Details of the Target
General Information of Target
| Target ID | LDTP04327 | |||||
|---|---|---|---|---|---|---|
| Target Name | Cytosolic purine 5'-nucleotidase (NT5C2) | |||||
| Gene Name | NT5C2 | |||||
| Gene ID | 22978 | |||||
| Synonyms |
NT5B; NT5CP; PNT5; Cytosolic purine 5'-nucleotidase; EC 3.1.3.5; EC 3.1.3.99; Cytosolic 5'-nucleotidase II; cN-II; Cytosolic IMP/GMP-specific 5'-nucleotidase; Cytosolic nucleoside phosphotransferase 5'N; EC 2.7.1.77; High Km 5'-nucleotidase
|
|||||
| 3D Structure | ||||||
| Sequence |
MSTSWSDRLQNAADMPANMDKHALKKYRREAYHRVFVNRSLAMEKIKCFGFDMDYTLAVY
KSPEYESLGFELTVERLVSIGYPQELLSFAYDSTFPTRGLVFDTLYGNLLKVDAYGNLLV CAHGFNFIRGPETREQYPNKFIQRDDTERFYILNTLFNLPETYLLACLVDFFTNCPRYTS CETGFKDGDLFMSYRSMFQDVRDAVDWVHYKGSLKEKTVENLEKYVVKDGKLPLLLSRMK EVGKVFLATNSDYKYTDKIMTYLFDFPHGPKPGSSHRPWQSYFDLILVDARKPLFFGEGT VLRQVDTKTGKLKIGTYTGPLQHGIVYSGGSSDTICDLLGAKGKDILYIGDHIFGDILKS KKRQGWRTFLVIPELAQELHVWTDKSSLFEELQSLDIFLAELYKHLDSSSNERPDISSIQ RRIKKVTHDMDMCYGMMGSLFRSGSRQTLFASQVMRYADLYAASFINLLYYPFSYLFRAA HVLMPHESTVEHTHVDINEMESPLATRNRTSVDFKDTDYKRHQLTRSISEIKPPNLFPLA PQEITHCHDEDDDEEEEEEEE |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
5'(3')-deoxyribonucleotidase family
|
|||||
| Subcellular location |
Cytoplasm, cytosol
|
|||||
| Function |
Broad specificity cytosolic 5'-nucleotidase that catalyzes the dephosphorylation of 6-hydroxypurine nucleoside 5'-monophosphates. In addition, possesses a phosphotransferase activity by which it can transfer a phosphate from a donor nucleoside monophosphate to an acceptor nucleoside, preferably inosine, deoxyinosine and guanosine. Has the highest activities for IMP and GMP followed by dIMP, dGMP and XMP. Could also catalyze the transfer of phosphates from pyrimidine monophosphates but with lower efficiency. Through these activities regulates the purine nucleoside/nucleotide pools within the cell.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| A549 | SNV: p.N250K | DBIA Probe Info | |||
| C4II | SNV: p.V78I | DBIA Probe Info | |||
| HCT15 | Deletion: p.Q542RfsTer71 | DBIA Probe Info | |||
| HT | SNV: p.Y60H; p.T316A | . | |||
| KASUMI1 | SNV: p.H123R | . | |||
| NCIH2172 | SNV: p.F49S | DBIA Probe Info | |||
| RKO | Deletion: p.F354LfsTer5 | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
14.62 | LDD0402 | [1] | |
|
AZ-5 Probe Info |
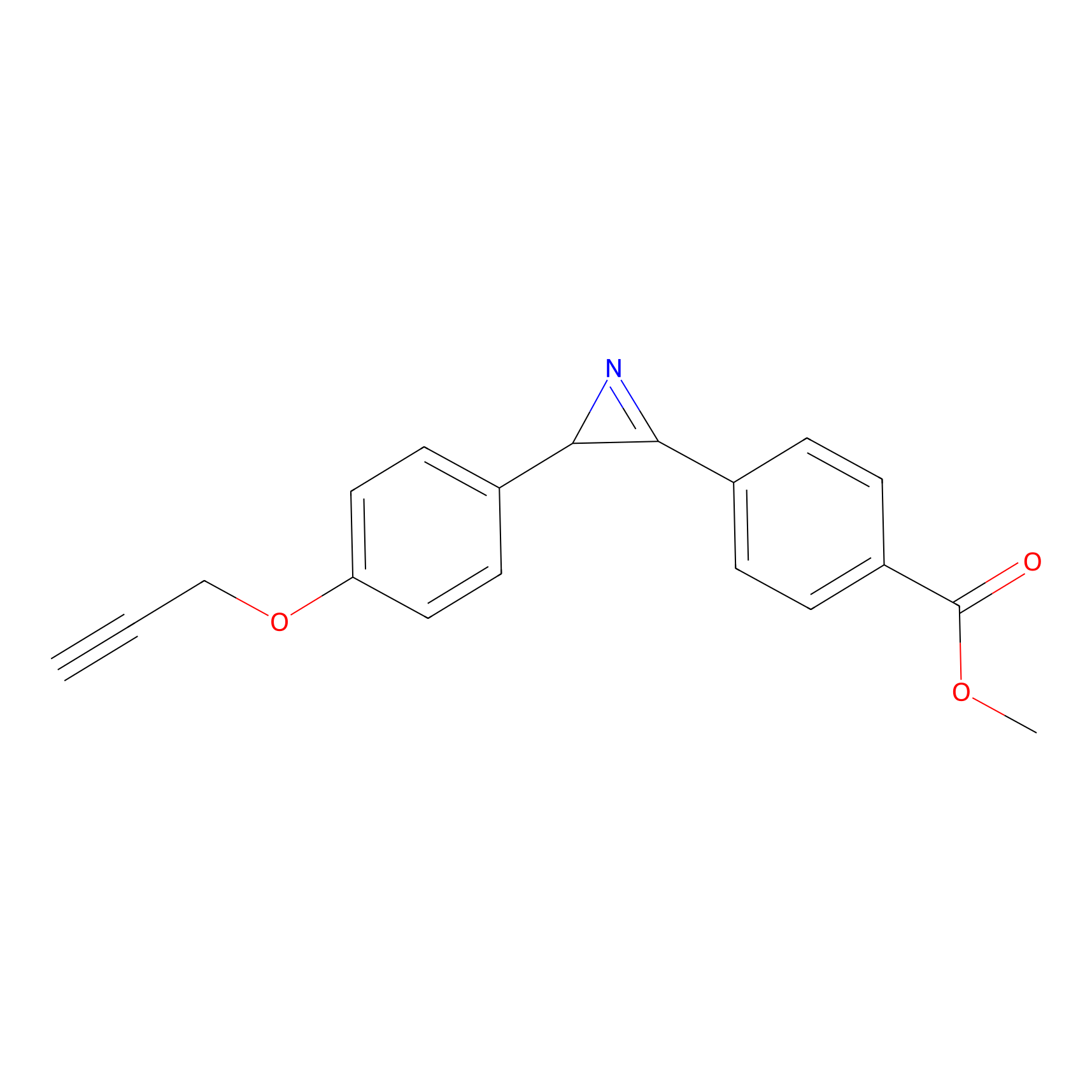 |
10.00 | LDD0394 | [2] | |
|
STPyne Probe Info |
 |
K140(7.14); K21(9.11); K231(10.00); K240(3.55) | LDD0277 | [3] | |
|
BTD Probe Info |
 |
C181(5.09) | LDD1699 | [4] | |
|
Probe 1 Probe Info |
 |
Y210(41.07) | LDD3495 | [5] | |
|
HHS-475 Probe Info |
 |
Y210(0.87) | LDD0264 | [6] | |
|
DBIA Probe Info |
 |
C181(0.89) | LDD0078 | [7] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [8] | |
|
ATP probe Probe Info |
 |
K211(0.00); K215(0.00); K140(0.00); K224(0.00) | LDD0199 | [9] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C336(0.00); C121(0.00); C433(0.00); C181(0.00) | LDD0038 | [10] | |
|
IA-alkyne Probe Info |
 |
C121(0.00); C433(0.00); C181(0.00); C336(0.00) | LDD0036 | [10] | |
|
IPIAA_L Probe Info |
 |
N.A. | LDD0031 | [11] | |
|
Lodoacetamide azide Probe Info |
 |
C336(0.00); C121(0.00); C433(0.00); C181(0.00) | LDD0037 | [10] | |
|
ATP probe Probe Info |
 |
K140(0.00); K244(0.00); K313(0.00) | LDD0035 | [12] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [13] | |
