Details of the Target
General Information of Target
| Target ID | LDTP12851 | |||||
|---|---|---|---|---|---|---|
| Target Name | UDP-glucose:glycoprotein glucosyltransferase 1 (UGGT1) | |||||
| Gene Name | UGGT1 | |||||
| Gene ID | 56886 | |||||
| Synonyms |
GT; UGCGL1; UGGT; UGT1; UGTR; UDP-glucose:glycoprotein glucosyltransferase 1; UGT1; hUGT1; EC 2.4.1.-; UDP--Glc:glycoprotein glucosyltransferase; UDP-glucose ceramide glucosyltransferase-like 1 |
|||||
| 3D Structure | ||||||
| Sequence |
MAPAKATNVVRLLLGSTALWLSQLGSGTVAASKSVTAHLAAKWPETPLLLEASEFMAEES
NEKFWQFLETVQELAIYKQTESDYSYYNLILKKAGQFLDNLHINLLKFAFSIRAYSPAIQ MFQQIAADEPPPDGCNAFVVIHKKHTCKINEIKKLLKKAASRTRPYLFKGDHKFPTNKEN LPVVILYAEMGTRTFSAFHKVLSEKAQNEEILYVLRHYIQKPSSRKMYLSGYGVELAIKS TEYKALDDTQVKTVTNTTVEDETETNEVQGFLFGKLKEIYSDLRDNLTAFQKYLIESNKQ MMPLKVWELQDLSFQAASQIMSAPVYDSIKLMKDISQNFPIKARSLTRIAVNQHMREEIK ENQKDLQVRFKIQPGDARLFINGLRVDMDVYDAFSILDMLKLEGKMMNGLRNLGINGEDM SKFLKLNSHIWEYTYVLDIRHSSIMWINDLENDDLYITWPTSCQKLLKPVFPGSVPSIRR NFHNLVLFIDPAQEYTLDFIKLADVFYSHEVPLRIGFVFILNTDDEVDGANDAGVALWRA FNYIAEEFDISEAFISIVHMYQKVKKDQNILTVDNVKSVLQNTFPHANIWDILGIHSKYD EERKAGASFYKMTGLGPLPQALYNGEPFKHEEMNIKELKMAVLQRMMDASVYLQREVFLG TLNDRTNAIDFLMDRNNVVPRINTLILRTNQQYLNLISTSVTADVEDFSTFFFLDSQDKS AVIAKNMYYLTQDDESIISAVTLWIIADFDKPSGRKLLFNALKHMKTSVHSRLGIIYNPT SKINEENTAISRGILAAFLTQKNMFLRSFLGQLAKEEIATAIYSGDKIKTFLIEGMDKNA FEKKYNTVGVNIFRTHQLFCQDVLKLRPGEMGIVSNGRFLGPLDEDFYAEDFYLLEKITF SNLGEKIKGIVENMGINANNMSDFIMKVDALMSSVPKRASRYDVTFLRENHSVIKTNPQE NDMFFNVIAIVDPLTREAQKMAQLLVVLGKIINMKIKLFMNCRGRLSEAPLESFYRFVLE PELMSGANDVSSLGPVAKFLDIPESPLLILNMITPEGWLVETVHSNCDLDNIHLKDTEKT VTAEYELEYLLLEGQCFDKVTEQPPRGLQFTLGTKNKPAVVDTIVMAHHGYFQLKANPGA WILRLHQGKSEDIYQIVGHEGTDSQADLEDIIVVLNSFKSKILKVKVKKETDKIKEDILT DEDEKTKGLWDSIKSFTVSLHKENKKEKDVLNIFSVASGHLYERFLRIMMLSVLRNTKTP VKFWLLKNYLSPTFKEVIPHMAKEYGFRYELVQYRWPRWLRQQTERQRIIWGYKILFLDV LFPLAVDKIIFVDADQIVRHDLKELRDFDLDGAPYGYTPFCDSRREMDGYRFWKTGYWAS HLLRRKYHISALYVVDLKKFRRIGAGDRLRSQYQALSQDPNSLSNLDQDLPNNMIYQVAI KSLPQDWLWCETWCDDESKQRAKTIDLCNNPKTKESKLKAAARIVPEWVEYDAEIRQLLD HLENKKQDTILTHDEL |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Glycosyltransferase 8 family
|
|||||
