Details of the Target
General Information of Target
| Target ID | LDTP07002 | |||||
|---|---|---|---|---|---|---|
| Target Name | Transport and Golgi organization protein 1 homolog (MIA3) | |||||
| Gene Name | MIA3 | |||||
| Gene ID | 375056 | |||||
| Synonyms |
KIAA0268; TANGO; Transport and Golgi organization protein 1 homolog; TANGO1; C219-reactive peptide; D320; Melanoma inhibitory activity protein 3 |
|||||
| 3D Structure | ||||||
| Sequence |
MAAAPGLLVWLLVLRLPWRVPGQLDPSTGRRFSEHKLCADDECSMLMYRGEALEDFTGPD
CRFVNFKKGDPVYVYYKLARGWPEVWAGSVGRTFGYFPKDLIQVVHEYTKEELQVPTDET DFVCFDGGRDDFHNYNVEELLGFLELYNSAATDSEKAVEKTLQDMEKNPELSKEREPEPE PVEANSEESDSVFSENTEDLQEQFTTQKHHSHANSQANHAQGEQASFESFEEMLQDKLKV PESENNKTSNSSQVSNEQDKIDAYKLLKKEMTLDLKTKFGSTADALVSDDETTRLVTSLE DDFDEELDTEYYAVGKEDEENQEDFDELPLLTFTDGEDMKTPAKSGVEKYPTDKEQNSNE EDKVQLTVPPGIKNDDKNILTTWGDTIFSIVTGGEETRDTMDLESSSSEEEKEDDDDALV PDSKQGKPQSATDYSDPDNVDDGLFIVDIPKTNNDKEVNAEHHIKGKGRGVQESKRGLVQ DKTELEDENQEGMTVHSSVHSNNLNSMPAAEKGKDTLKSAYDDTENDLKGAAIHISKGML HEEKPGEQILEGGSESESAQKAAGNQMNDRKIQQESLGSAPLMGDDHPNASRDSVEGDAL VNGAKLHTLSVEHQREELKEELVLKTQNQPRFSSPDEIDLPRELEDEVPILGRNLPWQQE RDVAATASKQMSEKIRLSEGEAKEDSLDEEFFHHKAMQGTEVGQTDQTDSTGGPAFLSKV EEDDYPSEELLEDENAINAKRSKEKNPGNQGRQFDVNLQVPDRAVLGTIHPDPEIEESKQ ETSMILDSEKTSETAAKGVNTGGREPNTMVEKERPLADKKAQRPFERSDFSDSIKIQTPE LGEVFQNKDSDYLKNDNPEEHLKTSGLAGEPEGELSKEDHENTEKYMGTESQGSAAAEPE DDSFHWTPHTSVEPGHSDKREDLLIISSFFKEQQSLQRFQKYFNVHELEALLQEMSSKLK SAQQESLPYNMEKVLDKVFRASESQILSIAEKMLDTRVAENRDLGMNENNIFEEAAVLDD IQDLIYFVRYKHSTAEETATLVMAPPLEEGLGGAMEEMQPLHEDNFSREKTAELNVQVPE EPTHLDQRVIGDTHASEVSQKPNTEKDLDPGPVTTEDTPMDAIDANKQPETAAEEPASVT PLENAILLIYSFMFYLTKSLVATLPDDVQPGPDFYGLPWKPVFITAFLGIASFAIFLWRT VLVVKDRVYQVTEQQISEKLKTIMKENTELVQKLSNYEQKIKESKKHVQETRKQNMILSD EAIKYKDKIKTLEKNQEILDDTAKNLRVMLESEREQNVKNQDLISENKKSIEKLKDVISM NASEFSEVQIALNEAKLSEEKVKSECHRVQEENARLKKKKEQLQQEIEDWSKLHAELSEQ IKSFEKSQKDLEVALTHKDDNINALTNCITQLNLLECESESEGQNKGGNDSDELANGEVG GDRNEKMKNQIKQMMDVSRTQTAISVVEEDLKLLQLKLRASVSTKCNLEDQVKKLEDDRN SLQAAKAGLEDECKTLRQKVEILNELYQQKEMALQKKLSQEEYERQEREHRLSAADEKAV SAAEEVKTYKRRIEEMEDELQKTERSFKNQIATHEKKAHENWLKARAAERAIAEEKREAA NLRHKLLELTQKMAMLQEEPVIVKPMPGKPNTQNPPRRGPLSQNGSFGPSPVSGGECSPP LTVEPPVRPLSATLNRRDMPRSEFGSVDGPLPHPRWSAEASGKPSPSDPGSGTATMMNSS SRGSSPTRVLDEGKVNMAPKGPPPFPGVPLMSTPMGGPVPPPIRYGPPPQLCGPFGPRPL PPPFGPGMRPPLGLREFAPGVPPGRRDLPLHPRGFLPGHAPFRPLGSLGPREYFIPGTRL PPPTHGPQEYPPPPAVRDLLPSGSRDEPPPASQSTSQDCSQALKQSP |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
MIA/OTOR family, Tango1 subfamily
|
|||||
| Subcellular location |
Endoplasmic reticulum membrane
|
|||||
| Function |
Plays a role in the transport of cargos that are too large to fit into COPII-coated vesicles and require specific mechanisms to be incorporated into membrane-bound carriers and exported from the endoplasmic reticulum. This protein is required for collagen VII (COL7A1) secretion by loading COL7A1 into transport carriers. It may participate in cargo loading of COL7A1 at endoplasmic reticulum exit sites by binding to COPII coat subunits Sec23/24 and guiding SH3-bound COL7A1 into a growing carrier. Does not play a role in global protein secretion and is apparently specific to COL7A1 cargo loading. However, it may participate in secretion of other proteins in cells that do not secrete COL7A1. It is also specifically required for the secretion of lipoproteins by participating in their export from the endoplasmic reticulum. Required for correct assembly of COPII coat components at endoplasmic reticulum exit sites (ERES) and for the localization of SEC16A and membrane-bound ER-resident complexes consisting of MIA2 and PREB/SEC12 to ERES.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| CCK81 | SNV: p.F1193L | DBIA Probe Info | |||
| GB2 | SNV: p.E139K | DBIA Probe Info | |||
| HT | SNV: p.N505I | DBIA Probe Info | |||
| JHH4 | SNV: p.G280V | . | |||
| LN229 | SNV: p.V1641M | . | |||
| MEWO | SNV: p.P1060S | DBIA Probe Info | |||
| NCIH1793 | SNV: p.G578C | . | |||
| NCIH2172 | SNV: p.L841V | . | |||
| NCIH446 | SNV: p.E187Ter | . | |||
| OVK18 | SNV: p.E701D | DBIA Probe Info | |||
| PC3 | SNV: p.L1085V | . | |||