|
WYneO Probe Info |
 |
N.A. | LDD0022 | [13] | |
|
ENE Probe Info |
 |
N.A. | LDD0006 | [13] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [13] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [13] | |
|
Acrolein Probe Info |
 |
H352(0.00); C181(0.00) | LDD0217 | [14] | |
|
AOyne Probe Info |
 |
8.70 | LDD0443 | [15] | |
|
NAIA_5 Probe Info |
 |
C121(0.00); C433(0.00); C547(0.00); C181(0.00) | LDD2223 | [16] | |
|
HHS-465 Probe Info |
 |
Y210(0.00); K211(0.00) | LDD2240 | [17] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C004 Probe Info |
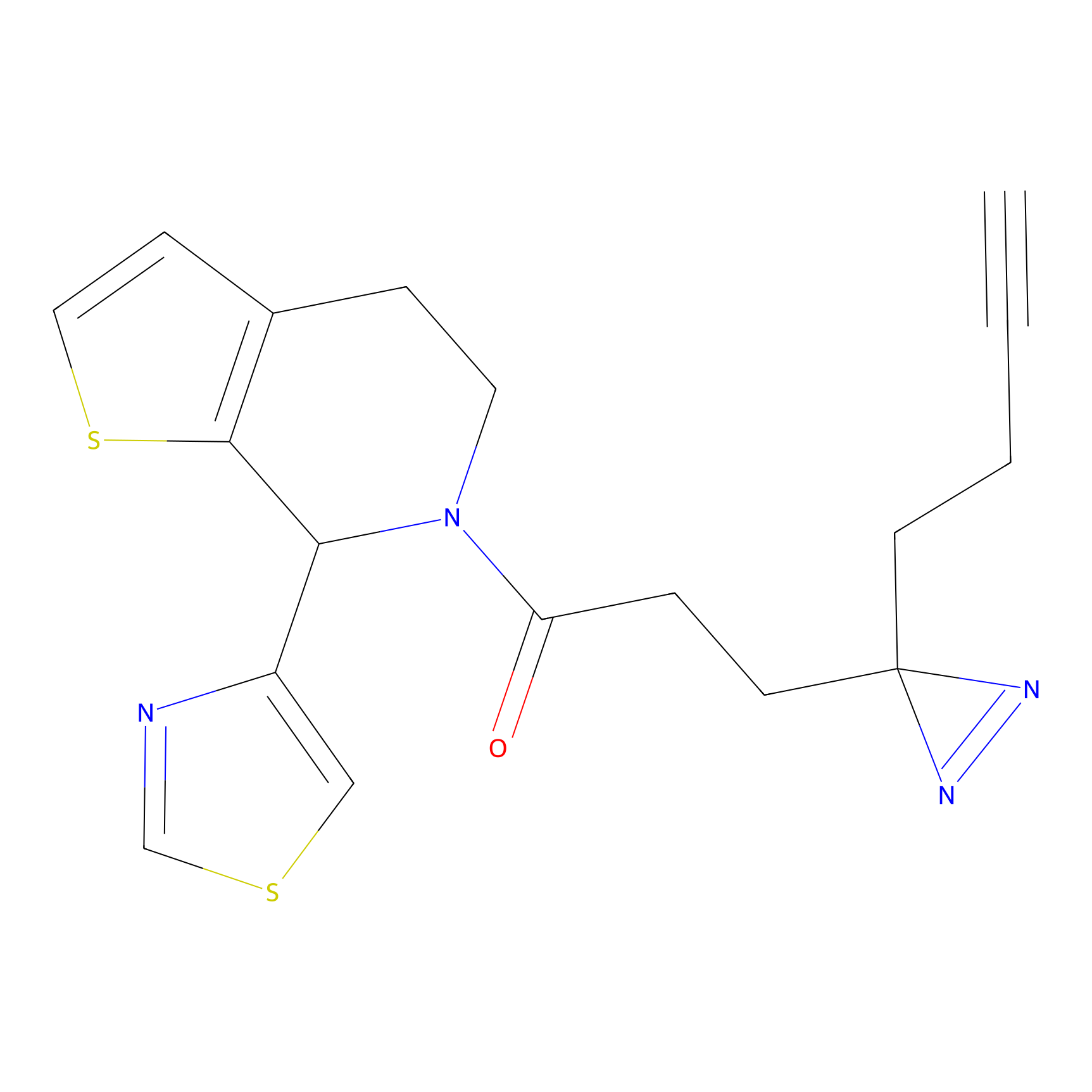 |
7.16 | LDD1714 | [18] | |
|
C092 Probe Info |
 |
22.47 | LDD1783 | [18] | |
|
C160 Probe Info |
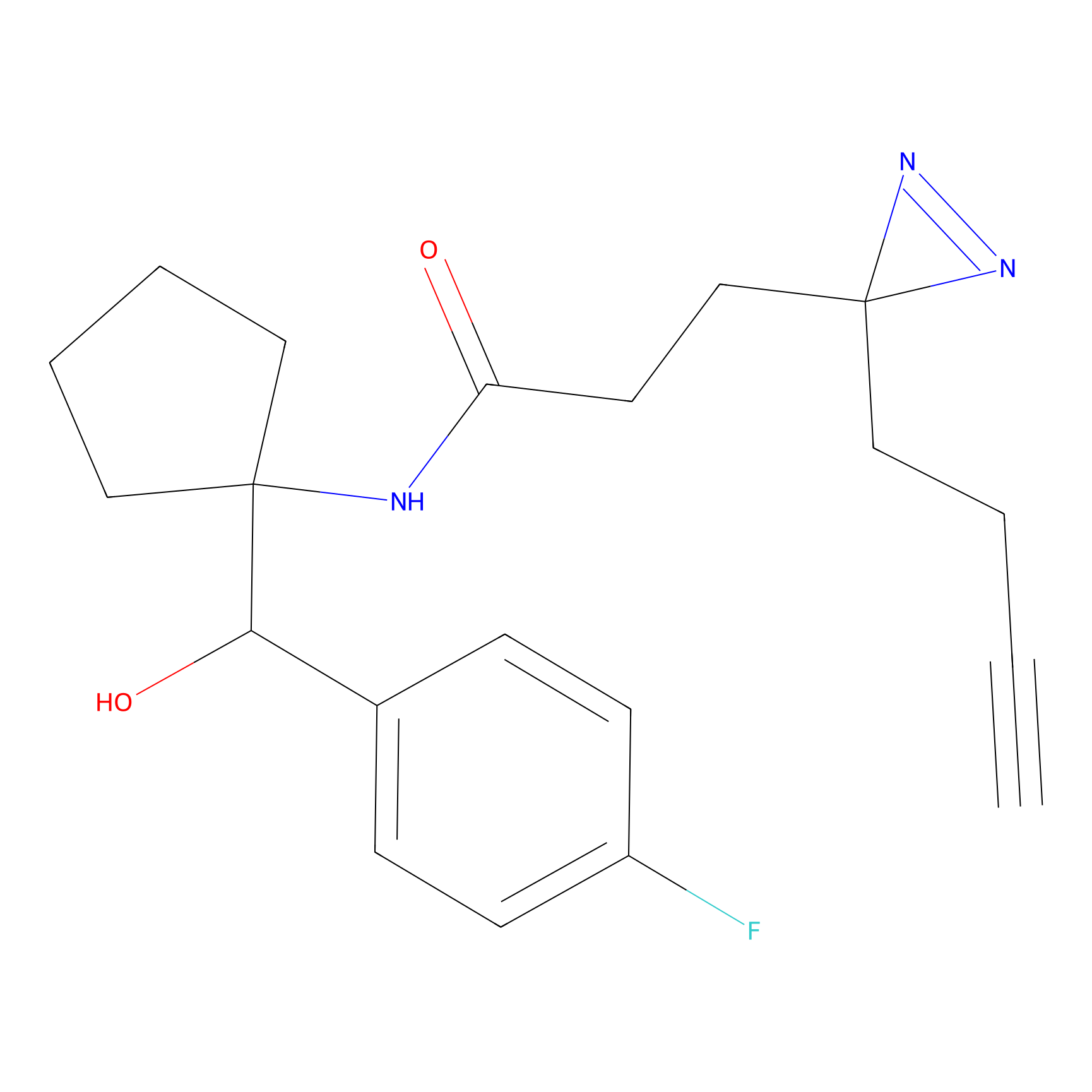 |
35.02 | LDD1840 | [18] | |
|
C177 Probe Info |
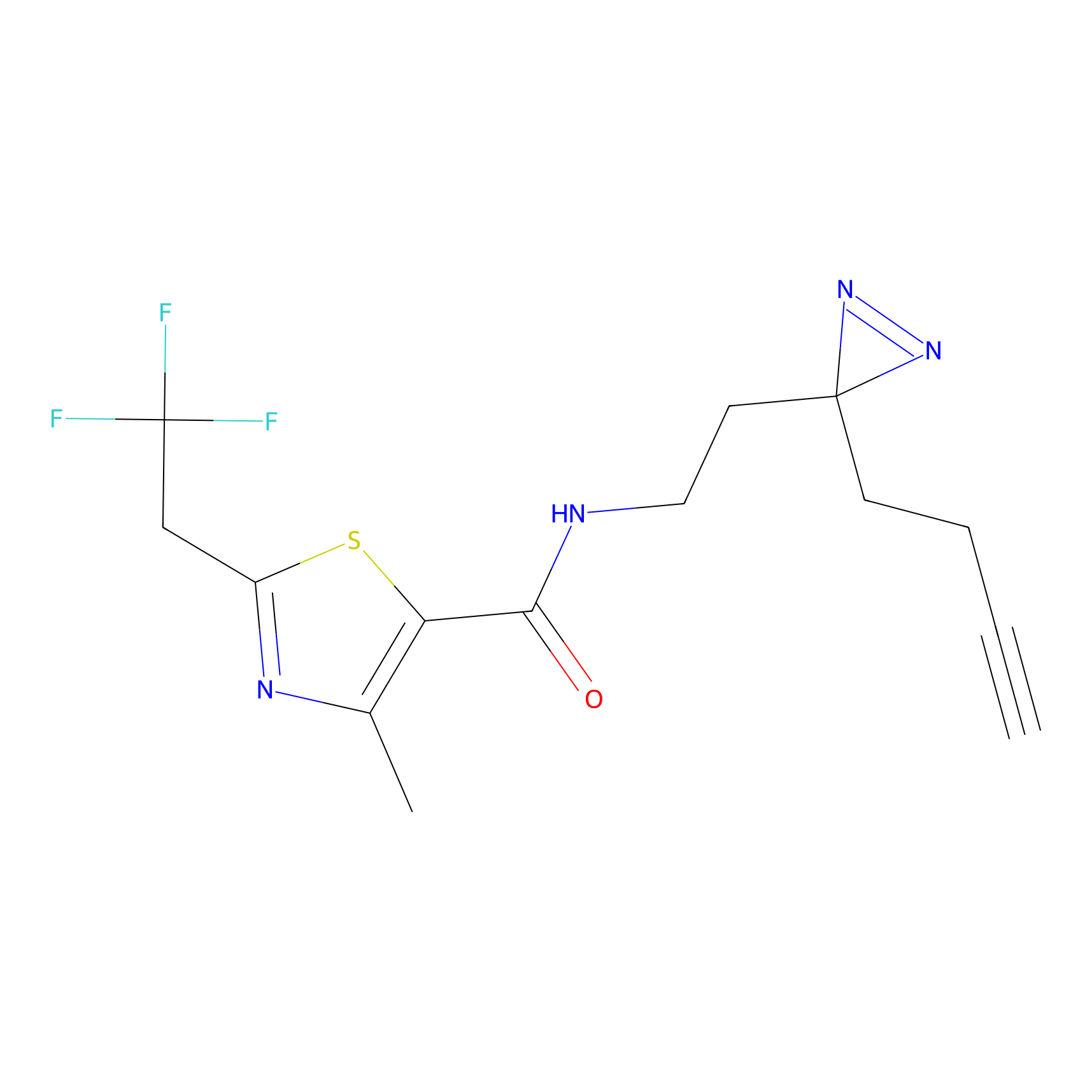 |
8.69 | LDD1856 | [18] | |
|
C232 Probe Info |
 |
35.26 | LDD1905 | [18] | |
|
C361 Probe Info |
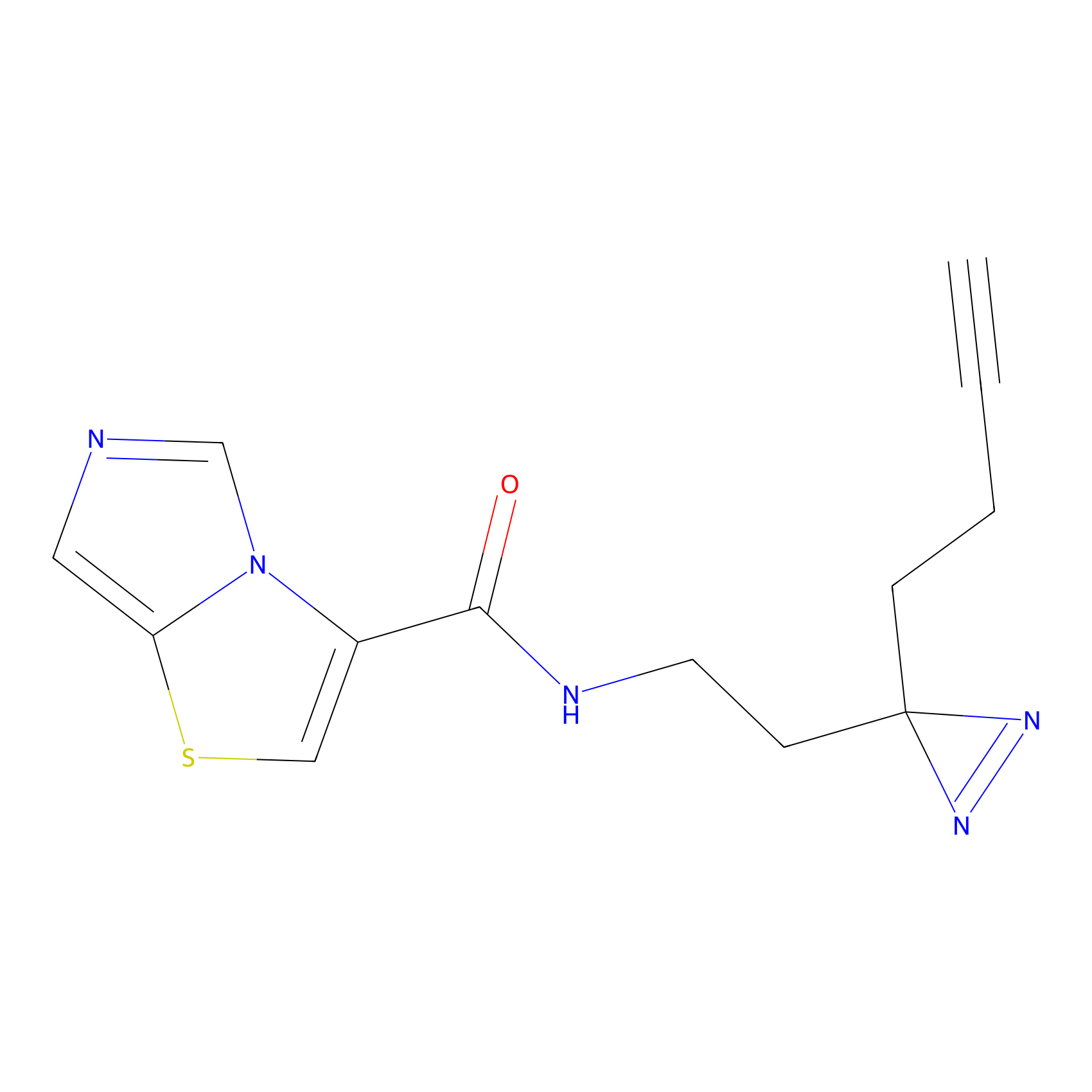 |
26.91 | LDD2022 | [18] | |
|
C422 Probe Info |
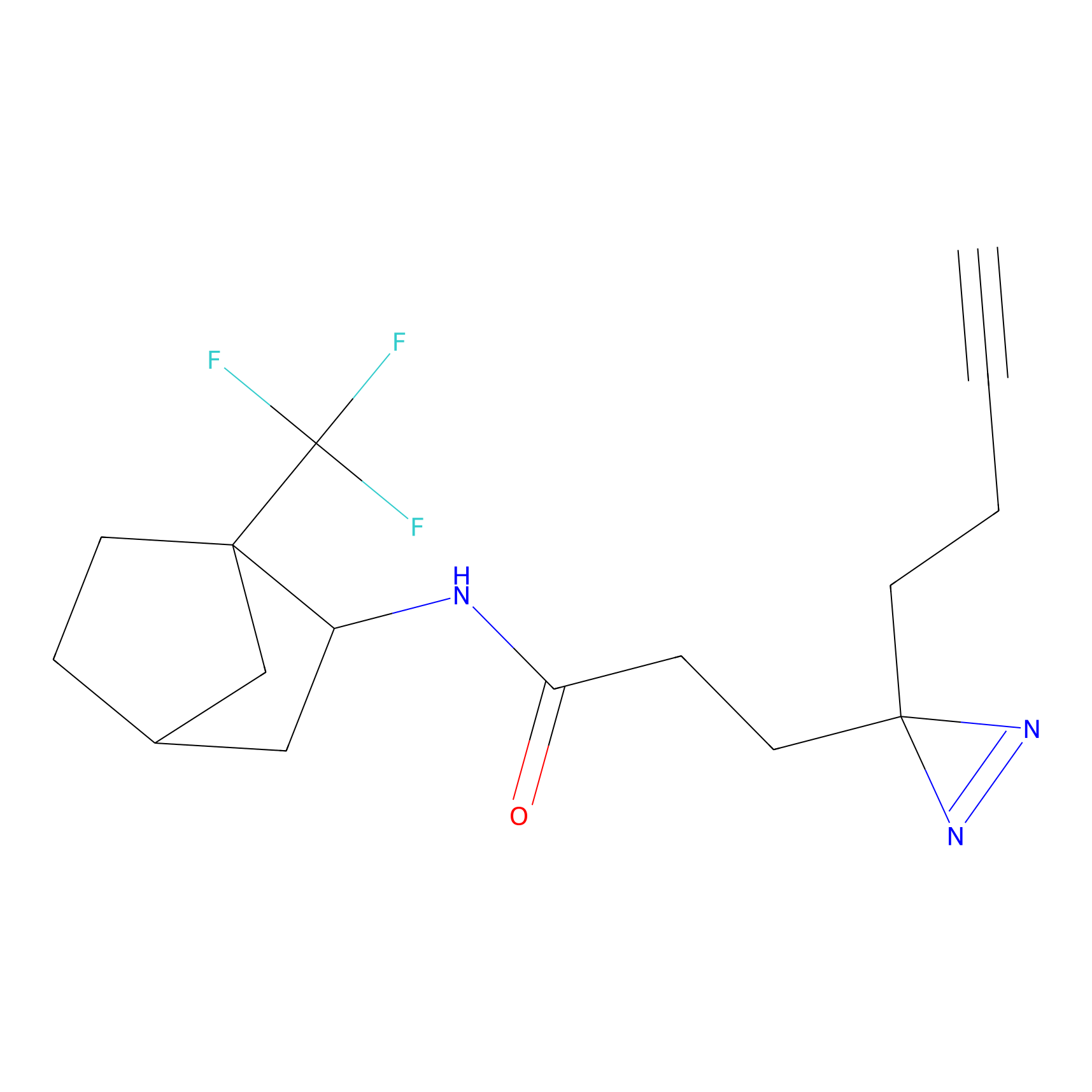 |
8.22 | LDD2077 | [18] | |
|
FFF probe13 Probe Info |
 |
13.29 | LDD0475 | [19] | |
|
FFF probe14 Probe Info |
 |
20.00 | LDD0477 | [19] | |
|
FFF probe15 Probe Info |
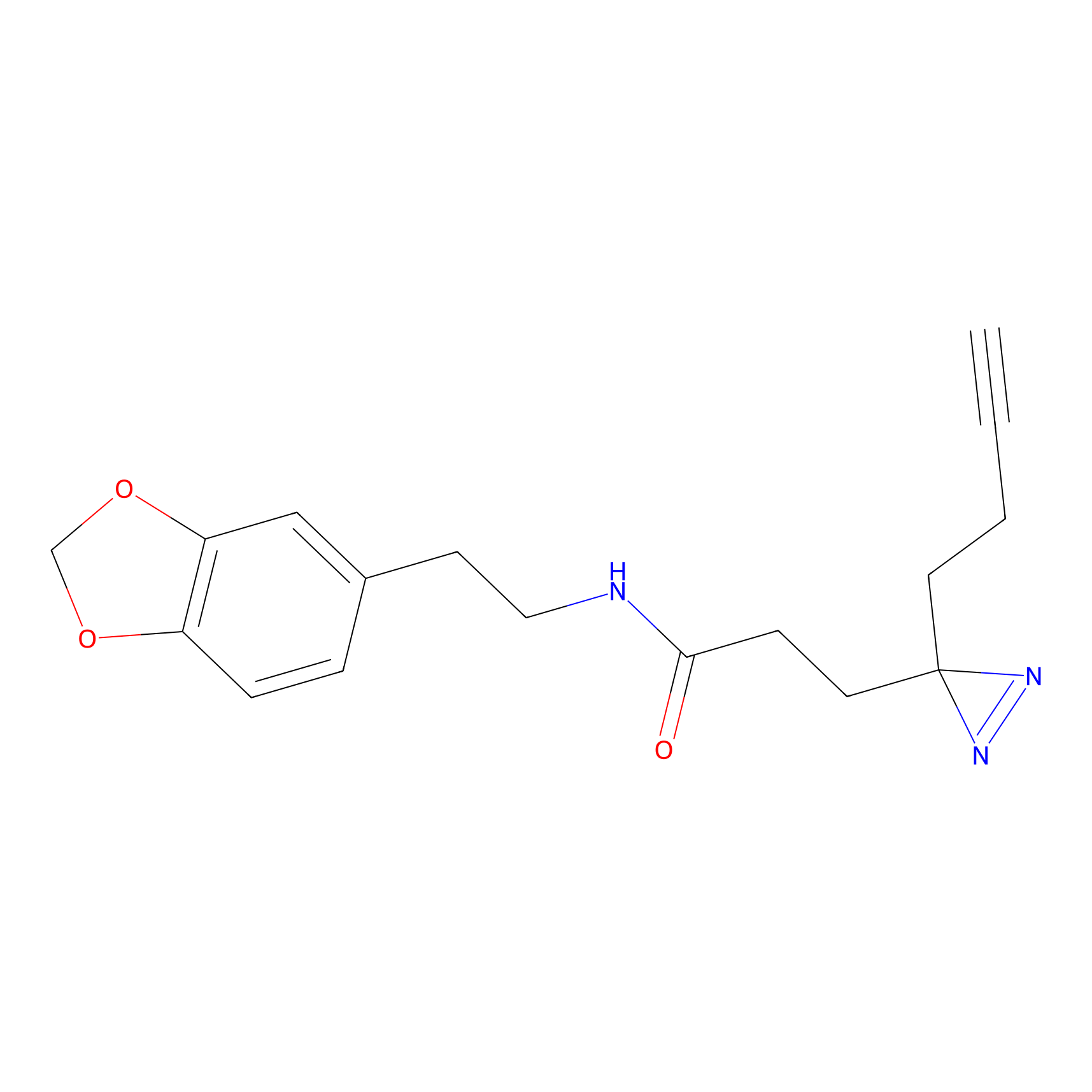 |
5.66 | LDD0478 | [19] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0214 | AC1 | HCT 116 | C181(1.34) | LDD0531 | [7] |
| LDCM0215 | AC10 | HCT 116 | C181(1.19); C433(0.94) | LDD0532 | [7] |
| LDCM0216 | AC100 | HCT 116 | C181(1.03); C433(1.30) | LDD0533 | [7] |
| LDCM0217 | AC101 | HCT 116 | C181(0.95); C433(1.21) | LDD0534 | [7] |
| LDCM0218 | AC102 | HCT 116 | C181(1.15); C433(0.96) | LDD0535 | [7] |
| LDCM0219 | AC103 | HCT 116 | C181(1.35); C433(1.07) | LDD0536 | [7] |
| LDCM0220 | AC104 | HCT 116 | C181(1.09); C433(1.53) | LDD0537 | [7] |
| LDCM0221 | AC105 | HCT 116 | C181(1.05); C433(1.09) | LDD0538 | [7] |
| LDCM0222 | AC106 | HCT 116 | C181(1.24); C433(1.09) | LDD0539 | [7] |
| LDCM0223 | AC107 | HCT 116 | C181(1.15); C433(1.16) | LDD0540 | [7] |
| LDCM0224 | AC108 | HCT 116 | C181(1.12); C433(1.13) | LDD0541 | [7] |
| LDCM0225 | AC109 | HCT 116 | C181(0.94); C433(1.09) | LDD0542 | [7] |
| LDCM0226 | AC11 | HCT 116 | C181(1.33); C433(0.72) | LDD0543 | [7] |
| LDCM0227 | AC110 | HCT 116 | C181(1.04); C433(1.27) | LDD0544 | [7] |
| LDCM0228 | AC111 | HCT 116 | C181(0.91); C433(0.75) | LDD0545 | [7] |
| LDCM0229 | AC112 | HCT 116 | C181(1.18); C433(1.22) | LDD0546 | [7] |
| LDCM0230 | AC113 | HCT 116 | C181(1.05); C433(0.98) | LDD0547 | [7] |
| LDCM0231 | AC114 | HCT 116 | C181(1.17); C433(1.04) | LDD0548 | [7] |
| LDCM0232 | AC115 | HCT 116 | C181(1.37); C433(1.00) | LDD0549 | [7] |
| LDCM0233 | AC116 | HCT 116 | C181(1.25); C433(1.02) | LDD0550 | [7] |
| LDCM0234 | AC117 | HCT 116 | C181(1.13); C433(1.23) | LDD0551 | [7] |
| LDCM0235 | AC118 | HCT 116 | C181(1.13); C433(1.02) | LDD0552 | [7] |
| LDCM0236 | AC119 | HCT 116 | C181(1.26); C433(1.15) | LDD0553 | [7] |
| LDCM0237 | AC12 | HCT 116 | C181(1.31); C433(0.96) | LDD0554 | [7] |
| LDCM0238 | AC120 | HCT 116 | C181(1.30); C433(1.09) | LDD0555 | [7] |
| LDCM0239 | AC121 | HCT 116 | C181(1.03); C433(1.33) | LDD0556 | [7] |
| LDCM0240 | AC122 | HCT 116 | C181(1.23); C433(1.00) | LDD0557 | [7] |