| Subcellular location |
Endoplasmic reticulum lumen
|
|||||
| Function |
Recognizes glycoproteins with minor folding defects. Reglucosylates single N-glycans near the misfolded part of the protein, thus providing quality control for protein folding in the endoplasmic reticulum. Reglucosylated proteins are recognized by calreticulin for recycling to the endoplasmic reticulum and refolding or degradation.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| CAL51 | SNV: p.P1496L | DBIA Probe Info | |||
| CHL1 | SNV: p.V600G | DBIA Probe Info | |||
| DU145 | SNV: p.V33I | DBIA Probe Info | |||
| GB1 | Deletion: p.E836GfsTer10 | DBIA Probe Info | |||
| HCT15 | SNV: p.V558I | DBIA Probe Info | |||
| HELA | SNV: p.S126C | . | |||
| HEP3B217 | SNV: p.G1337S | DBIA Probe Info | |||
| HUH7 | SNV: p.A1158T | DBIA Probe Info | |||
| Ishikawa (Heraklio) 02 ER | SNV: p.D1185E | DBIA Probe Info | |||
| MOLT4 | SNV: p.D1223N | IA-alkyne Probe Info | |||
| NALM6 | SNV: p.A825T | DBIA Probe Info | |||
| OCIAML5 | Insertion: . | DBIA Probe Info | |||
| RCC10RGB | SNV: p.G837R | . | |||
| SHP77 | SNV: p.K306Q; p.W321Ter | . | |||
| SNU245 | SNV: p.R482Q | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
P1 Probe Info |
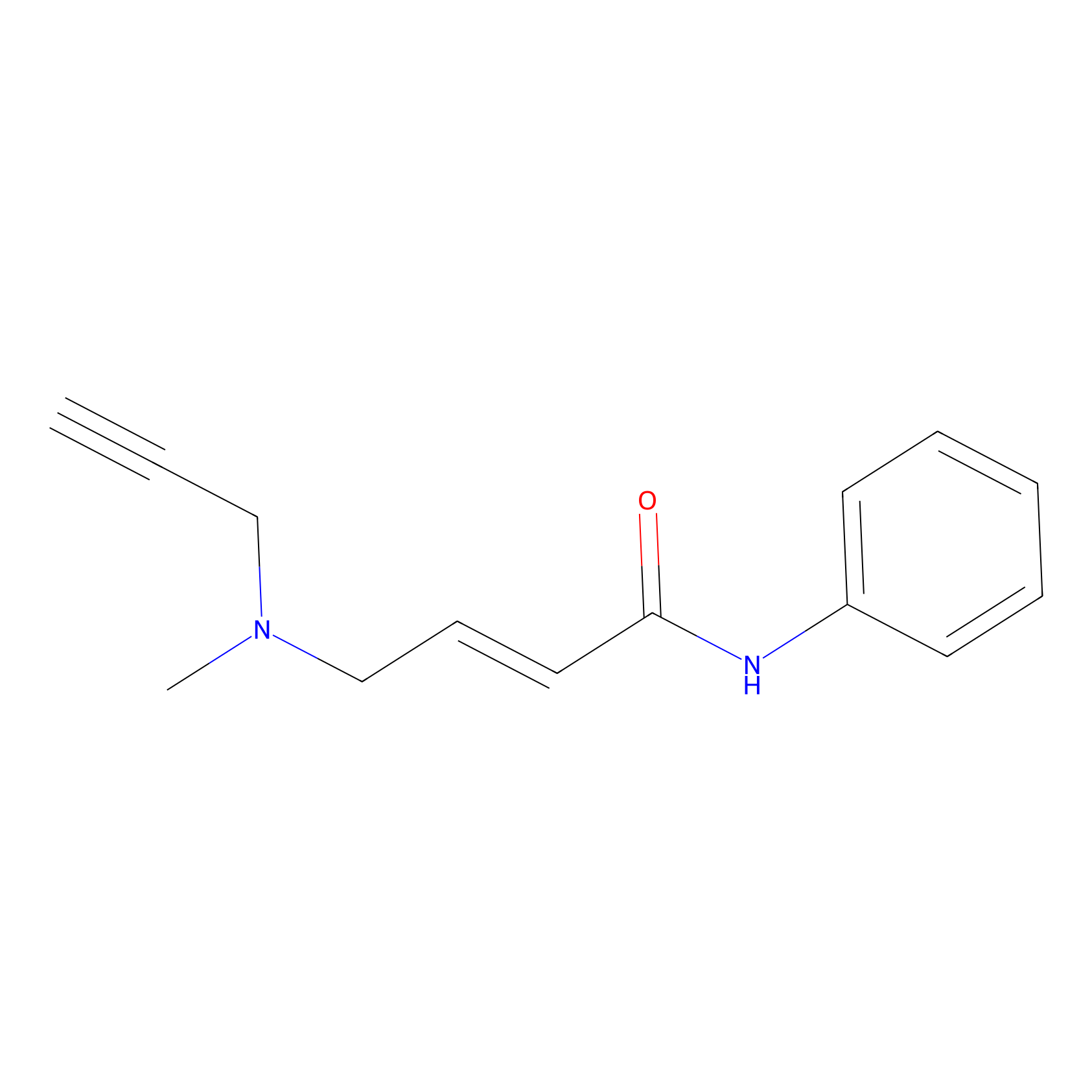 |
10.00 | LDD0448 | [1] | |
|
P2 Probe Info |
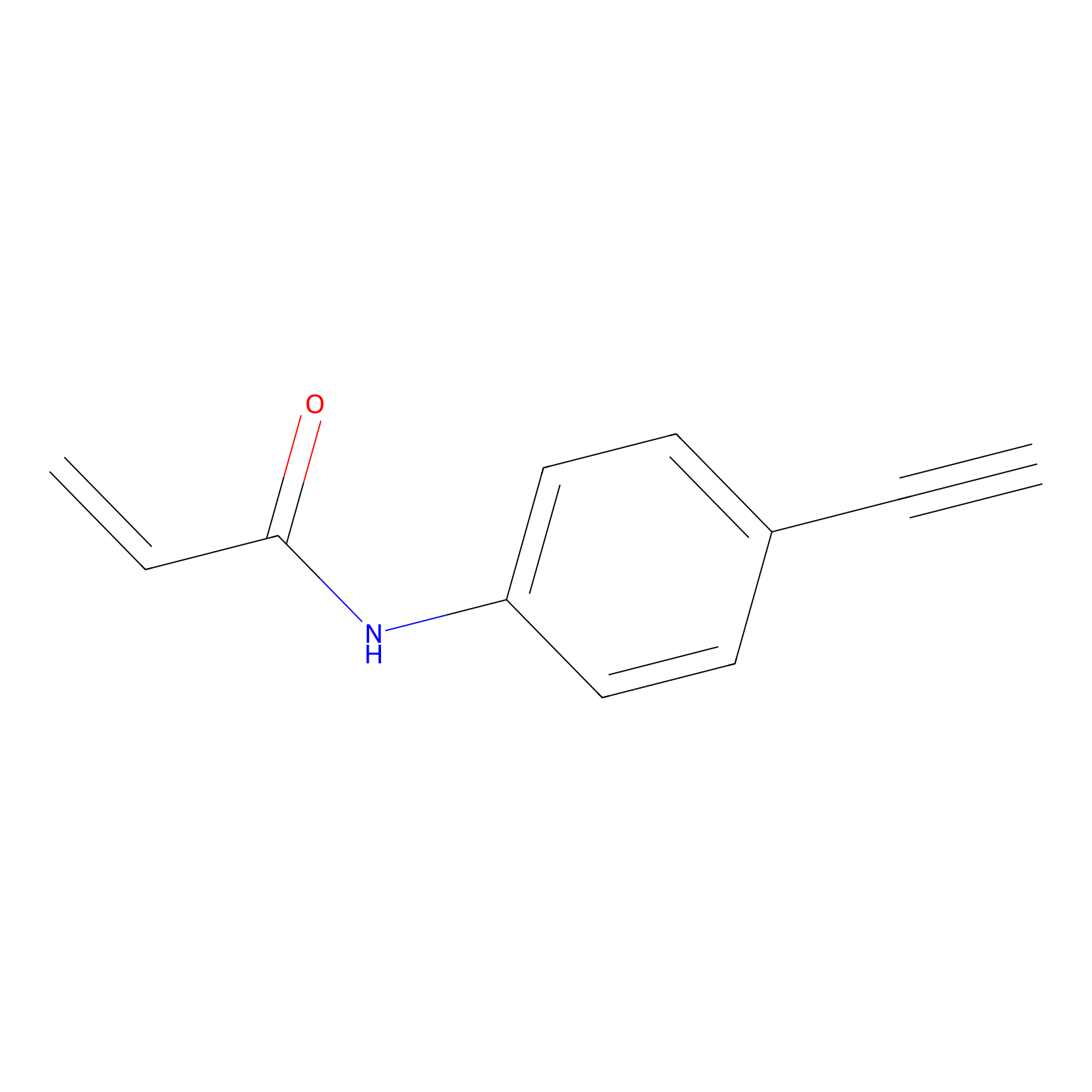 |
10.00 | LDD0449 | [1] | |
|
P3 Probe Info |
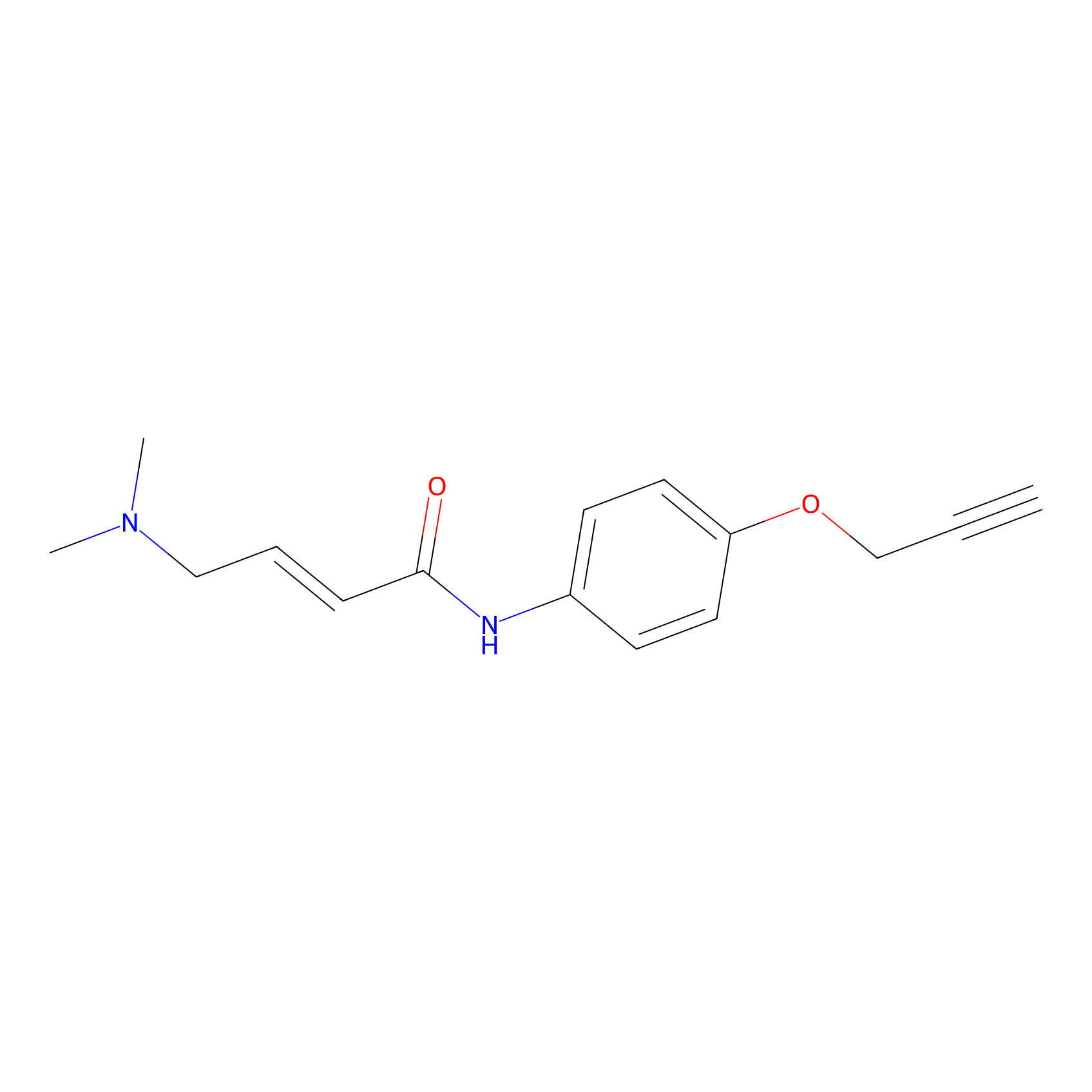 |
10.00 | LDD0450 | [1] | |
|
C-Sul Probe Info |
 |
5.39 | LDD0066 | [2] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [3] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [3] | |
|
STPyne Probe Info |
 |
K306(10.00); K356(4.35); K362(5.75); K744(1.89) | LDD0277 | [4] | |
|
ONAyne Probe Info |
 |
K217(5.00) | LDD0275 | [4] | |
|
IPM Probe Info |
 |
C880(0.00); C1024(0.00) | LDD0241 | [5] | |
|
OPA-S-S-alkyne Probe Info |
 |
K356(1.45); K362(3.31) | LDD3494 | [6] | |
|
Probe 1 Probe Info |
 |
Y449(9.36); Y805(22.87); Y966(11.71) | LDD3495 | [7] | |
|
BTD Probe Info |
 |
C1024(3.38) | LDD1700 | [8] | |
|
HHS-475 Probe Info |
 |
Y599(0.05) | LDD0264 | [9] | |
|
HHS-465 Probe Info |
 |