| SKOV3 | SNV: p.P83T | DBIA Probe Info | |||
| TOV21G | Deletion: p.N168TfsTer15 | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
DA-P2 Probe Info |
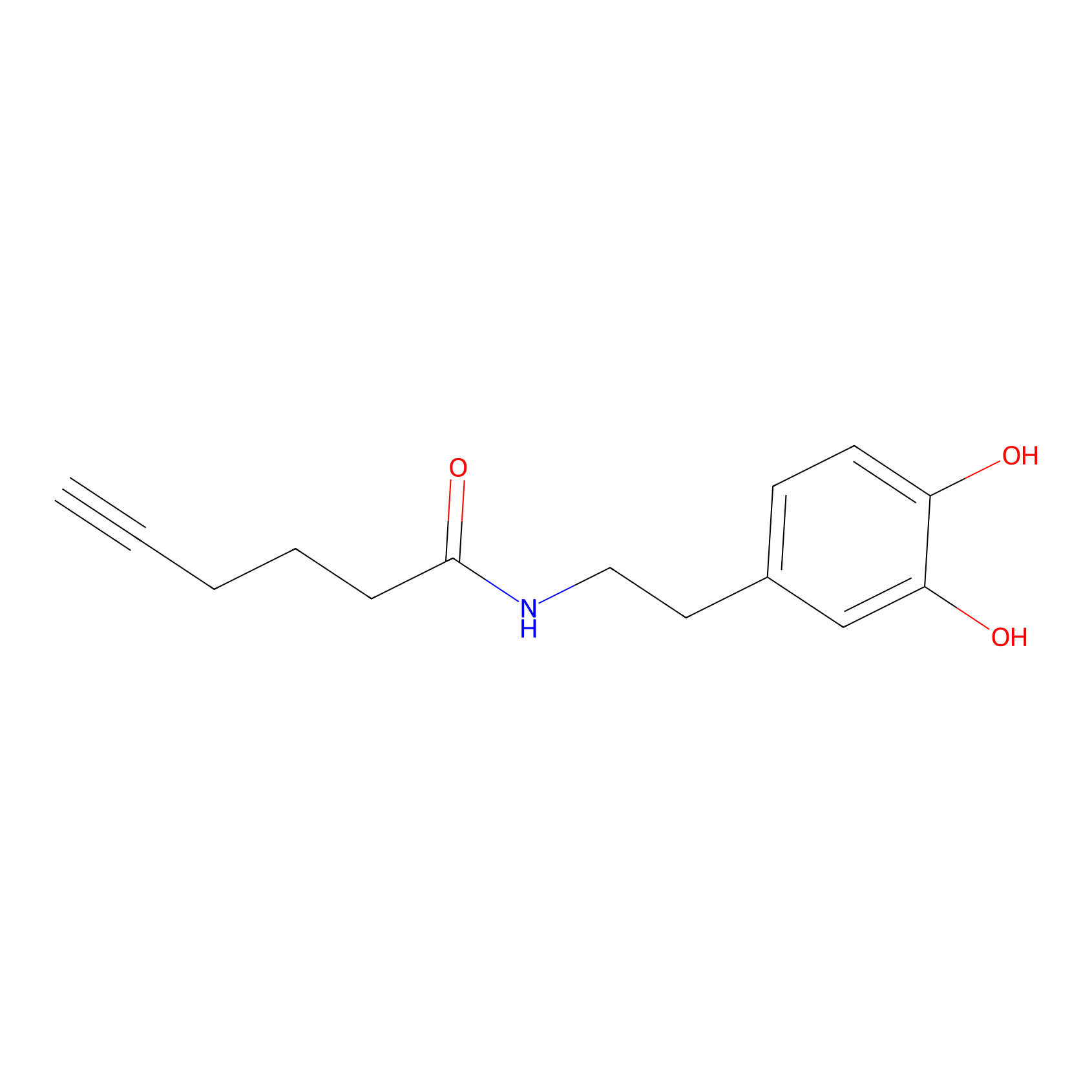 |
5.66 | LDD0348 | [2] | |
|
HRP Probe Info |
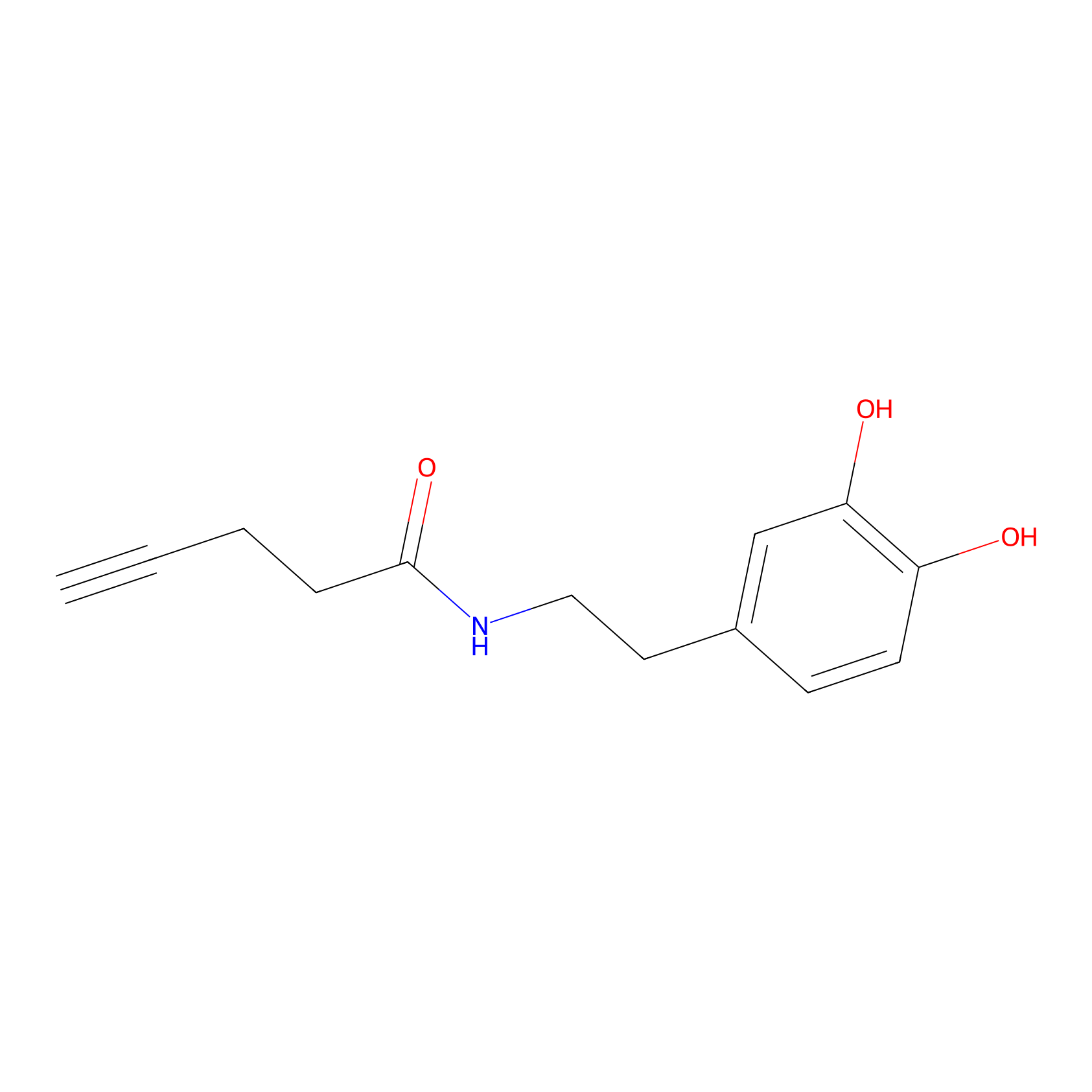 |
4.63 | LDD0347 | [2] | |
|
TG42 Probe Info |
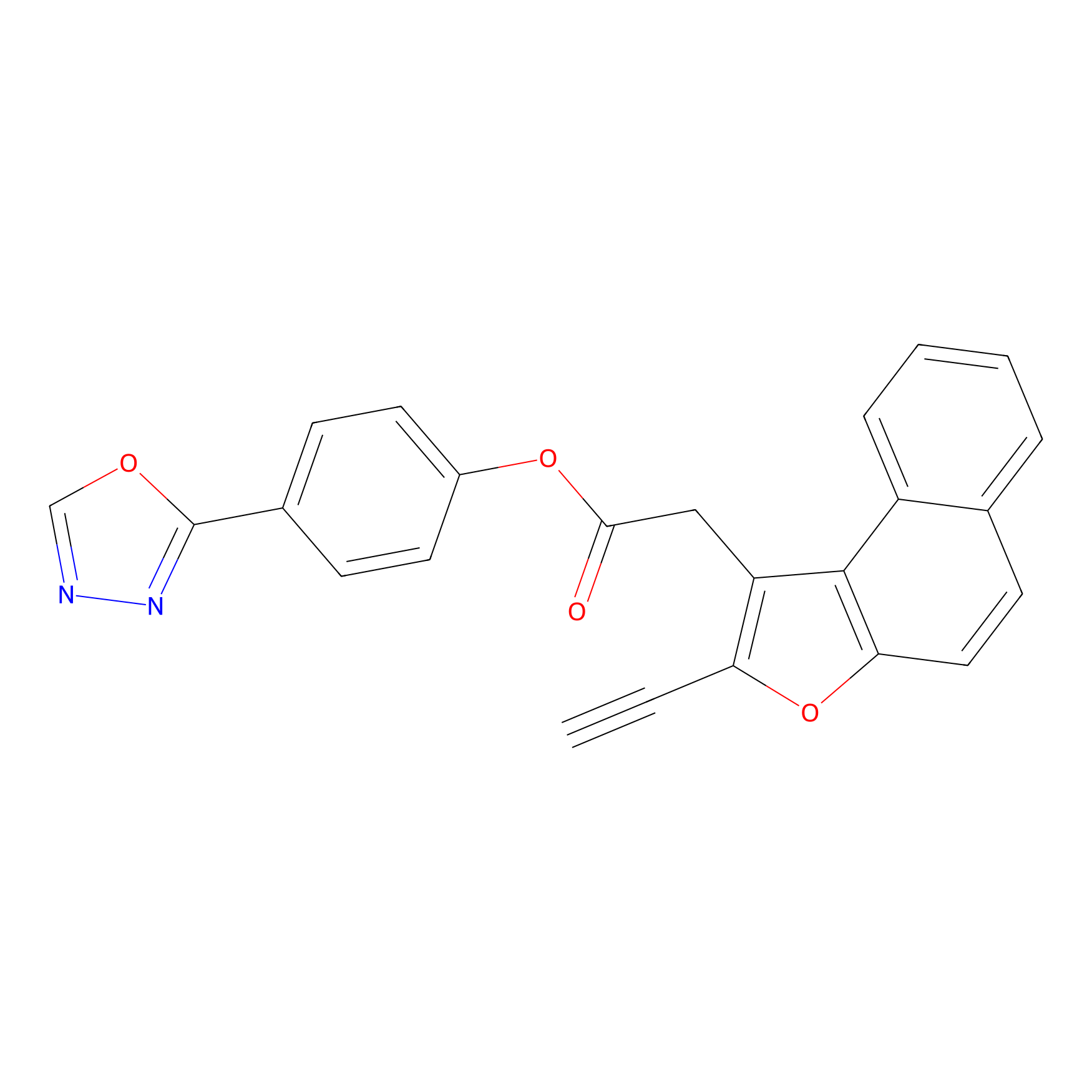 |
4.92 | LDD0326 | [3] | |
|
TH211 Probe Info |
 |
Y108(20.00) | LDD0260 | [4] | |
|
FBP2 Probe Info |
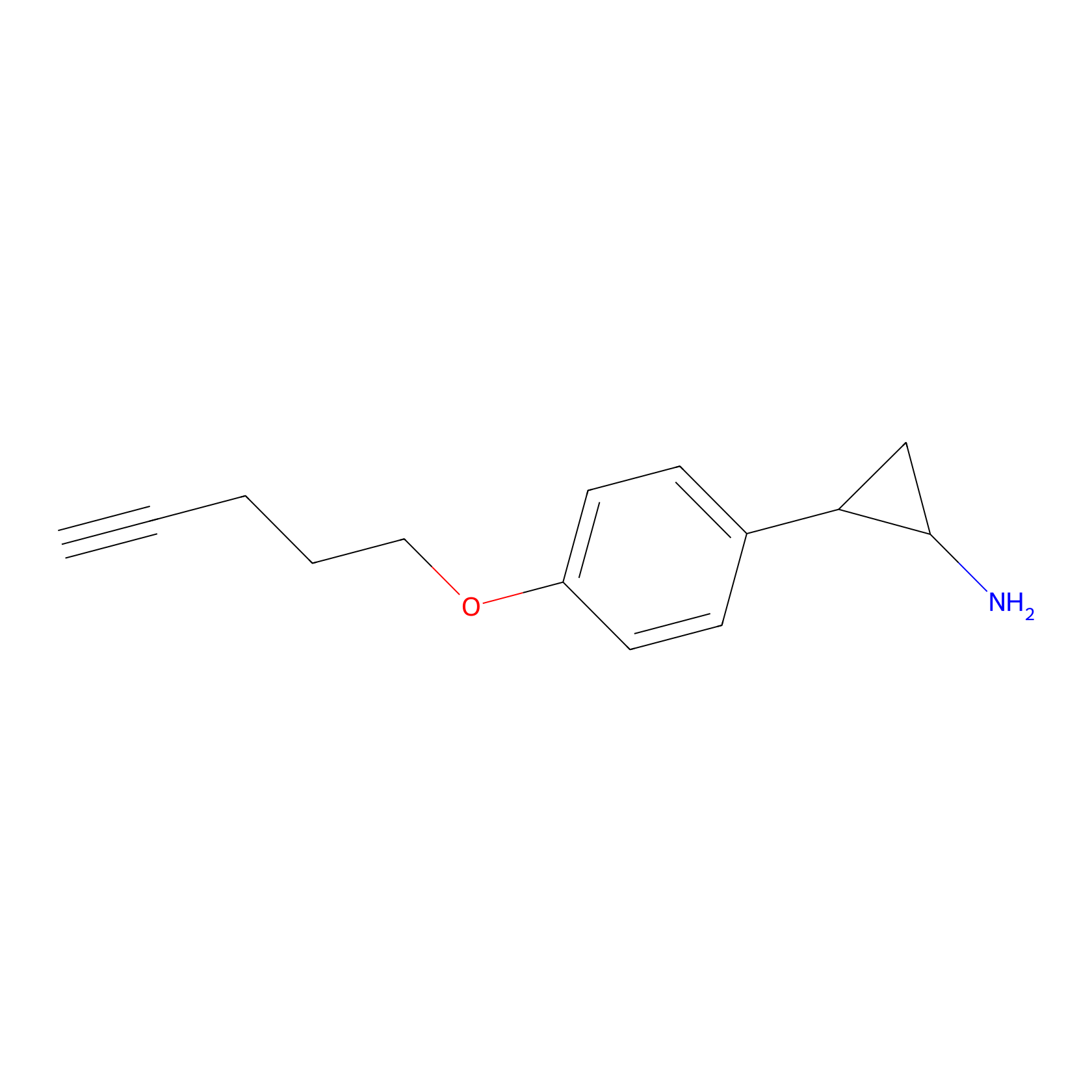 |
2.38 | LDD0323 | [5] | |
|
Probe 1 Probe Info |
 |
Y73(7.42); Y1237(23.21); Y1543(7.44) | LDD3495 | [6] | |
|
P12 Probe Info |
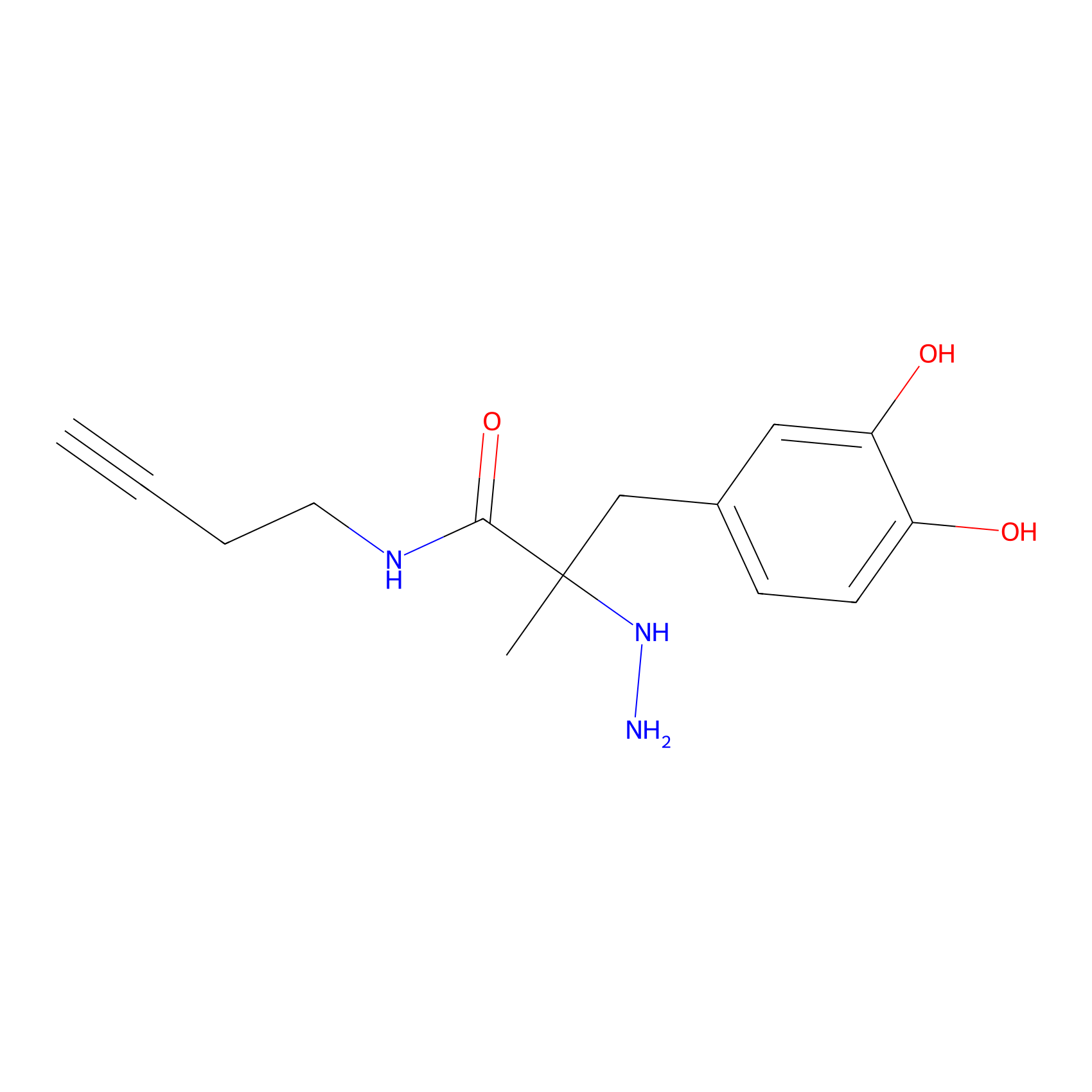 |
13.36 | LDD0202 | [7] | |
|
BTD Probe Info |
 |
C1486(1.14) | LDD1700 | [8] | |
|
DA-P3 Probe Info |
 |
5.52 | LDD0179 | [2] | |
|
AHL-Pu-1 Probe Info |
 |
C1417(10.03) | LDD0168 | [9] | |
|
DBIA Probe Info |
 |
C124(1.07) | LDD0078 | [10] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [11] | |
|
IA-alkyne Probe Info |
 |
C1346(0.00); C61(0.00) | LDD0162 | [12] | |
|
Lodoacetamide azide Probe Info |
 |
C1513(0.00); C1486(0.00) | LDD0037 | [13] | |
|
IPM Probe Info |
 |
C1513(0.00); C1899(0.00); C1486(0.00) | LDD0147 | [14] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [15] | |
|
PPMS Probe Info |
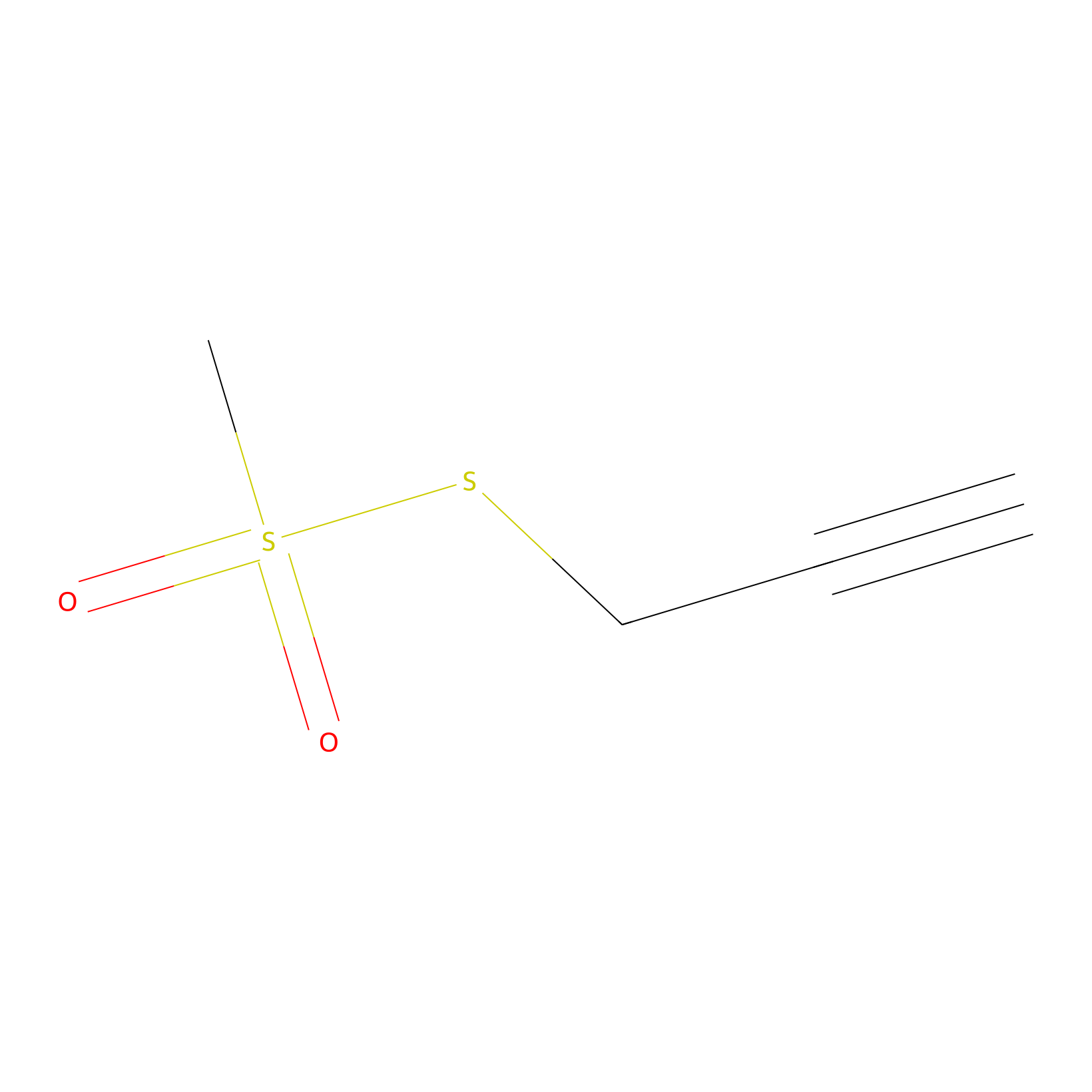 |
N.A. | LDD0008 | [15] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [16] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [15] | |
|
TFBX Probe Info |
 |
C1486(0.00); C1513(0.00); C1346(0.00) | LDD0148 | [14] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [15] | |
|
Phosphinate-6 Probe Info |
 |
C1899(0.00); C1346(0.00); C1486(0.00) | LDD0018 | [17] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [18] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [19] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [19] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [19] | |
|
W1 Probe Info |
 |
N.A. | LDD0236 | [20] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C201 Probe Info |
 |
27.47 | LDD1877 | [21] | |
|
C349 Probe Info |
 |
8.75 | LDD2010 | [21] | |
|
FFF probe11 Probe Info |
 |
15.08 | LDD0472 | [22] | |
|
FFF probe13 Probe Info |
 |
20.00 | LDD0475 | [22] | |
|
FFF probe2 Probe Info |
 |
13.56 | LDD0463 | [22] | |
|
OEA-DA Probe Info |
 |
8.18 | LDD0046 | [23] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C1486(0.69) | LDD2112 | [8] |
| LDCM0025 | 4SU-RNA | HEK-293T | C1417(10.03) | LDD0168 | [9] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C1417(2.32) | LDD0169 | [9] |
| LDCM0214 | AC1 | HCT 116 | C1408(1.06); C1417(1.06); C1486(0.96) | LDD0531 | [10] |
| LDCM0215 | AC10 | HCT 116 | C1899(1.57) | LDD0532 | [10] |
| LDCM0216 | AC100 | HCT 116 | C1899(0.94) | LDD0533 | [10] |
| LDCM0217 | AC101 | HCT 116 | C1899(0.97) | LDD0534 | [10] |
| LDCM0218 | AC102 | HCT 116 | C1899(0.83) | LDD0535 | [10] |
| LDCM0219 | AC103 | HCT 116 | C1899(1.20) | LDD0536 | [10] |
| LDCM0220 | AC104 | HCT 116 | C1899(1.11) | LDD0537 | [10] |
| LDCM0221 | AC105 | HCT 116 | C1899(1.17) | LDD0538 | [10] |
| LDCM0222 | AC106 | HCT 116 | C1899(1.11) | LDD0539 | [10] |
| LDCM0223 | AC107 | HCT 116 | C1899(0.98) | LDD0540 | [10] |
| LDCM0224 | AC108 | HCT 116 | C1899(1.29) | LDD0541 | [10] |
| LDCM0225 | AC109 | HCT 116 | C1899(1.17) | LDD0542 | [10] |
| LDCM0226 | AC11 | HCT 116 | C1899(1.28) | LDD0543 | [10] |
| LDCM0227 | AC110 | HCT 116 | C1899(0.82) | LDD0544 | [10] |
| LDCM0228 | AC111 | HCT 116 | C1899(0.97) | LDD0545 | [10] |
| LDCM0229 | AC112 | HCT 116 | C1899(0.83) | LDD0546 | [10] |
| LDCM0230 | AC113 | HCT 116 | C1486(0.88); C61(1.07) | LDD0547 | [10] |
| LDCM0231 | AC114 | HCT 116 | C1486(0.80); C61(0.89) | LDD0548 | [10] |
| LDCM0232 | AC115 | HCT 116 | C1486(0.83); C61(1.02) | LDD0549 | [10] |
| LDCM0233 | AC116 | HCT 116 | C1486(0.82); C61(1.15) | LDD0550 | [10] |
| LDCM0234 | AC117 | HCT 116 | C1486(0.81); C61(1.08) | LDD0551 | [10] |
| LDCM0235 | AC118 | HCT 116 | C1486(0.87); C61(1.00) | LDD0552 | [10] |
| LDCM0236 | AC119 | HCT 116 | C1486(0.77); C61(1.12) | LDD0553 | [10] |