| LDCM0241 | AC123 | HCT 116 | C181(1.16); C433(1.32) | LDD0558 | [7] |
| LDCM0242 | AC124 | HCT 116 | C181(0.98); C433(1.13) | LDD0559 | [7] |
| LDCM0243 | AC125 | HCT 116 | C181(1.09); C433(0.94) | LDD0560 | [7] |
| LDCM0244 | AC126 | HCT 116 | C181(1.26); C433(1.03) | LDD0561 | [7] |
| LDCM0245 | AC127 | HCT 116 | C181(1.22); C433(1.18) | LDD0562 | [7] |
| LDCM0246 | AC128 | HCT 116 | C181(0.91) | LDD0563 | [7] |
| LDCM0247 | AC129 | HCT 116 | C181(0.98) | LDD0564 | [7] |
| LDCM0249 | AC130 | HCT 116 | C181(0.94) | LDD0566 | [7] |
| LDCM0250 | AC131 | HCT 116 | C181(0.82) | LDD0567 | [7] |
| LDCM0251 | AC132 | HCT 116 | C181(1.00) | LDD0568 | [7] |
| LDCM0252 | AC133 | HCT 116 | C181(0.97) | LDD0569 | [7] |
| LDCM0253 | AC134 | HCT 116 | C181(1.00) | LDD0570 | [7] |
| LDCM0254 | AC135 | HCT 116 | C181(1.02) | LDD0571 | [7] |
| LDCM0255 | AC136 | HCT 116 | C181(1.03) | LDD0572 | [7] |
| LDCM0256 | AC137 | HCT 116 | C181(0.99) | LDD0573 | [7] |
| LDCM0257 | AC138 | HCT 116 | C181(1.16) | LDD0574 | [7] |
| LDCM0258 | AC139 | HCT 116 | C181(1.10) | LDD0575 | [7] |
| LDCM0259 | AC14 | HCT 116 | C181(1.16); C433(0.83) | LDD0576 | [7] |
| LDCM0260 | AC140 | HCT 116 | C181(1.22) | LDD0577 | [7] |
| LDCM0261 | AC141 | HCT 116 | C181(1.07) | LDD0578 | [7] |
| LDCM0262 | AC142 | HCT 116 | C181(1.11) | LDD0579 | [7] |
| LDCM0263 | AC143 | HCT 116 | C181(1.09) | LDD0580 | [7] |
| LDCM0264 | AC144 | HCT 116 | C181(1.21) | LDD0581 | [7] |
| LDCM0265 | AC145 | HCT 116 | C181(1.10) | LDD0582 | [7] |
| LDCM0266 | AC146 | HCT 116 | C181(1.33) | LDD0583 | [7] |
| LDCM0267 | AC147 | HCT 116 | C181(1.16) | LDD0584 | [7] |
| LDCM0268 | AC148 | HCT 116 | C181(1.40) | LDD0585 | [7] |
| LDCM0269 | AC149 | HCT 116 | C181(1.29) | LDD0586 | [7] |
| LDCM0270 | AC15 | HCT 116 | C433(0.60); C181(1.18) | LDD0587 | [7] |
| LDCM0271 | AC150 | HCT 116 | C181(1.09) | LDD0588 | [7] |
| LDCM0272 | AC151 | HCT 116 | C181(1.18) | LDD0589 | [7] |
| LDCM0273 | AC152 | HCT 116 | C181(1.48) | LDD0590 | [7] |
| LDCM0274 | AC153 | HCT 116 | C181(1.72) | LDD0591 | [7] |
| LDCM0621 | AC154 | HCT 116 | C181(1.20) | LDD2158 | [7] |
| LDCM0622 | AC155 | HCT 116 | C181(1.11) | LDD2159 | [7] |
| LDCM0623 | AC156 | HCT 116 | C181(1.11) | LDD2160 | [7] |
| LDCM0624 | AC157 | HCT 116 | C181(0.94) | LDD2161 | [7] |
| LDCM0276 | AC17 | HCT 116 | C433(0.53); C181(1.12) | LDD0593 | [7] |
| LDCM0277 | AC18 | HCT 116 | C433(0.46); C181(1.33) | LDD0594 | [7] |
| LDCM0278 | AC19 | HCT 116 | C433(1.00); C181(1.21) | LDD0595 | [7] |
| LDCM0279 | AC2 | HCT 116 | C181(1.21) | LDD0596 | [7] |
| LDCM0280 | AC20 | HCT 116 | C433(0.68); C181(1.25) | LDD0597 | [7] |
| LDCM0281 | AC21 | HCT 116 | C433(0.94); C181(1.12) | LDD0598 | [7] |
| LDCM0282 | AC22 | HCT 116 | C433(0.73); C181(1.11) | LDD0599 | [7] |
| LDCM0283 | AC23 | HCT 116 | C433(0.79); C181(1.10) | LDD0600 | [7] |
| LDCM0284 | AC24 | HCT 116 | C181(0.89); C433(0.96) | LDD0601 | [7] |
| LDCM0285 | AC25 | HCT 116 | C181(1.06); C433(1.15) | LDD0602 | [7] |
| LDCM0286 | AC26 | HCT 116 | C433(0.86); C181(1.28) | LDD0603 | [7] |
| LDCM0287 | AC27 | HCT 116 | C433(0.91); C181(1.17) | LDD0604 | [7] |
| LDCM0288 | AC28 | HCT 116 | C181(1.17); C433(1.29) | LDD0605 | [7] |
| LDCM0289 | AC29 | HCT 116 | C181(1.04); C433(1.26) | LDD0606 | [7] |
| LDCM0290 | AC3 | HCT 116 | C181(1.21) | LDD0607 | [7] |