Y671(4.99) | LDD2237 | [10] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0221 | [11] | |
|
DBIA Probe Info |
 |
C1493(1.10) | LDD0080 | [12] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [13] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C1386(0.00); C880(0.00); C147(0.00); C159(0.00) | LDD0038 | [14] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0032 | [15] | |
|
IPIAA_H Probe Info |
 |
N.A. | LDD0030 | [16] | |
|
IPIAA_L Probe Info |
 |
N.A. | LDD0031 | [16] | |
|
Lodoacetamide azide Probe Info |
 |
C1386(0.00); C880(0.00); C147(0.00); C159(0.00) | LDD0037 | [14] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [17] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [11] | |
|
NAIA_5 Probe Info |
 |
C1493(0.00); C1386(0.00); C159(0.00); C1118(0.00) | LDD2223 | [18] | |
|
HHS-482 Probe Info |
 |
Y1267(0.67) | LDD2239 | [10] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C349 Probe Info |
 |
13.45 | LDD2010 | [19] | |
|
FFF probe12 Probe Info |
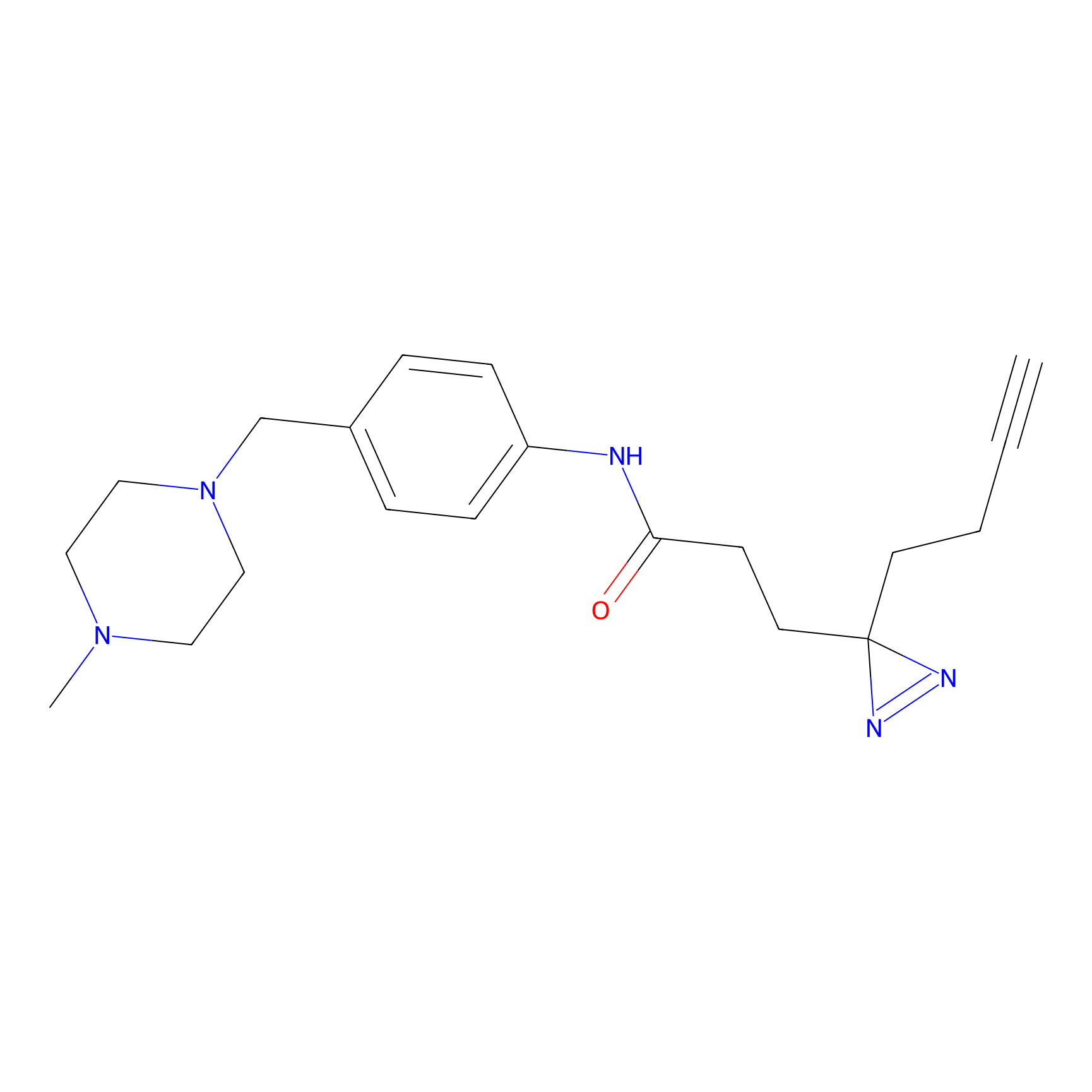 |
13.25 | LDD0473 | [20] | |
|
FFF probe13 Probe Info |
 |
20.00 | LDD0475 | [20] | |
|
FFF probe14 Probe Info |
 |
20.00 | LDD0477 | [20] | |
|
FFF probe3 Probe Info |
 |
20.00 | LDD0464 | [20] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0138 | [21] | |
|
DA-2 Probe Info |
 |
N.A. | LDD0070 | [22] | |
|
OEA-DA Probe Info |
 |
4.28 | LDD0046 | [23] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0068 | [24] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C1024(0.87) | LDD2117 | [8] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C1024(1.32) | LDD2152 | [8] |
| LDCM0214 | AC1 | HCT 116 | C1386(1.02) | LDD0531 | [12] |
| LDCM0215 | AC10 | PaTu 8988t | C1479(1.14); C1475(0.47) | LDD1094 | [12] |
| LDCM0216 | AC100 | HCT 116 | C1386(0.77) | LDD0533 | [12] |
| LDCM0217 | AC101 | HCT 116 | C1386(1.08) | LDD0534 | [12] |
| LDCM0218 | AC102 | HCT 116 | C1386(0.80) | LDD0535 | [12] |
| LDCM0219 | AC103 | HCT 116 | C1386(0.92) | LDD0536 | [12] |
| LDCM0220 | AC104 | HCT 116 | C1386(1.02) | LDD0537 | [12] |
| LDCM0221 | AC105 | HCT 116 | C1386(0.97) | LDD0538 | [12] |
