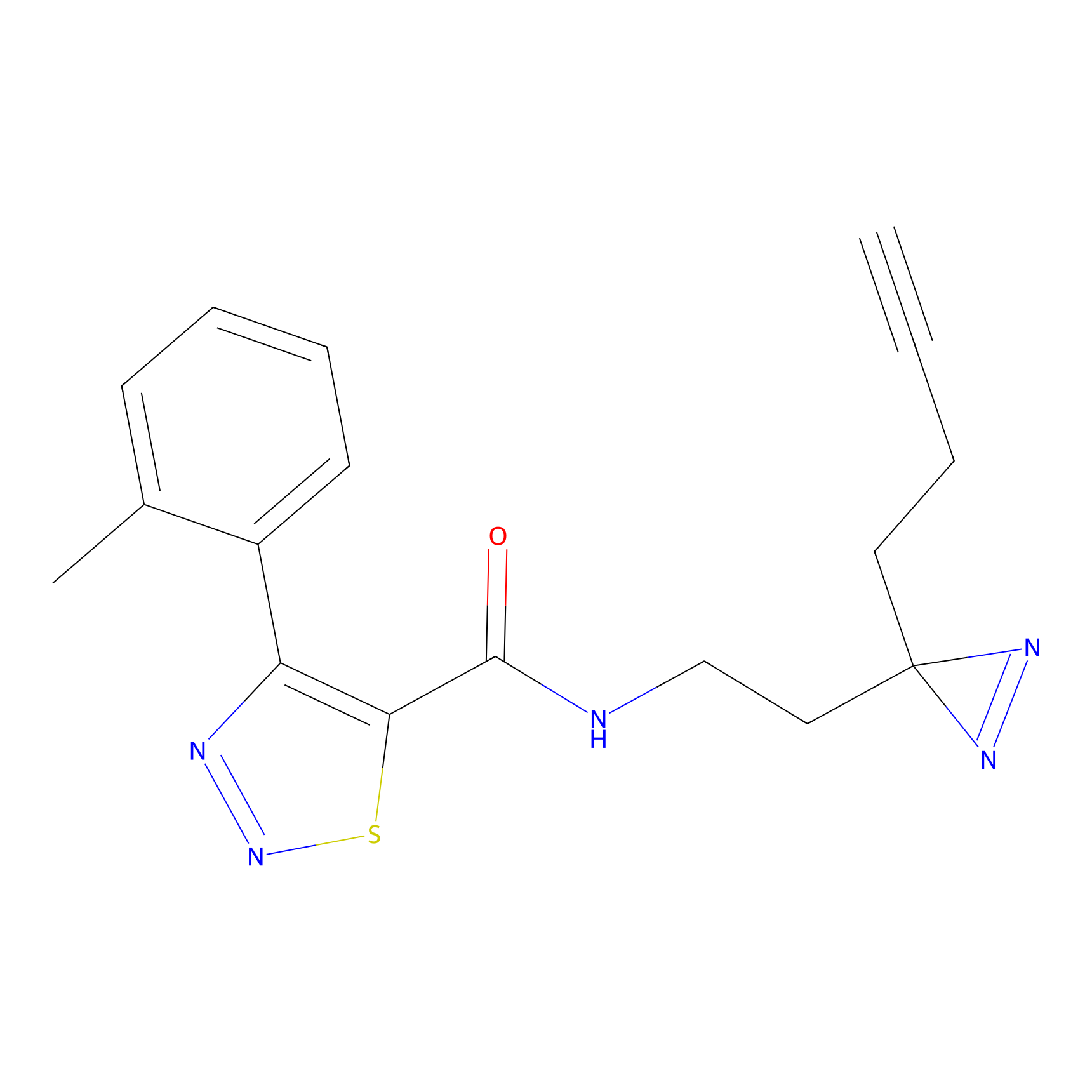 |
6.77 | LDD2058 | [6] | |
|
C403 Probe Info |
 |
30.48 | LDD2061 | [6] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0033 | Curcusone1d | MCF-7 | 2.02 | LDD0188 | [2] |
| LDCM0625 | F8 | Ramos | C322(1.64) | LDD2187 | [7] |
| LDCM0573 | Fragment11 | Ramos | C322(6.80) | LDD2190 | [7] |
| LDCM0576 | Fragment14 | Ramos | C322(0.74) | LDD2193 | [7] |
| LDCM0566 | Fragment4 | Ramos | C322(1.89) | LDD2184 | [7] |
| LDCM0614 | Fragment56 | Ramos | C322(0.90) | LDD2205 | [7] |
| LDCM0022 | KB02 | A101D | C249(2.23) | LDD2250 | [1] |
| LDCM0023 | KB03 | Ramos | C322(1.31) | LDD2183 | [7] |
| LDCM0024 | KB05 | G361 | C249(3.63) | LDD3311 | [1] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C322(15.00) | LDD2206 | [8] |
References