| LDCM0291 | AC30 | HCT 116 | C181(1.20); C433(1.93) | LDD0608 | [7] |
| LDCM0292 | AC31 | HCT 116 | C181(1.15); C433(1.69) | LDD0609 | [7] |
| LDCM0293 | AC32 | HCT 116 | C433(0.90); C181(1.63) | LDD0610 | [7] |
| LDCM0294 | AC33 | HCT 116 | C433(1.00); C181(1.24) | LDD0611 | [7] |
| LDCM0295 | AC34 | HCT 116 | C433(1.29); C181(1.32) | LDD0612 | [7] |
| LDCM0296 | AC35 | HCT 116 | C181(0.84) | LDD0613 | [7] |
| LDCM0297 | AC36 | HCT 116 | C181(0.95) | LDD0614 | [7] |
| LDCM0298 | AC37 | HCT 116 | C181(0.85) | LDD0615 | [7] |
| LDCM0299 | AC38 | HCT 116 | C181(1.00) | LDD0616 | [7] |
| LDCM0300 | AC39 | HCT 116 | C181(1.01) | LDD0617 | [7] |
| LDCM0301 | AC4 | HCT 116 | C181(1.29) | LDD0618 | [7] |
| LDCM0302 | AC40 | HCT 116 | C181(0.87) | LDD0619 | [7] |
| LDCM0303 | AC41 | HCT 116 | C181(0.84) | LDD0620 | [7] |
| LDCM0304 | AC42 | HCT 116 | C181(0.90) | LDD0621 | [7] |
| LDCM0305 | AC43 | HCT 116 | C181(0.91) | LDD0622 | [7] |
| LDCM0306 | AC44 | HCT 116 | C181(0.83) | LDD0623 | [7] |
| LDCM0307 | AC45 | HCT 116 | C181(0.82) | LDD0624 | [7] |
| LDCM0308 | AC46 | HCT 116 | C433(0.97); C181(1.09) | LDD0625 | [7] |
| LDCM0309 | AC47 | HCT 116 | C433(1.08); C181(1.09) | LDD0626 | [7] |
| LDCM0310 | AC48 | HCT 116 | C433(0.63); C181(1.26) | LDD0627 | [7] |
| LDCM0311 | AC49 | HCT 116 | C433(0.54); C181(1.26) | LDD0628 | [7] |
| LDCM0312 | AC5 | HCT 116 | C181(1.38) | LDD0629 | [7] |
| LDCM0313 | AC50 | HCT 116 | C433(0.60); C181(1.21) | LDD0630 | [7] |
| LDCM0314 | AC51 | HCT 116 | C433(0.87); C181(1.13) | LDD0631 | [7] |
| LDCM0315 | AC52 | HCT 116 | C433(1.20); C181(1.30) | LDD0632 | [7] |
| LDCM0316 | AC53 | HCT 116 | C433(0.77); C181(1.03) | LDD0633 | [7] |
| LDCM0317 | AC54 | HCT 116 | C433(0.68); C181(1.16) | LDD0634 | [7] |
| LDCM0318 | AC55 | HCT 116 | C433(0.46); C181(1.14) | LDD0635 | [7] |
| LDCM0319 | AC56 | HCT 116 | C433(0.38); C181(1.24) | LDD0636 | [7] |
| LDCM0320 | AC57 | HCT 116 | C181(1.19) | LDD0637 | [7] |
| LDCM0321 | AC58 | HCT 116 | C181(1.26) | LDD0638 | [7] |
| LDCM0322 | AC59 | HCT 116 | C181(1.44) | LDD0639 | [7] |
| LDCM0323 | AC6 | HCT 116 | C433(0.70); C181(1.22) | LDD0640 | [7] |
| LDCM0324 | AC60 | HCT 116 | C181(1.28) | LDD0641 | [7] |
| LDCM0325 | AC61 | HCT 116 | C181(1.28) | LDD0642 | [7] |
| LDCM0326 | AC62 | HCT 116 | C181(1.43) | LDD0643 | [7] |
| LDCM0327 | AC63 | HCT 116 | C181(1.37) | LDD0644 | [7] |
| LDCM0328 | AC64 | HCT 116 | C181(1.32) | LDD0645 | [7] |
| LDCM0329 | AC65 | HCT 116 | C181(1.59) | LDD0646 | [7] |
| LDCM0330 | AC66 | HCT 116 | C181(1.56) | LDD0647 | [7] |
| LDCM0331 | AC67 | HCT 116 | C181(1.53) | LDD0648 | [7] |
| LDCM0332 | AC68 | HCT 116 | C433(0.59); C181(1.11) | LDD0649 | [7] |
| LDCM0333 | AC69 | HCT 116 | C433(0.46); C181(1.12) | LDD0650 | [7] |
| LDCM0334 | AC7 | HCT 116 | C181(1.13); C433(1.17) | LDD0651 | [7] |
| LDCM0335 | AC70 | HCT 116 | C433(0.42); C181(1.08) | LDD0652 | [7] |
| LDCM0336 | AC71 | HCT 116 | C433(0.53); C181(0.80) | LDD0653 | [7] |
| LDCM0337 | AC72 | HCT 116 | C433(0.63); C181(1.23) | LDD0654 | [7] |
| LDCM0338 | AC73 | HCT 116 | C433(0.24); C181(1.33) | LDD0655 | [7] |
| LDCM0339 | AC74 | HCT 116 | C433(0.22); C181(1.00) | LDD0656 | [7] |
| LDCM0340 | AC75 | HCT 116 | C433(0.19); C181(1.28) | LDD0657 | [7] |
| LDCM0341 | AC76 | HCT 116 | C433(0.48); C181(1.12) | LDD0658 | [7] |