| LDCM0222 | AC106 | HCT 116 | C1386(0.74) | LDD0539 | [12] |
| LDCM0223 | AC107 | HCT 116 | C1386(0.99) | LDD0540 | [12] |
| LDCM0224 | AC108 | HCT 116 | C1386(0.82) | LDD0541 | [12] |
| LDCM0225 | AC109 | HCT 116 | C1386(1.09) | LDD0542 | [12] |
| LDCM0226 | AC11 | PaTu 8988t | C1479(1.50); C1475(0.67) | LDD1105 | [12] |
| LDCM0227 | AC110 | HCT 116 | C1386(0.92) | LDD0544 | [12] |
| LDCM0228 | AC111 | HCT 116 | C1386(1.51) | LDD0545 | [12] |
| LDCM0229 | AC112 | HCT 116 | C1386(0.97) | LDD0546 | [12] |
| LDCM0230 | AC113 | HCT 116 | C1386(0.72) | LDD0547 | [12] |
| LDCM0231 | AC114 | HCT 116 | C1386(0.70) | LDD0548 | [12] |
| LDCM0232 | AC115 | HCT 116 | C1386(0.61) | LDD0549 | [12] |
| LDCM0233 | AC116 | HCT 116 | C1386(0.64) | LDD0550 | [12] |
| LDCM0234 | AC117 | HCT 116 | C1386(0.73) | LDD0551 | [12] |
| LDCM0235 | AC118 | HCT 116 | C1386(0.67) | LDD0552 | [12] |
| LDCM0236 | AC119 | HCT 116 | C1386(0.75) | LDD0553 | [12] |
| LDCM0237 | AC12 | PaTu 8988t | C1479(0.83); C1475(0.60) | LDD1116 | [12] |
| LDCM0238 | AC120 | HCT 116 | C1386(1.17) | LDD0555 | [12] |
| LDCM0239 | AC121 | HCT 116 | C1386(1.17) | LDD0556 | [12] |
| LDCM0240 | AC122 | HCT 116 | C1386(0.68) | LDD0557 | [12] |
| LDCM0241 | AC123 | HCT 116 | C1386(0.94) | LDD0558 | [12] |
| LDCM0242 | AC124 | HCT 116 | C1386(0.78) | LDD0559 | [12] |
| LDCM0243 | AC125 | HCT 116 | C1386(0.85) | LDD0560 | [12] |
| LDCM0244 | AC126 | HCT 116 | C1386(0.89) | LDD0561 | [12] |
| LDCM0245 | AC127 | HCT 116 | C1386(0.69) | LDD0562 | [12] |
| LDCM0246 | AC128 | HEK-293T | C1475(0.81) | LDD0844 | [12] |
| LDCM0247 | AC129 | HEK-293T | C1475(1.55) | LDD0845 | [12] |
| LDCM0249 | AC130 | HEK-293T | C1475(1.15) | LDD0847 | [12] |
| LDCM0250 | AC131 | HEK-293T | C1475(1.19) | LDD0848 | [12] |
| LDCM0251 | AC132 | HEK-293T | C1475(0.86) | LDD0849 | [12] |
| LDCM0252 | AC133 | HEK-293T | C1475(0.85) | LDD0850 | [12] |
| LDCM0253 | AC134 | HEK-293T | C1475(1.47) | LDD0851 | [12] |
| LDCM0254 | AC135 | HEK-293T | C1475(1.12) | LDD0852 | [12] |
| LDCM0255 | AC136 | HEK-293T | C1475(1.09) | LDD0853 | [12] |
| LDCM0256 | AC137 | HEK-293T | C1475(0.66) | LDD0854 | [12] |