| LDCM0237 | AC12 | HCT 116 | C1899(0.95) | LDD0554 | [10] |
| LDCM0238 | AC120 | HCT 116 | C1486(0.84); C61(0.86) | LDD0555 | [10] |
| LDCM0239 | AC121 | HCT 116 | C1486(0.73); C61(0.95) | LDD0556 | [10] |
| LDCM0240 | AC122 | HCT 116 | C1486(0.78); C61(1.11) | LDD0557 | [10] |
| LDCM0241 | AC123 | HCT 116 | C1486(0.84); C61(0.81) | LDD0558 | [10] |
| LDCM0242 | AC124 | HCT 116 | C1486(0.79); C61(0.88) | LDD0559 | [10] |
| LDCM0243 | AC125 | HCT 116 | C1486(0.81); C61(0.91) | LDD0560 | [10] |
| LDCM0244 | AC126 | HCT 116 | C1486(0.83); C61(1.10) | LDD0561 | [10] |
| LDCM0245 | AC127 | HCT 116 | C1486(0.86); C61(1.01) | LDD0562 | [10] |
| LDCM0246 | AC128 | HCT 116 | C1408(1.65); C1417(1.65); C1486(0.71); C1899(1.14) | LDD0563 | [10] |
| LDCM0247 | AC129 | HCT 116 | C1408(1.09); C1417(1.09); C1486(0.80); C1899(0.74) | LDD0564 | [10] |
| LDCM0249 | AC130 | HCT 116 | C1408(1.35); C1417(1.35); C1486(0.79); C1899(1.40) | LDD0566 | [10] |
| LDCM0250 | AC131 | HCT 116 | C1408(1.10); C1417(1.10); C1486(0.78); C1899(0.73) | LDD0567 | [10] |
| LDCM0251 | AC132 | HCT 116 | C1408(0.99); C1417(0.99); C1486(0.79); C1899(0.90) | LDD0568 | [10] |
| LDCM0252 | AC133 | HCT 116 | C1408(1.86); C1417(1.86); C1486(0.81); C1899(0.90) | LDD0569 | [10] |
| LDCM0253 | AC134 | HCT 116 | C1408(1.44); C1417(1.44); C1486(0.71); C1899(0.89) | LDD0570 | [10] |
| LDCM0254 | AC135 | HCT 116 | C1408(1.12); C1417(1.12); C1486(0.89); C1899(0.83) | LDD0571 | [10] |
| LDCM0255 | AC136 | HCT 116 | C1408(1.04); C1417(1.04); C1486(0.79); C1899(1.01) | LDD0572 | [10] |
| LDCM0256 | AC137 | HCT 116 | C1408(1.32); C1417(1.32); C1486(0.81); C1899(1.00) | LDD0573 | [10] |
| LDCM0257 | AC138 | HCT 116 | C1408(1.63); C1417(1.63); C1486(0.82); C1899(0.80) | LDD0574 | [10] |
| LDCM0258 | AC139 | HCT 116 | C1408(1.14); C1417(1.14); C1486(0.89); C1899(0.93) | LDD0575 | [10] |
| LDCM0259 | AC14 | HCT 116 | C1899(1.30) | LDD0576 | [10] |
| LDCM0260 | AC140 | HCT 116 | C1408(1.12); C1417(1.12); C1486(0.86); C1899(1.09) | LDD0577 | [10] |
| LDCM0261 | AC141 | HCT 116 | C1408(0.97); C1417(0.97); C1486(0.90); C1899(0.98) | LDD0578 | [10] |
| LDCM0262 | AC142 | HCT 116 | C1408(1.13); C1417(1.13); C1486(0.84); C1899(0.78) | LDD0579 | [10] |
| LDCM0263 | AC143 | HCT 116 | C1408(0.99); C1417(0.99) | LDD0580 | [10] |
| LDCM0264 | AC144 | HCT 116 | C1408(1.29); C1417(1.29) | LDD0581 | [10] |
| LDCM0265 | AC145 | HCT 116 | C1408(1.11); C1417(1.11) | LDD0582 | [10] |
| LDCM0266 | AC146 | HCT 116 | C1408(1.41); C1417(1.41) | LDD0583 | [10] |
| LDCM0267 | AC147 | HCT 116 | C1408(1.29); C1417(1.29) | LDD0584 | [10] |
| LDCM0268 | AC148 | HCT 116 | C1408(1.88); C1417(1.88) | LDD0585 | [10] |
| LDCM0269 | AC149 | HCT 116 | C1408(1.47); C1417(1.47) | LDD0586 | [10] |
| LDCM0270 | AC15 | HCT 116 | C1899(1.25) | LDD0587 | [10] |
| LDCM0271 | AC150 | HCT 116 | C1408(0.92); C1417(0.92) | LDD0588 | [10] |
| LDCM0272 | AC151 | HCT 116 | C1408(1.30); C1417(1.30) | LDD0589 | [10] |
| LDCM0273 | AC152 | HCT 116 | C1408(1.54); C1417(1.54) | LDD0590 | [10] |
| LDCM0274 | AC153 | HCT 116 | C1408(2.14); C1417(2.14) | LDD0591 | [10] |
| LDCM0621 | AC154 | HCT 116 | C1408(1.33); C1417(1.33) | LDD2158 | [10] |
| LDCM0622 | AC155 | HCT 116 | C1408(1.26); C1417(1.26) | LDD2159 | [10] |
| LDCM0623 | AC156 | HCT 116 | C1408(0.98); C1417(0.98) | LDD2160 | [10] |
| LDCM0624 | AC157 | HCT 116 | C1408(1.10); C1417(1.10) | LDD2161 | [10] |
| LDCM0276 | AC17 | HCT 116 | C1486(1.04); C61(1.13); C1899(1.16) | LDD0593 | [10] |
| LDCM0277 | AC18 | HCT 116 | C1486(0.85); C61(1.01); C1899(1.22) | LDD0594 | [10] |
| LDCM0278 | AC19 | HCT 116 | C1486(0.86); C61(1.07); C1899(1.35) | LDD0595 | [10] |
| LDCM0279 | AC2 | HCT 116 | C1408(0.95); C1417(0.95); C1486(1.10) | LDD0596 | [10] |
| LDCM0280 | AC20 | HCT 116 | C61(0.85); C1486(1.08); C1899(1.14) | LDD0597 | [10] |
| LDCM0281 | AC21 | HCT 116 | C1486(0.84); C61(1.10); C1899(1.55) | LDD0598 | [10] |
| LDCM0282 | AC22 | HCT 116 | C1486(0.98); C61(1.06); C1899(1.24) | LDD0599 | [10] |
| LDCM0283 | AC23 | HCT 116 | C1486(0.84); C61(0.92); C1899(1.33) | LDD0600 | [10] |