| LDCM0342 | AC77 | HCT 116 | C433(0.48); C181(0.92) | LDD0659 | [7] |
| LDCM0343 | AC78 | HCT 116 | C433(0.48); C181(0.96) | LDD0660 | [7] |
| LDCM0344 | AC79 | HCT 116 | C433(0.80); C181(1.13) | LDD0661 | [7] |
| LDCM0345 | AC8 | HCT 116 | C433(0.86); C181(1.39) | LDD0662 | [7] |
| LDCM0346 | AC80 | HCT 116 | C433(0.52); C181(1.08) | LDD0663 | [7] |
| LDCM0347 | AC81 | HCT 116 | C433(0.62); C181(1.05) | LDD0664 | [7] |
| LDCM0348 | AC82 | HCT 116 | C433(0.19); C181(1.18) | LDD0665 | [7] |
| LDCM0349 | AC83 | HCT 116 | C433(1.15); C181(1.19) | LDD0666 | [7] |
| LDCM0350 | AC84 | HCT 116 | C433(1.00); C181(1.28) | LDD0667 | [7] |
| LDCM0351 | AC85 | HCT 116 | C433(1.01); C181(1.30) | LDD0668 | [7] |
| LDCM0352 | AC86 | HCT 116 | C433(0.94); C181(1.05) | LDD0669 | [7] |
| LDCM0353 | AC87 | HCT 116 | C433(0.89); C181(1.15) | LDD0670 | [7] |
| LDCM0354 | AC88 | HCT 116 | C181(1.22); C433(1.24) | LDD0671 | [7] |
| LDCM0355 | AC89 | HCT 116 | C433(0.99); C181(1.21) | LDD0672 | [7] |
| LDCM0357 | AC90 | HCT 116 | C433(0.94); C181(1.16) | LDD0674 | [7] |
| LDCM0358 | AC91 | HCT 116 | C181(1.11); C433(1.26) | LDD0675 | [7] |
| LDCM0359 | AC92 | HCT 116 | C433(1.21); C181(1.29) | LDD0676 | [7] |
| LDCM0360 | AC93 | HCT 116 | C433(1.02); C181(1.37) | LDD0677 | [7] |
| LDCM0361 | AC94 | HCT 116 | C181(1.08); C433(1.23) | LDD0678 | [7] |
| LDCM0362 | AC95 | HCT 116 | C181(1.23); C433(1.41) | LDD0679 | [7] |
| LDCM0363 | AC96 | HCT 116 | C181(1.08); C433(1.22) | LDD0680 | [7] |
| LDCM0364 | AC97 | HCT 116 | C433(1.05); C181(1.27) | LDD0681 | [7] |
| LDCM0365 | AC98 | HCT 116 | C433(1.26); C181(1.27) | LDD0682 | [7] |
| LDCM0366 | AC99 | HCT 116 | C181(1.07); C433(1.25) | LDD0683 | [7] |
| LDCM0248 | AKOS034007472 | HCT 116 | C181(1.03); C433(0.78) | LDD0565 | [7] |
| LDCM0356 | AKOS034007680 | HCT 116 | C433(0.84); C181(1.17) | LDD0673 | [7] |
| LDCM0275 | AKOS034007705 | HCT 116 | C433(0.44); C181(1.54) | LDD0592 | [7] |
| LDCM0020 | ARS-1620 | HCC44 | C181(0.89) | LDD0078 | [7] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [14] |
| LDCM0632 | CL-Sc | Hep-G2 | C181(1.46) | LDD2227 | [16] |
| LDCM0367 | CL1 | HCT 116 | C181(0.91) | LDD0684 | [7] |
| LDCM0368 | CL10 | HCT 116 | C181(1.09) | LDD0685 | [7] |
| LDCM0369 | CL100 | HCT 116 | C181(1.32) | LDD0686 | [7] |
| LDCM0370 | CL101 | HCT 116 | C433(0.96); C181(1.13) | LDD0687 | [7] |
| LDCM0371 | CL102 | HCT 116 | C433(0.82); C181(1.01) | LDD0688 | [7] |
| LDCM0372 | CL103 | HCT 116 | C433(0.65); C181(0.92) | LDD0689 | [7] |
| LDCM0373 | CL104 | HCT 116 | C433(0.82); C181(1.00) | LDD0690 | [7] |
| LDCM0374 | CL105 | HCT 116 | C433(0.81); C181(1.20) | LDD0691 | [7] |
| LDCM0375 | CL106 | HCT 116 | C433(0.51); C181(1.52) | LDD0692 | [7] |
| LDCM0376 | CL107 | HCT 116 | C433(0.67); C181(1.55) | LDD0693 | [7] |
| LDCM0377 | CL108 | HCT 116 | C433(0.86); C181(1.38) | LDD0694 | [7] |
| LDCM0378 | CL109 | HCT 116 | C433(1.09); C181(1.17) | LDD0695 | [7] |
| LDCM0379 | CL11 | HCT 116 | C181(1.18) | LDD0696 | [7] |
| LDCM0380 | CL110 | HCT 116 | C433(0.77); C181(1.27) | LDD0697 | [7] |
| LDCM0381 | CL111 | HCT 116 | C433(0.79); C181(1.29) | LDD0698 | [7] |
| LDCM0382 | CL112 | HCT 116 | C433(0.88); C181(1.10) | LDD0699 | [7] |
| LDCM0383 | CL113 | HCT 116 | C433(1.12); C181(1.38) | LDD0700 | [7] |
| LDCM0384 | CL114 | HCT 116 | C181(1.14); C433(1.42) | LDD0701 | [7] |