| LDCM0257 | AC138 | HEK-293T | C1475(0.78) | LDD0855 | [12] |
| LDCM0258 | AC139 | HEK-293T | C1475(0.84) | LDD0856 | [12] |
| LDCM0259 | AC14 | PaTu 8988t | C1479(1.16); C1475(0.60) | LDD1138 | [12] |
| LDCM0260 | AC140 | HEK-293T | C1475(0.92) | LDD0858 | [12] |
| LDCM0261 | AC141 | HEK-293T | C1475(0.73) | LDD0859 | [12] |
| LDCM0262 | AC142 | HEK-293T | C1475(0.97) | LDD0860 | [12] |
| LDCM0263 | AC143 | HEK-293T | C1475(1.03) | LDD0861 | [12] |
| LDCM0264 | AC144 | HEK-293T | C1475(1.08) | LDD0862 | [12] |
| LDCM0265 | AC145 | HEK-293T | C1475(1.08) | LDD0863 | [12] |
| LDCM0266 | AC146 | HEK-293T | C1475(0.94) | LDD0864 | [12] |
| LDCM0267 | AC147 | HEK-293T | C1475(0.86) | LDD0865 | [12] |
| LDCM0268 | AC148 | HEK-293T | C1475(0.52) | LDD0866 | [12] |
| LDCM0269 | AC149 | HEK-293T | C1475(0.78) | LDD0867 | [12] |
| LDCM0270 | AC15 | PaTu 8988t | C1479(1.66); C1475(0.50) | LDD1149 | [12] |
| LDCM0271 | AC150 | HEK-293T | C1475(1.10) | LDD0869 | [12] |
| LDCM0272 | AC151 | HEK-293T | C1475(0.73) | LDD0870 | [12] |
| LDCM0273 | AC152 | HEK-293T | C1475(0.77) | LDD0871 | [12] |
| LDCM0274 | AC153 | HEK-293T | C1475(1.02) | LDD0872 | [12] |
| LDCM0621 | AC154 | HEK-293T | C1475(0.92) | LDD2162 | [12] |
| LDCM0622 | AC155 | HEK-293T | C1475(0.79) | LDD2163 | [12] |
| LDCM0623 | AC156 | HEK-293T | C1475(0.75) | LDD2164 | [12] |
| LDCM0624 | AC157 | HEK-293T | C1475(0.74) | LDD2165 | [12] |
| LDCM0276 | AC17 | PaTu 8988t | C1479(1.24); C1475(1.24) | LDD1155 | [12] |
| LDCM0277 | AC18 | PaTu 8988t | C1479(0.86); C1475(0.86) | LDD1156 | [12] |
| LDCM0278 | AC19 | PaTu 8988t | C1479(0.99); C1475(0.99) | LDD1157 | [12] |
| LDCM0279 | AC2 | HCT 116 | C1386(0.98) | LDD0596 | [12] |
| LDCM0280 | AC20 | PaTu 8988t | C1479(1.09); C1475(1.09) | LDD1159 | [12] |
| LDCM0281 | AC21 | PaTu 8988t | C1479(0.66); C1475(0.66) | LDD1160 | [12] |
| LDCM0282 | AC22 | PaTu 8988t | C1479(0.68); C1475(0.68) | LDD1161 | [12] |
| LDCM0283 | AC23 | PaTu 8988t | C1479(0.68); C1475(0.68) | LDD1162 | [12] |
| LDCM0284 | AC24 | PaTu 8988t | C1479(0.78); C1475(0.78) | LDD1163 | [12] |
| LDCM0285 | AC25 | HEK-293T | C1479(1.65) | LDD0883 | [12] |