| LDCM0284 | AC24 | HCT 116 | C61(0.88); C1486(1.06); C1899(1.11) | LDD0601 | [10] |
| LDCM0285 | AC25 | HCT 116 | C1486(1.05); C1899(1.08) | LDD0602 | [10] |
| LDCM0286 | AC26 | HCT 116 | C1486(1.12); C1899(1.32) | LDD0603 | [10] |
| LDCM0287 | AC27 | HCT 116 | C1486(1.21); C1899(1.73) | LDD0604 | [10] |
| LDCM0288 | AC28 | HCT 116 | C1486(1.25); C1899(1.36) | LDD0605 | [10] |
| LDCM0289 | AC29 | HCT 116 | C1899(1.24); C1486(1.72) | LDD0606 | [10] |
| LDCM0290 | AC3 | HCT 116 | C1486(0.87); C1408(1.02); C1417(1.02) | LDD0607 | [10] |
| LDCM0291 | AC30 | HCT 116 | C1899(1.47); C1486(1.97) | LDD0608 | [10] |
| LDCM0292 | AC31 | HCT 116 | C1899(1.23); C1486(1.29) | LDD0609 | [10] |
| LDCM0293 | AC32 | HCT 116 | C1899(1.70); C1486(1.73) | LDD0610 | [10] |
| LDCM0294 | AC33 | HCT 116 | C1486(1.08); C1899(1.48) | LDD0611 | [10] |
| LDCM0295 | AC34 | HCT 116 | C1486(1.12); C1899(1.64) | LDD0612 | [10] |
| LDCM0296 | AC35 | HCT 116 | C1899(0.63); C1486(0.91) | LDD0613 | [10] |
| LDCM0297 | AC36 | HCT 116 | C1899(0.66); C1486(1.04) | LDD0614 | [10] |
| LDCM0298 | AC37 | HCT 116 | C1899(0.78); C1486(1.11) | LDD0615 | [10] |
| LDCM0299 | AC38 | HCT 116 | C1899(0.77); C1486(1.21) | LDD0616 | [10] |
| LDCM0300 | AC39 | HCT 116 | C1899(0.96); C1486(1.08) | LDD0617 | [10] |
| LDCM0301 | AC4 | HCT 116 | C1486(1.01); C1408(1.13); C1417(1.13) | LDD0618 | [10] |
| LDCM0302 | AC40 | HCT 116 | C1899(1.02); C1486(1.04) | LDD0619 | [10] |
| LDCM0303 | AC41 | HCT 116 | C1899(0.87); C1486(1.14) | LDD0620 | [10] |
| LDCM0304 | AC42 | HCT 116 | C1899(1.22); C1486(1.27) | LDD0621 | [10] |
| LDCM0305 | AC43 | HCT 116 | C1899(0.94); C1486(1.16) | LDD0622 | [10] |
| LDCM0306 | AC44 | HCT 116 | C1899(0.98); C1486(1.15) | LDD0623 | [10] |
| LDCM0307 | AC45 | HCT 116 | C1899(1.09); C1486(1.12) | LDD0624 | [10] |
| LDCM0308 | AC46 | HEK-293T | C124(0.83); C1408(0.98); C1417(0.85); C1486(0.85) | LDD0906 | [10] |
| LDCM0309 | AC47 | HEK-293T | C124(0.76); C1408(0.88); C1417(1.02); C1486(0.86) | LDD0907 | [10] |
| LDCM0310 | AC48 | HEK-293T | C124(0.85); C1408(0.91); C1417(1.06); C1486(0.86) | LDD0908 | [10] |
| LDCM0311 | AC49 | HEK-293T | C124(0.92); C1408(1.21); C1417(1.01); C1486(1.01) | LDD0909 | [10] |
| LDCM0312 | AC5 | HCT 116 | C1408(0.90); C1417(0.90); C1486(1.00) | LDD0629 | [10] |
| LDCM0313 | AC50 | HEK-293T | C124(0.82); C1408(0.98); C1417(0.90); C1486(0.76) | LDD0911 | [10] |
| LDCM0314 | AC51 | HEK-293T | C124(0.74); C1408(1.06); C1417(1.10); C1486(0.75) | LDD0912 | [10] |
| LDCM0315 | AC52 | HEK-293T | C124(0.73); C1408(1.45); C1417(1.18); C1486(0.82) | LDD0913 | [10] |
| LDCM0316 | AC53 | HEK-293T | C124(0.67); C1408(0.96); C1417(1.05); C1486(0.82) | LDD0914 | [10] |
| LDCM0317 | AC54 | HEK-293T | C124(0.80); C1408(1.10); C1417(1.07); C1486(0.93) | LDD0915 | [10] |
| LDCM0318 | AC55 | HEK-293T | C124(0.91); C1408(1.18); C1417(1.05); C1486(0.87) | LDD0916 | [10] |
| LDCM0319 | AC56 | HEK-293T | C124(0.88); C1408(1.12); C1417(1.14); C1486(0.81) | LDD0917 | [10] |
| LDCM0320 | AC57 | HCT 116 | C1486(0.43) | LDD0637 | [10] |
| LDCM0321 | AC58 | HCT 116 | C1486(0.69) | LDD0638 | [10] |
| LDCM0322 | AC59 | HCT 116 | C1486(0.61) | LDD0639 | [10] |
| LDCM0323 | AC6 | HCT 116 | C1899(1.57) | LDD0640 | [10] |
| LDCM0324 | AC60 | HCT 116 | C1486(0.48) | LDD0641 | [10] |
| LDCM0325 | AC61 | HCT 116 | C1486(0.59) | LDD0642 | [10] |
| LDCM0326 | AC62 | HCT 116 | C1486(0.74) | LDD0643 | [10] |
| LDCM0327 | AC63 | HCT 116 | C1486(0.50) | LDD0644 | [10] |
| LDCM0328 | AC64 | HCT 116 | C1486(0.42) | LDD0645 | [10] |
| LDCM0329 | AC65 | HCT 116 | C1486(0.61) | LDD0646 | [10] |
| LDCM0330 | AC66 | HCT 116 | C1486(0.62) | LDD0647 | [10] |
| LDCM0331 | AC67 | HCT 116 | C1486(0.57) | LDD0648 | [10] |
| LDCM0332 | AC68 | HCT 116 | C1486(0.80); C1899(1.09) | LDD0649 | [10] |
| LDCM0333 | AC69 | HCT 116 | C1486(0.75); C1899(1.54) | LDD0650 | [10] |
| LDCM0334 | AC7 | HCT 116 | C1899(1.12) | LDD0651 | [10] |
| LDCM0335 | AC70 | HCT 116 | C1486(0.90); C1899(1.40) | LDD0652 | [10] |
| LDCM0336 | AC71 | HCT 116 | C1899(1.15); C1486(1.15) | LDD0653 | [10] |