| LDCM0385 | CL115 | HCT 116 | C433(0.96); C181(1.04) | LDD0702 | [7] |
| LDCM0386 | CL116 | HCT 116 | C181(1.22); C433(1.28) | LDD0703 | [7] |
| LDCM0387 | CL117 | HCT 116 | C181(0.96) | LDD0704 | [7] |
| LDCM0388 | CL118 | HCT 116 | C181(0.84) | LDD0705 | [7] |
| LDCM0389 | CL119 | HCT 116 | C181(0.94) | LDD0706 | [7] |
| LDCM0390 | CL12 | HCT 116 | C181(1.18) | LDD0707 | [7] |
| LDCM0391 | CL120 | HCT 116 | C181(1.02) | LDD0708 | [7] |
| LDCM0392 | CL121 | HCT 116 | C433(0.88); C181(1.26) | LDD0709 | [7] |
| LDCM0393 | CL122 | HCT 116 | C433(0.80); C181(1.09) | LDD0710 | [7] |
| LDCM0394 | CL123 | HCT 116 | C433(0.61); C181(1.16) | LDD0711 | [7] |
| LDCM0395 | CL124 | HCT 116 | C433(0.60); C181(1.29) | LDD0712 | [7] |
| LDCM0396 | CL125 | HCT 116 | C181(1.14) | LDD0713 | [7] |
| LDCM0397 | CL126 | HCT 116 | C181(1.19) | LDD0714 | [7] |
| LDCM0398 | CL127 | HCT 116 | C181(1.19) | LDD0715 | [7] |
| LDCM0399 | CL128 | HCT 116 | C181(1.25) | LDD0716 | [7] |
| LDCM0400 | CL13 | HCT 116 | C181(1.10) | LDD0717 | [7] |
| LDCM0401 | CL14 | HCT 116 | C181(0.93) | LDD0718 | [7] |
| LDCM0402 | CL15 | HCT 116 | C181(1.05) | LDD0719 | [7] |
| LDCM0403 | CL16 | HCT 116 | C181(1.18) | LDD0720 | [7] |
| LDCM0404 | CL17 | HCT 116 | C181(1.05) | LDD0721 | [7] |
| LDCM0405 | CL18 | HCT 116 | C181(1.25) | LDD0722 | [7] |
| LDCM0406 | CL19 | HCT 116 | C181(1.18) | LDD0723 | [7] |
| LDCM0407 | CL2 | HCT 116 | C181(1.01) | LDD0724 | [7] |
| LDCM0408 | CL20 | HCT 116 | C181(1.39) | LDD0725 | [7] |
| LDCM0409 | CL21 | HCT 116 | C181(1.27) | LDD0726 | [7] |
| LDCM0410 | CL22 | HCT 116 | C181(1.63) | LDD0727 | [7] |
| LDCM0411 | CL23 | HCT 116 | C181(1.25) | LDD0728 | [7] |
| LDCM0412 | CL24 | HCT 116 | C181(1.39) | LDD0729 | [7] |
| LDCM0413 | CL25 | HCT 116 | C181(1.37) | LDD0730 | [7] |
| LDCM0414 | CL26 | HCT 116 | C181(1.27) | LDD0731 | [7] |
| LDCM0415 | CL27 | HCT 116 | C181(1.14) | LDD0732 | [7] |
| LDCM0416 | CL28 | HCT 116 | C181(1.29) | LDD0733 | [7] |
| LDCM0417 | CL29 | HCT 116 | C181(1.28) | LDD0734 | [7] |
| LDCM0418 | CL3 | HCT 116 | C181(1.02) | LDD0735 | [7] |
| LDCM0419 | CL30 | HCT 116 | C181(1.20) | LDD0736 | [7] |
| LDCM0420 | CL31 | HCT 116 | C181(0.98); C433(1.03) | LDD0737 | [7] |
| LDCM0421 | CL32 | HCT 116 | C181(1.18); C433(0.48) | LDD0738 | [7] |
| LDCM0422 | CL33 | HCT 116 | C181(1.02); C433(0.44) | LDD0739 | [7] |
| LDCM0423 | CL34 | HCT 116 | C181(1.14); C433(0.72) | LDD0740 | [7] |
| LDCM0424 | CL35 | HCT 116 | C181(1.17); C433(0.68) | LDD0741 | [7] |
| LDCM0425 | CL36 | HCT 116 | C181(1.06); C433(0.54) | LDD0742 | [7] |
| LDCM0426 | CL37 | HCT 116 | C181(1.19); C433(0.51) | LDD0743 | [7] |
| LDCM0428 | CL39 | HCT 116 | C181(1.27); C433(0.48) | LDD0745 | [7] |
| LDCM0429 | CL4 | HCT 116 | C181(0.90) | LDD0746 | [7] |
| LDCM0430 | CL40 | HCT 116 | C181(1.17); C433(0.78) | LDD0747 | [7] |
| LDCM0431 | CL41 | HCT 116 | C181(1.05); C433(0.63) | LDD0748 | [7] |
| LDCM0432 | CL42 | HCT 116 | C181(1.44); C433(0.42) | LDD0749 | [7] |
| LDCM0433 | CL43 | HCT 116 | C181(1.22); C433(0.46) | LDD0750 | [7] |
| LDCM0434 | CL44 | HCT 116 | C181(1.11); C433(0.57) | LDD0751 | [7] |
| LDCM0435 | CL45 | HCT 116 | C181(1.29); C433(0.59) | LDD0752 | [7] |
| LDCM0436 | CL46 | HCT 116 | C181(0.97); C433(1.32) | LDD0753 | [7] |