| LDCM0286 | AC26 | HEK-293T | C1479(1.66) | LDD0884 | [12] |
| LDCM0287 | AC27 | HEK-293T | C1479(1.48) | LDD0885 | [12] |
| LDCM0288 | AC28 | HEK-293T | C1479(1.75) | LDD0886 | [12] |
| LDCM0289 | AC29 | HEK-293T | C1479(1.44) | LDD0887 | [12] |
| LDCM0290 | AC3 | HCT 116 | C1386(1.05) | LDD0607 | [12] |
| LDCM0291 | AC30 | HEK-293T | C1479(1.53) | LDD0889 | [12] |
| LDCM0292 | AC31 | HEK-293T | C1479(1.38) | LDD0890 | [12] |
| LDCM0293 | AC32 | HEK-293T | C1479(1.53) | LDD0891 | [12] |
| LDCM0294 | AC33 | HEK-293T | C1479(1.47) | LDD0892 | [12] |
| LDCM0295 | AC34 | HEK-293T | C1479(1.57) | LDD0893 | [12] |
| LDCM0296 | AC35 | PaTu 8988t | C1479(1.16); C1475(1.16) | LDD1175 | [12] |
| LDCM0297 | AC36 | PaTu 8988t | C1479(1.15); C1475(1.15) | LDD1176 | [12] |
| LDCM0298 | AC37 | PaTu 8988t | C1479(0.95); C1475(0.95) | LDD1177 | [12] |
| LDCM0299 | AC38 | PaTu 8988t | C1479(1.18); C1475(1.18) | LDD1178 | [12] |
| LDCM0300 | AC39 | PaTu 8988t | C1479(1.18); C1475(1.18) | LDD1179 | [12] |
| LDCM0301 | AC4 | HCT 116 | C1386(1.11) | LDD0618 | [12] |
| LDCM0302 | AC40 | PaTu 8988t | C1479(1.14); C1475(1.14) | LDD1181 | [12] |
| LDCM0303 | AC41 | PaTu 8988t | C1479(1.06); C1475(1.06) | LDD1182 | [12] |
| LDCM0304 | AC42 | PaTu 8988t | C1479(1.16); C1475(1.16) | LDD1183 | [12] |
| LDCM0305 | AC43 | PaTu 8988t | C1479(1.50); C1475(1.50) | LDD1184 | [12] |
| LDCM0306 | AC44 | PaTu 8988t | C1479(0.87); C1475(0.87) | LDD1185 | [12] |
| LDCM0307 | AC45 | PaTu 8988t | C1479(1.01); C1475(1.01) | LDD1186 | [12] |
| LDCM0308 | AC46 | HCT 116 | C1386(0.71) | LDD0625 | [12] |
| LDCM0309 | AC47 | HCT 116 | C1386(0.87) | LDD0626 | [12] |
| LDCM0310 | AC48 | HCT 116 | C1386(0.86) | LDD0627 | [12] |
| LDCM0311 | AC49 | HCT 116 | C1386(0.85) | LDD0628 | [12] |
| LDCM0312 | AC5 | HCT 116 | C1386(1.04) | LDD0629 | [12] |
| LDCM0313 | AC50 | HCT 116 | C1386(0.94) | LDD0630 | [12] |
| LDCM0314 | AC51 | HCT 116 | C1386(0.99) | LDD0631 | [12] |
| LDCM0315 | AC52 | HCT 116 | C1386(0.78) | LDD0632 | [12] |
| LDCM0316 | AC53 | HCT 116 | C1386(0.91) | LDD0633 | [12] |
| LDCM0317 | AC54 | HCT 116 | C1386(0.90) | LDD0634 | [12] |