| LDCM0337 | AC72 | HCT 116 | C1486(0.98); C1899(1.60) | LDD0654 | [10] |
| LDCM0338 | AC73 | HCT 116 | C1486(0.91); C1899(2.54) | LDD0655 | [10] |
| LDCM0339 | AC74 | HCT 116 | C1486(0.80); C1899(1.92) | LDD0656 | [10] |
| LDCM0340 | AC75 | HCT 116 | C1486(1.01); C1899(2.57) | LDD0657 | [10] |
| LDCM0341 | AC76 | HCT 116 | C1486(0.96); C1899(1.48) | LDD0658 | [10] |
| LDCM0342 | AC77 | HCT 116 | C1486(0.87); C1899(1.14) | LDD0659 | [10] |
| LDCM0343 | AC78 | HCT 116 | C1486(0.78); C1899(1.52) | LDD0660 | [10] |
| LDCM0344 | AC79 | HCT 116 | C1486(1.19); C1899(1.37) | LDD0661 | [10] |
| LDCM0345 | AC8 | HCT 116 | C1899(1.25) | LDD0662 | [10] |
| LDCM0346 | AC80 | HCT 116 | C1486(1.04); C1899(1.50) | LDD0663 | [10] |
| LDCM0347 | AC81 | HCT 116 | C1486(0.88); C1899(1.17) | LDD0664 | [10] |
| LDCM0348 | AC82 | HCT 116 | C1486(1.06); C1899(2.63) | LDD0665 | [10] |
| LDCM0349 | AC83 | HCT 116 | C1899(1.84) | LDD0666 | [10] |
| LDCM0350 | AC84 | HCT 116 | C1899(1.51) | LDD0667 | [10] |
| LDCM0351 | AC85 | HCT 116 | C1899(1.26) | LDD0668 | [10] |
| LDCM0352 | AC86 | HCT 116 | C1899(1.44) | LDD0669 | [10] |
| LDCM0353 | AC87 | HCT 116 | C1899(1.08) | LDD0670 | [10] |
| LDCM0354 | AC88 | HCT 116 | C1899(1.33) | LDD0671 | [10] |
| LDCM0355 | AC89 | HCT 116 | C1899(1.47) | LDD0672 | [10] |
| LDCM0357 | AC90 | HCT 116 | C1899(1.13) | LDD0674 | [10] |
| LDCM0358 | AC91 | HCT 116 | C1899(2.34) | LDD0675 | [10] |
| LDCM0359 | AC92 | HCT 116 | C1899(2.02) | LDD0676 | [10] |
| LDCM0360 | AC93 | HCT 116 | C1899(1.16) | LDD0677 | [10] |
| LDCM0361 | AC94 | HCT 116 | C1899(1.09) | LDD0678 | [10] |
| LDCM0362 | AC95 | HCT 116 | C1899(1.49) | LDD0679 | [10] |
| LDCM0363 | AC96 | HCT 116 | C1899(1.54) | LDD0680 | [10] |
| LDCM0364 | AC97 | HCT 116 | C1899(1.66) | LDD0681 | [10] |
| LDCM0365 | AC98 | HCT 116 | C1899(1.55) | LDD0682 | [10] |
| LDCM0366 | AC99 | HCT 116 | C1899(0.77) | LDD0683 | [10] |
| LDCM0248 | AKOS034007472 | HCT 116 | C1899(1.15) | LDD0565 | [10] |
| LDCM0356 | AKOS034007680 | HCT 116 | C1899(1.60) | LDD0673 | [10] |
| LDCM0275 | AKOS034007705 | HCT 116 | C1899(2.02) | LDD0592 | [10] |
| LDCM0156 | Aniline | NCI-H1299 | 11.78 | LDD0403 | [1] |
| LDCM0020 | ARS-1620 | HCC44 | C124(1.07) | LDD0078 | [10] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C1486(0.57) | LDD2091 | [8] |
| LDCM0088 | C45 | HEK-293T | 13.36 | LDD0202 | [7] |
| LDCM0087 | Capsaicin | HEK-293T | 100.66 | LDD0185 | [2] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [19] |
| LDCM0367 | CL1 | HCT 116 | C1408(0.82); C1417(0.82); C1486(0.97) | LDD0684 | [10] |
| LDCM0368 | CL10 | HCT 116 | C1486(0.91); C1408(1.24); C1417(1.24) | LDD0685 | [10] |
| LDCM0369 | CL100 | HCT 116 | C1486(1.16); C1408(1.25); C1417(1.25) | LDD0686 | [10] |
| LDCM0370 | CL101 | HCT 116 | C1899(1.26) | LDD0687 | [10] |
| LDCM0371 | CL102 | HCT 116 | C1899(1.08) | LDD0688 | [10] |
| LDCM0372 | CL103 | HCT 116 | C1899(1.44) | LDD0689 | [10] |
| LDCM0373 | CL104 | HCT 116 | C1899(1.04) | LDD0690 | [10] |
| LDCM0374 | CL105 | HCT 116 | C1486(0.88); C61(1.11); C1899(1.77) | LDD0691 | [10] |
| LDCM0375 | CL106 | HCT 116 | C1486(0.87); C61(1.12); C1899(1.80) | LDD0692 | [10] |
| LDCM0376 | CL107 | HCT 116 | C1486(0.90); C61(1.17); C1899(1.75) | LDD0693 | [10] |
| LDCM0377 | CL108 | HCT 116 | C1486(1.02); C61(1.16); C1899(1.55) | LDD0694 | [10] |
| LDCM0378 | CL109 | HCT 116 | C1486(0.94); C61(1.23); C1899(1.56) | LDD0695 | [10] |
| LDCM0379 | CL11 | HCT 116 | C1486(0.85); C1408(1.76); C1417(1.76) | LDD0696 | [10] |
| LDCM0380 | CL110 | HCT 116 | C1486(0.84); C61(1.27); C1899(1.71) | LDD0697 | [10] |
| LDCM0381 | CL111 | HCT 116 | C1486(0.79); C61(1.15); C1899(1.47) | LDD0698 | [10] |
| LDCM0382 | CL112 | HCT 116 | C1899(1.03); C1486(1.13) | LDD0699 | [10] |
| LDCM0383 | CL113 | HCT 116 | C1899(1.17); C1486(1.36) | LDD0700 | [10] |
| LDCM0384 | CL114 | HCT 116 | C1899(1.16); C1486(1.32) | LDD0701 | [10] |
| LDCM0385 | CL115 | HCT 116 | C1899(1.20); C1486(1.51) | LDD0702 | [10] |
| LDCM0386 | CL116 | HCT 116 | C1899(1.19); C1486(1.25) | LDD0703 | [10] |