| LDCM0437 | CL47 | HCT 116 | C181(0.97); C433(0.94) | LDD0754 | [7] |
| LDCM0438 | CL48 | HCT 116 | C181(0.93); C433(1.05) | LDD0755 | [7] |
| LDCM0439 | CL49 | HCT 116 | C181(1.01); C433(1.72) | LDD0756 | [7] |
| LDCM0440 | CL5 | HCT 116 | C181(1.01) | LDD0757 | [7] |
| LDCM0441 | CL50 | HCT 116 | C181(0.84); C433(1.13) | LDD0758 | [7] |
| LDCM0442 | CL51 | HCT 116 | C181(0.95); C433(1.31) | LDD0759 | [7] |
| LDCM0443 | CL52 | HCT 116 | C181(0.98); C433(1.16) | LDD0760 | [7] |
| LDCM0444 | CL53 | HCT 116 | C181(0.87); C433(0.97) | LDD0761 | [7] |
| LDCM0445 | CL54 | HCT 116 | C181(0.87); C433(1.02) | LDD0762 | [7] |
| LDCM0446 | CL55 | HCT 116 | C181(0.94); C433(1.27) | LDD0763 | [7] |
| LDCM0447 | CL56 | HCT 116 | C181(1.06); C433(1.40) | LDD0764 | [7] |
| LDCM0448 | CL57 | HCT 116 | C181(0.87); C433(1.39) | LDD0765 | [7] |
| LDCM0449 | CL58 | HCT 116 | C181(0.93); C433(1.16) | LDD0766 | [7] |
| LDCM0450 | CL59 | HCT 116 | C181(0.95); C433(1.12) | LDD0767 | [7] |
| LDCM0451 | CL6 | HCT 116 | C181(1.03) | LDD0768 | [7] |
| LDCM0452 | CL60 | HCT 116 | C181(0.99); C433(1.40) | LDD0769 | [7] |
| LDCM0453 | CL61 | HCT 116 | C181(0.79) | LDD0770 | [7] |
| LDCM0454 | CL62 | HCT 116 | C181(0.76) | LDD0771 | [7] |
| LDCM0455 | CL63 | HCT 116 | C181(0.89) | LDD0772 | [7] |
| LDCM0456 | CL64 | HCT 116 | C181(0.87) | LDD0773 | [7] |
| LDCM0457 | CL65 | HCT 116 | C181(0.79) | LDD0774 | [7] |
| LDCM0458 | CL66 | HCT 116 | C181(0.75) | LDD0775 | [7] |
| LDCM0459 | CL67 | HCT 116 | C181(0.70) | LDD0776 | [7] |
| LDCM0460 | CL68 | HCT 116 | C181(0.76) | LDD0777 | [7] |
| LDCM0461 | CL69 | HCT 116 | C181(0.75) | LDD0778 | [7] |
| LDCM0462 | CL7 | HCT 116 | C181(1.16) | LDD0779 | [7] |
| LDCM0463 | CL70 | HCT 116 | C181(0.75) | LDD0780 | [7] |
| LDCM0464 | CL71 | HCT 116 | C181(0.78) | LDD0781 | [7] |
| LDCM0465 | CL72 | HCT 116 | C181(0.76) | LDD0782 | [7] |
| LDCM0466 | CL73 | HCT 116 | C181(0.77) | LDD0783 | [7] |
| LDCM0467 | CL74 | HCT 116 | C181(0.69) | LDD0784 | [7] |
| LDCM0469 | CL76 | HCT 116 | C181(0.90) | LDD0786 | [7] |
| LDCM0470 | CL77 | HCT 116 | C181(0.96) | LDD0787 | [7] |
| LDCM0471 | CL78 | HCT 116 | C181(1.10) | LDD0788 | [7] |
| LDCM0472 | CL79 | HCT 116 | C181(1.06) | LDD0789 | [7] |
| LDCM0473 | CL8 | HCT 116 | C181(1.07) | LDD0790 | [7] |
| LDCM0474 | CL80 | HCT 116 | C181(0.94) | LDD0791 | [7] |
| LDCM0475 | CL81 | HCT 116 | C181(0.83) | LDD0792 | [7] |
| LDCM0476 | CL82 | HCT 116 | C181(1.15) | LDD0793 | [7] |
| LDCM0477 | CL83 | HCT 116 | C181(1.18) | LDD0794 | [7] |
| LDCM0478 | CL84 | HCT 116 | C181(1.25) | LDD0795 | [7] |
| LDCM0479 | CL85 | HCT 116 | C181(1.10) | LDD0796 | [7] |
| LDCM0480 | CL86 | HCT 116 | C181(1.11) | LDD0797 | [7] |
| LDCM0481 | CL87 | HCT 116 | C181(0.83) | LDD0798 | [7] |
| LDCM0482 | CL88 | HCT 116 | C181(1.33) | LDD0799 | [7] |
| LDCM0483 | CL89 | HCT 116 | C181(1.22) | LDD0800 | [7] |
| LDCM0484 | CL9 | HCT 116 | C181(1.07) | LDD0801 | [7] |
| LDCM0485 | CL90 | HCT 116 | C181(0.70) | LDD0802 | [7] |
| LDCM0486 | CL91 | HCT 116 | C181(1.16) | LDD0803 | [7] |
| LDCM0487 | CL92 | HCT 116 | C181(1.05) | LDD0804 | [7] |
| LDCM0488 | CL93 | HCT 116 | C181(1.05) | LDD0805 | [7] |
| LDCM0489 | CL94 | HCT 116 | C181(1.00) | LDD0806 | [7] |
| LDCM0490 | CL95 | HCT 116 | C181(1.10) | LDD0807 | [7] |
| LDCM0491 | CL96 | HCT 116 | C181(1.14) | LDD0808 | [7] |