| LDCM0318 | AC55 | HCT 116 | C1386(0.83) | LDD0635 | [12] |
| LDCM0319 | AC56 | HCT 116 | C1386(0.94) | LDD0636 | [12] |
| LDCM0320 | AC57 | HCT 116 | C1386(1.09) | LDD0637 | [12] |
| LDCM0321 | AC58 | HCT 116 | C1386(1.36) | LDD0638 | [12] |
| LDCM0322 | AC59 | HCT 116 | C1386(1.08) | LDD0639 | [12] |
| LDCM0323 | AC6 | PaTu 8988t | C1479(1.17); C1475(0.73) | LDD1202 | [12] |
| LDCM0324 | AC60 | HCT 116 | C1386(1.09) | LDD0641 | [12] |
| LDCM0325 | AC61 | HCT 116 | C1386(0.95) | LDD0642 | [12] |
| LDCM0326 | AC62 | HCT 116 | C1386(0.99) | LDD0643 | [12] |
| LDCM0327 | AC63 | HCT 116 | C1386(1.07) | LDD0644 | [12] |
| LDCM0328 | AC64 | HCT 116 | C1386(1.01) | LDD0645 | [12] |
| LDCM0329 | AC65 | HCT 116 | C1386(1.16) | LDD0646 | [12] |
| LDCM0330 | AC66 | HCT 116 | C1386(1.35) | LDD0647 | [12] |
| LDCM0331 | AC67 | HCT 116 | C1386(0.94) | LDD0648 | [12] |
| LDCM0332 | AC68 | HCT 116 | C1386(1.16) | LDD0649 | [12] |
| LDCM0333 | AC69 | HCT 116 | C1386(1.12) | LDD0650 | [12] |
| LDCM0334 | AC7 | PaTu 8988t | C1479(0.93); C1475(0.69) | LDD1213 | [12] |
| LDCM0335 | AC70 | HCT 116 | C1386(0.69) | LDD0652 | [12] |
| LDCM0336 | AC71 | HCT 116 | C1386(0.95) | LDD0653 | [12] |
| LDCM0337 | AC72 | HCT 116 | C1386(0.71) | LDD0654 | [12] |
| LDCM0338 | AC73 | HCT 116 | C1386(0.81) | LDD0655 | [12] |
| LDCM0339 | AC74 | HCT 116 | C1386(0.89) | LDD0656 | [12] |
| LDCM0340 | AC75 | HCT 116 | C1386(0.85) | LDD0657 | [12] |
| LDCM0341 | AC76 | HCT 116 | C1386(0.81) | LDD0658 | [12] |
| LDCM0342 | AC77 | HCT 116 | C1386(1.14) | LDD0659 | [12] |
| LDCM0343 | AC78 | HCT 116 | C1386(0.93) | LDD0660 | [12] |
| LDCM0344 | AC79 | HCT 116 | C1386(0.89) | LDD0661 | [12] |
| LDCM0345 | AC8 | PaTu 8988t | C1479(1.09); C1475(0.56) | LDD1224 | [12] |
| LDCM0346 | AC80 | HCT 116 | C1386(0.88) | LDD0663 | [12] |
| LDCM0347 | AC81 | HCT 116 | C1386(0.80) | LDD0664 | [12] |
| LDCM0348 | AC82 | HCT 116 | C1386(0.71) | LDD0665 | [12] |
| LDCM0349 | AC83 | HCT 116 | C1386(1.07) | LDD0666 | [12] |
| LDCM0350 | AC84 | HCT 116 | C1386(1.19) | LDD0667 | [12] |
| LDCM0351 | AC85 | HCT 116 | C1386(1.03) | LDD0668 | [12] |
| LDCM0352 | AC86 | HCT 116 | C1386(1.08) | LDD0669 | [12] |