| LDCM0387 | CL117 | HCT 116 | C1486(0.94); C1899(1.63) | LDD0704 | [10] |
| LDCM0388 | CL118 | HCT 116 | C1899(1.03); C1486(1.17) | LDD0705 | [10] |
| LDCM0389 | CL119 | HCT 116 | C1899(1.08); C1486(1.09) | LDD0706 | [10] |
| LDCM0390 | CL12 | HCT 116 | C1486(1.00); C1408(1.54); C1417(1.54) | LDD0707 | [10] |
| LDCM0391 | CL120 | HCT 116 | C1899(0.90); C1486(1.24) | LDD0708 | [10] |
| LDCM0392 | CL121 | HEK-293T | C124(0.81); C1408(1.26); C1417(1.10); C1486(0.84) | LDD0990 | [10] |
| LDCM0393 | CL122 | HEK-293T | C124(0.93); C1408(1.04); C1417(0.97); C1486(0.85) | LDD0991 | [10] |
| LDCM0394 | CL123 | HEK-293T | C124(0.75); C1408(0.75); C1417(0.83); C1486(0.75) | LDD0992 | [10] |
| LDCM0395 | CL124 | HEK-293T | C124(0.79); C1408(1.13); C1417(1.00); C1486(0.88) | LDD0993 | [10] |
| LDCM0396 | CL125 | HCT 116 | C1486(0.52) | LDD0713 | [10] |
| LDCM0397 | CL126 | HCT 116 | C1486(0.78) | LDD0714 | [10] |
| LDCM0398 | CL127 | HCT 116 | C1486(0.62) | LDD0715 | [10] |
| LDCM0399 | CL128 | HCT 116 | C1486(0.59) | LDD0716 | [10] |
| LDCM0400 | CL13 | HCT 116 | C1486(0.97); C1408(1.19); C1417(1.19) | LDD0717 | [10] |
| LDCM0401 | CL14 | HCT 116 | C1486(0.89); C1408(1.01); C1417(1.01) | LDD0718 | [10] |
| LDCM0402 | CL15 | HCT 116 | C1486(0.89); C1408(1.39); C1417(1.39) | LDD0719 | [10] |
| LDCM0403 | CL16 | HCT 116 | C1408(1.19); C1417(1.19) | LDD0720 | [10] |
| LDCM0404 | CL17 | HCT 116 | C1408(0.98); C1417(0.98); C1486(1.12) | LDD0721 | [10] |
| LDCM0405 | CL18 | HCT 116 | C1486(1.03); C1408(1.49); C1417(1.49) | LDD0722 | [10] |
| LDCM0406 | CL19 | HCT 116 | C1486(1.04); C1408(1.28); C1417(1.28) | LDD0723 | [10] |
| LDCM0407 | CL2 | HCT 116 | C1408(0.87); C1417(0.87); C1486(0.92) | LDD0724 | [10] |
| LDCM0408 | CL20 | HCT 116 | C1486(1.06); C1408(1.79); C1417(1.79) | LDD0725 | [10] |
| LDCM0409 | CL21 | HCT 116 | C1486(1.03); C1408(1.69); C1417(1.69) | LDD0726 | [10] |
| LDCM0410 | CL22 | HCT 116 | C1486(0.89); C1408(2.66); C1417(2.66) | LDD0727 | [10] |
| LDCM0411 | CL23 | HCT 116 | C1486(0.93); C1408(1.49); C1417(1.49) | LDD0728 | [10] |
| LDCM0412 | CL24 | HCT 116 | C1486(1.04); C1408(1.84); C1417(1.84) | LDD0729 | [10] |
| LDCM0413 | CL25 | HCT 116 | C1408(1.66); C1417(1.66); C1486(0.97) | LDD0730 | [10] |
| LDCM0414 | CL26 | HCT 116 | C1408(1.42); C1417(1.42); C1486(1.05) | LDD0731 | [10] |
| LDCM0415 | CL27 | HCT 116 | C1408(1.65); C1417(1.65); C1486(0.95) | LDD0732 | [10] |
| LDCM0416 | CL28 | HCT 116 | C1408(1.79); C1417(1.79); C1486(1.08) | LDD0733 | [10] |
| LDCM0417 | CL29 | HCT 116 | C1408(1.68); C1417(1.68); C1486(0.98) | LDD0734 | [10] |
| LDCM0418 | CL3 | HCT 116 | C1408(1.07); C1417(1.07); C1486(0.89) | LDD0735 | [10] |
| LDCM0419 | CL30 | HCT 116 | C1408(1.32); C1417(1.32); C1486(0.86) | LDD0736 | [10] |
| LDCM0420 | CL31 | HCT 116 | C1486(0.96); C61(0.85) | LDD0737 | [10] |
| LDCM0421 | CL32 | HCT 116 | C61(0.86) | LDD0738 | [10] |
| LDCM0422 | CL33 | HCT 116 | C61(1.05) | LDD0739 | [10] |
| LDCM0423 | CL34 | HCT 116 | C61(0.90) | LDD0740 | [10] |
| LDCM0424 | CL35 | HCT 116 | C61(0.93) | LDD0741 | [10] |
| LDCM0425 | CL36 | HCT 116 | C61(1.15) | LDD0742 | [10] |
| LDCM0426 | CL37 | HCT 116 | C61(1.02) | LDD0743 | [10] |
| LDCM0428 | CL39 | HCT 116 | C61(0.93) | LDD0745 | [10] |
| LDCM0429 | CL4 | HCT 116 | C1408(1.09); C1417(1.09); C1486(0.95) | LDD0746 | [10] |
| LDCM0430 | CL40 | HCT 116 | C61(0.72) | LDD0747 | [10] |
| LDCM0431 | CL41 | HCT 116 | C61(0.95) | LDD0748 | [10] |
| LDCM0432 | CL42 | HCT 116 | C61(0.90) | LDD0749 | [10] |
| LDCM0433 | CL43 | HCT 116 | C61(1.07) | LDD0750 | [10] |
| LDCM0434 | CL44 | HCT 116 | C61(0.94) | LDD0751 | [10] |
| LDCM0435 | CL45 | HCT 116 | C61(0.88) | LDD0752 | [10] |
| LDCM0436 | CL46 | HCT 116 | C1408(0.64); C1417(0.64); C1486(0.99); C61(1.05) | LDD0753 | [10] |
| LDCM0437 | CL47 | HCT 116 | C1408(0.69); C1417(0.69); C1486(0.90); C61(1.08) | LDD0754 | [10] |
| LDCM0438 | CL48 | HCT 116 | C1408(0.67); C1417(0.67); C1486(1.08); C61(1.09) | LDD0755 | [10] |
| LDCM0439 | CL49 | HCT 116 | C1408(0.84); C1417(0.84); C1486(1.02); C61(0.92) | LDD0756 | [10] |