| LDCM0492 | CL97 | HCT 116 | C181(1.24) | LDD0809 | [7] |
| LDCM0493 | CL98 | HCT 116 | C181(1.26) | LDD0810 | [7] |
| LDCM0494 | CL99 | HCT 116 | C181(1.30) | LDD0811 | [7] |
| LDCM0634 | CY-0357 | Hep-G2 | C181(0.40) | LDD2228 | [16] |
| LDCM0495 | E2913 | HEK-293T | C181(0.80); C433(1.02); C336(1.01); C48(0.86) | LDD1698 | [20] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C181(1.10) | LDD1702 | [4] |
| LDCM0625 | F8 | Ramos | C181(0.98); C336(2.74) | LDD2187 | [21] |
| LDCM0572 | Fragment10 | Ramos | C336(4.31) | LDD2189 | [21] |
| LDCM0574 | Fragment12 | Ramos | C181(0.32); C336(6.25) | LDD2191 | [21] |
| LDCM0575 | Fragment13 | Ramos | C181(0.70) | LDD2192 | [21] |
| LDCM0576 | Fragment14 | Ramos | C181(1.43) | LDD2193 | [21] |
| LDCM0579 | Fragment20 | Ramos | C336(5.22) | LDD2194 | [21] |
| LDCM0580 | Fragment21 | Ramos | C181(0.88); C336(4.34) | LDD2195 | [21] |
| LDCM0582 | Fragment23 | Ramos | C181(0.58); C336(1.10) | LDD2196 | [21] |
| LDCM0578 | Fragment27 | Ramos | C181(3.90); C336(1.26) | LDD2197 | [21] |
| LDCM0586 | Fragment28 | Ramos | C181(0.61) | LDD2198 | [21] |
| LDCM0588 | Fragment30 | Ramos | C181(0.67); C336(5.69) | LDD2199 | [21] |
| LDCM0589 | Fragment31 | Ramos | C181(0.59); C336(1.04) | LDD2200 | [21] |
| LDCM0590 | Fragment32 | Ramos | C181(0.24); C336(6.93) | LDD2201 | [21] |
| LDCM0468 | Fragment33 | HCT 116 | C181(0.77) | LDD0785 | [7] |
| LDCM0596 | Fragment38 | Ramos | C181(1.27); C336(2.30) | LDD2203 | [21] |
| LDCM0566 | Fragment4 | Ramos | C181(0.82); C336(0.22) | LDD2184 | [21] |
| LDCM0427 | Fragment51 | HCT 116 | C181(1.23); C433(0.40) | LDD0744 | [7] |
| LDCM0610 | Fragment52 | Ramos | C181(1.74); C336(1.01) | LDD2204 | [21] |
| LDCM0614 | Fragment56 | Ramos | C181(0.66); C336(3.82) | LDD2205 | [21] |
| LDCM0569 | Fragment7 | Ramos | C181(0.78); C336(0.70) | LDD2186 | [21] |
| LDCM0571 | Fragment9 | Ramos | C181(0.57); C336(3.93) | LDD2188 | [21] |
| LDCM0116 | HHS-0101 | DM93 | Y210(0.87) | LDD0264 | [6] |
| LDCM0117 | HHS-0201 | DM93 | Y210(0.69) | LDD0265 | [6] |
| LDCM0118 | HHS-0301 | DM93 | Y210(0.72) | LDD0266 | [6] |
| LDCM0119 | HHS-0401 | DM93 | Y210(0.54) | LDD0267 | [6] |
| LDCM0120 | HHS-0701 | DM93 | Y210(0.60) | LDD0268 | [6] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [14] |
| LDCM0022 | KB02 | HCT 116 | C181(1.24) | LDD0080 | [7] |
| LDCM0023 | KB03 | HCT 116 | C181(0.96) | LDD0081 | [7] |
| LDCM0024 | KB05 | HCT 116 | C181(1.24) | LDD0082 | [7] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0232 | [14] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C181(1.07) | LDD2099 | [4] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C181(0.96) | LDD2123 | [4] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C181(1.04) | LDD2125 | [4] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C181(1.17) | LDD2127 | [4] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C181(1.20) | LDD2136 | [4] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C181(1.25) | LDD2137 | [4] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C181(2.09) | LDD2144 | [4] |
| LDCM0021 | THZ1 | HCT 116 | C181(0.98) | LDD2173 | [7] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| ATP synthase subunit O, mitochondrial (ATP5PO) | ATPase delta chain family | P48047 | |||
| Syntenin-1 (SDCBP) | . | O00560 | |||
Other
References