| LDCM0440 | CL5 | HCT 116 | C1408(1.08); C1417(1.08); C1486(1.02) | LDD0757 | [10] |
| LDCM0441 | CL50 | HCT 116 | C1408(0.82); C1417(0.82); C1486(1.02); C61(0.99) | LDD0758 | [10] |
| LDCM0442 | CL51 | HCT 116 | C1408(1.00); C1417(1.00); C1486(1.01); C61(1.09) | LDD0759 | [10] |
| LDCM0443 | CL52 | HCT 116 | C1408(0.79); C1417(0.79); C1486(1.03); C61(1.28) | LDD0760 | [10] |
| LDCM0444 | CL53 | HCT 116 | C1408(0.79); C1417(0.79); C1486(0.99); C61(1.20) | LDD0761 | [10] |
| LDCM0445 | CL54 | HCT 116 | C1408(0.74); C1417(0.74); C1486(1.01); C61(0.81) | LDD0762 | [10] |
| LDCM0446 | CL55 | HCT 116 | C1408(0.78); C1417(0.78); C1486(0.97); C61(1.00) | LDD0763 | [10] |
| LDCM0447 | CL56 | HCT 116 | C1408(0.76); C1417(0.76); C1486(0.86); C61(0.89) | LDD0764 | [10] |
| LDCM0448 | CL57 | HCT 116 | C1408(0.80); C1417(0.80); C1486(1.06); C61(1.27) | LDD0765 | [10] |
| LDCM0449 | CL58 | HCT 116 | C1408(0.70); C1417(0.70); C1486(1.08); C61(0.89) | LDD0766 | [10] |
| LDCM0450 | CL59 | HCT 116 | C1408(0.96); C1417(0.96); C1486(1.12); C61(0.95) | LDD0767 | [10] |
| LDCM0451 | CL6 | HCT 116 | C1408(1.01); C1417(1.01); C1486(1.06) | LDD0768 | [10] |
| LDCM0452 | CL60 | HCT 116 | C1408(0.76); C1417(0.76); C1486(1.07); C61(1.05) | LDD0769 | [10] |
| LDCM0453 | CL61 | HCT 116 | C1408(1.40); C1417(1.40); C1486(1.20) | LDD0770 | [10] |
| LDCM0454 | CL62 | HCT 116 | C1408(1.27); C1417(1.27); C1486(1.14) | LDD0771 | [10] |
| LDCM0455 | CL63 | HCT 116 | C1408(1.42); C1417(1.42); C1486(1.02) | LDD0772 | [10] |
| LDCM0456 | CL64 | HCT 116 | C1408(1.47); C1417(1.47); C1486(1.12) | LDD0773 | [10] |
| LDCM0457 | CL65 | HCT 116 | C1408(0.85); C1417(0.85); C1486(0.97) | LDD0774 | [10] |
| LDCM0458 | CL66 | HCT 116 | C1408(1.44); C1417(1.44); C1486(1.12) | LDD0775 | [10] |
| LDCM0459 | CL67 | HCT 116 | C1408(1.51); C1417(1.51); C1486(1.01) | LDD0776 | [10] |
| LDCM0460 | CL68 | HCT 116 | C1408(1.30); C1417(1.30); C1486(0.93) | LDD0777 | [10] |
| LDCM0461 | CL69 | HCT 116 | C1408(0.99); C1417(0.99); C1486(1.10) | LDD0778 | [10] |
| LDCM0462 | CL7 | HCT 116 | C1408(1.22); C1417(1.22); C1486(1.16) | LDD0779 | [10] |
| LDCM0463 | CL70 | HCT 116 | C1408(0.97); C1417(0.97); C1486(1.17) | LDD0780 | [10] |
| LDCM0464 | CL71 | HCT 116 | C1408(1.07); C1417(1.07); C1486(1.05) | LDD0781 | [10] |
| LDCM0465 | CL72 | HCT 116 | C1408(0.89); C1417(0.89); C1486(1.35) | LDD0782 | [10] |
| LDCM0466 | CL73 | HCT 116 | C1408(1.19); C1417(1.19); C1486(1.08) | LDD0783 | [10] |
| LDCM0467 | CL74 | HCT 116 | C1408(0.92); C1417(0.92); C1486(1.14) | LDD0784 | [10] |
| LDCM0469 | CL76 | HEK-293T | C1513(0.78); C1899(0.85); C61(1.22) | LDD1067 | [10] |
| LDCM0470 | CL77 | HEK-293T | C1513(1.13); C1899(0.76); C61(1.20) | LDD1068 | [10] |
| LDCM0471 | CL78 | HEK-293T | C1513(0.73); C1899(0.60); C61(1.70) | LDD1069 | [10] |
| LDCM0472 | CL79 | HEK-293T | C1513(0.99); C1899(0.57); C61(1.39) | LDD1070 | [10] |
| LDCM0473 | CL8 | HCT 116 | C1408(1.43); C1417(1.43); C1486(1.03) | LDD0790 | [10] |
| LDCM0474 | CL80 | HEK-293T | C1513(0.72); C1899(0.72); C61(1.37) | LDD1072 | [10] |
| LDCM0475 | CL81 | HEK-293T | C1513(1.03); C1899(0.67); C61(1.26) | LDD1073 | [10] |
| LDCM0476 | CL82 | HEK-293T | C1513(0.99); C1899(0.80); C61(0.94) | LDD1074 | [10] |
| LDCM0477 | CL83 | HEK-293T | C1513(0.94); C1899(0.99); C61(0.87) | LDD1075 | [10] |
| LDCM0478 | CL84 | HEK-293T | C1513(1.18); C1899(0.58); C61(1.38) | LDD1076 | [10] |
| LDCM0479 | CL85 | HEK-293T | C1513(0.79); C1899(0.72); C61(1.26) | LDD1077 | [10] |
| LDCM0480 | CL86 | HEK-293T | C1513(0.70); C1899(0.55); C61(1.61) | LDD1078 | [10] |
| LDCM0481 | CL87 | HEK-293T | C1513(1.29); C1899(0.66); C61(1.12) | LDD1079 | [10] |
| LDCM0482 | CL88 | HEK-293T | C1513(1.01); C1899(0.76); C61(0.92) | LDD1080 | [10] |
| LDCM0483 | CL89 | HEK-293T | C1513(0.86); C1899(0.68); C61(1.17) | LDD1081 | [10] |
| LDCM0484 | CL9 | HCT 116 | C1408(1.08); C1417(1.08); C1486(1.15) | LDD0801 | [10] |
| LDCM0485 | CL90 | HEK-293T | C1513(1.23); C1899(0.62); C61(0.84) | LDD1083 | [10] |
| LDCM0486 | CL91 | HCT 116 | C1408(1.00); C1417(1.00); C1486(1.46) | LDD0803 | [10] |
| LDCM0487 | CL92 | HCT 116 | C1408(0.77); C1417(0.77); C1486(1.19) | LDD0804 | [10] |
| LDCM0488 | CL93 | HCT 116 | C1408(1.01); C1417(1.01); C1486(0.92) | LDD0805 | [10] |
| LDCM0489 | CL94 | HCT 116 | C1408(1.11); C1417(1.11); C1486(0.78) | LDD0806 | [10] |
| LDCM0490 | CL95 | HCT 116 | C1408(1.33); C1417(1.33); C1486(1.07) | LDD0807 | [10] |
| LDCM0491 | CL96 | HCT 116 | C1408(1.24); C1417(1.24); C1486(1.54) | LDD0808 | [10] |
| LDCM0492 | CL97 | HCT 116 | C1408(1.06); C1417(1.06); C1486(1.20) | LDD0809 | [10] |
| LDCM0493 | CL98 | HCT 116 | C1408(1.02); C1417(1.02); C1486(1.16) | LDD0810 | [10] |
| LDCM0494 | CL99 | HCT 116 | C1408(1.11); C1417(1.11); C1486(1.21) | LDD0811 | [10] |
| LDCM0028 | Dobutamine | HEK-293T | 5.38 | LDD0180 | [2] |
| LDCM0027 | Dopamine | HEK-293T | 5.52 | LDD0179 | [2] |
| LDCM0495 | E2913 | HEK-293T | C1513(0.97); C1899(1.21); C1486(0.98); C1408(0.94) | LDD1698 | [24] |
| LDCM0468 | Fragment33 | HCT 116 | C1408(1.58); C1417(1.58); C1486(1.12) | LDD0785 | [10] |
| LDCM0427 | Fragment51 | HCT 116 | C61(0.73) | LDD0744 | [10] |
| LDCM0107 | IAA | HeLa | H613(0.00); H1374(0.00) | LDD0221 | [19] |
| LDCM0022 | KB02 | HCT 116 | C1408(1.93); C1417(1.93); C38(2.51); C43(1.68) | LDD0080 | [10] |
| LDCM0023 | KB03 | HCT 116 | C1408(2.69); C1417(2.69); C38(4.66); C43(2.00) | LDD0081 | [10] |
| LDCM0024 | KB05 | HCT 116 | C1408(5.82); C1417(5.82); C38(5.52); C43(2.33) | LDD0082 | [10] |
| LDCM0030 | Luteolin | HEK-293T | 8.91 | LDD0182 | [2] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C1486(0.59) | LDD2121 | [8] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [19] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C1486(0.65) | LDD2089 | [8] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C1486(0.89) | LDD2092 | [8] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C1486(1.28) | LDD2094 | [8] |
| LDCM0503 | Nucleophilic fragment 14b | MDA-MB-231 | C1486(0.94) | LDD2096 | [8] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C1486(0.65) | LDD2099 | [8] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C1486(0.58) | LDD2104 | [8] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C1486(1.69) | LDD2105 | [8] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C1486(0.50) | LDD2106 | [8] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C1486(0.47) | LDD2108 | [8] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C1486(0.60) | LDD2109 | [8] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C1486(0.76) | LDD2111 | [8] |
| LDCM0523 | Nucleophilic fragment 24b | MDA-MB-231 | C1486(1.02) | LDD2116 | [8] |
| LDCM0529 | Nucleophilic fragment 27b | MDA-MB-231 | C1486(0.87) | LDD2122 | [8] |
| LDCM0531 | Nucleophilic fragment 28b | MDA-MB-231 | C1486(1.04) | LDD2124 | [8] |
| LDCM0533 | Nucleophilic fragment 29b | MDA-MB-231 | C1486(1.02) | LDD2126 | [8] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C1486(0.76) | LDD2129 | [8] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C1486(0.64) | LDD2135 | [8] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C1486(1.02) | LDD2136 | [8] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C1486(0.72) | LDD2137 | [8] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C1486(1.14) | LDD1700 | [8] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C1486(0.64) | LDD2140 | [8] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C1486(0.65) | LDD2141 | [8] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C1486(0.70) | LDD2146 | [8] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C1486(0.41) | LDD2150 | [8] |
| LDCM0557 | Nucleophilic fragment 8b | MDA-MB-231 | C1486(0.99) | LDD2151 | [8] |
| LDCM0021 | THZ1 | HCT 116 | C1408(0.98); C1417(0.98) | LDD2173 | [10] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| PR domain zinc finger protein 14 (PRDM14) | Class V-like SAM-binding methyltransferase superfamily | Q9GZV8 | |||
| Zinc finger protein RFP (TRIM27) | TRIM/RBCC family | P14373 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Chloride intracellular channel protein 3 (CLIC3) | Chloride channel CLIC family | O95833 | |||
Other
References