| LDCM0353 | AC87 | HCT 116 | C1386(1.09) | LDD0670 | [12] |
| LDCM0354 | AC88 | HCT 116 | C1386(1.09) | LDD0671 | [12] |
| LDCM0355 | AC89 | HCT 116 | C1386(1.06) | LDD0672 | [12] |
| LDCM0357 | AC90 | HCT 116 | C1386(0.78) | LDD0674 | [12] |
| LDCM0358 | AC91 | HCT 116 | C1386(1.00) | LDD0675 | [12] |
| LDCM0359 | AC92 | HCT 116 | C1386(0.82) | LDD0676 | [12] |
| LDCM0360 | AC93 | HCT 116 | C1386(0.92) | LDD0677 | [12] |
| LDCM0361 | AC94 | HCT 116 | C1386(0.84) | LDD0678 | [12] |
| LDCM0362 | AC95 | HCT 116 | C1386(0.97) | LDD0679 | [12] |
| LDCM0363 | AC96 | HCT 116 | C1386(0.92) | LDD0680 | [12] |
| LDCM0364 | AC97 | HCT 116 | C1386(0.89) | LDD0681 | [12] |
| LDCM0365 | AC98 | HCT 116 | C1386(0.87) | LDD0682 | [12] |
| LDCM0366 | AC99 | HCT 116 | C1386(0.67) | LDD0683 | [12] |
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C1024(0.83) | LDD2113 | [8] |
| LDCM0248 | AKOS034007472 | PaTu 8988t | C1479(1.08); C1475(0.53) | LDD1127 | [12] |
| LDCM0356 | AKOS034007680 | PaTu 8988t | C1479(0.88); C1475(0.71) | LDD1235 | [12] |
| LDCM0275 | AKOS034007705 | PaTu 8988t | C1479(1.07); C1475(0.58) | LDD1154 | [12] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [11] |
| LDCM0367 | CL1 | HCT 116 | C1475(1.00) | LDD0684 | [12] |
| LDCM0368 | CL10 | HCT 116 | C1475(0.95) | LDD0685 | [12] |
| LDCM0369 | CL100 | HCT 116 | C1386(1.06) | LDD0686 | [12] |
| LDCM0370 | CL101 | PaTu 8988t | C1479(1.12); C1475(0.70) | LDD1249 | [12] |
| LDCM0371 | CL102 | PaTu 8988t | C1479(1.01); C1475(0.79) | LDD1250 | [12] |
| LDCM0372 | CL103 | PaTu 8988t | C1479(0.74); C1475(0.32) | LDD1251 | [12] |
| LDCM0373 | CL104 | PaTu 8988t | C1479(1.03); C1475(0.71) | LDD1252 | [12] |
| LDCM0374 | CL105 | PaTu 8988t | C1479(1.14); C1475(1.14) | LDD1253 | [12] |
| LDCM0375 | CL106 | PaTu 8988t | C1479(0.95); C1475(0.95) | LDD1254 | [12] |
| LDCM0376 | CL107 | PaTu 8988t | C1479(0.98); C1475(0.98) | LDD1255 | [12] |
| LDCM0377 | CL108 | PaTu 8988t | C1479(1.13); C1475(1.13) | LDD1256 | [12] |
| LDCM0378 | CL109 | PaTu 8988t | C1479(0.97); C1475(0.97) | LDD1257 | [12] |
| LDCM0379 | CL11 | HCT 116 | C1475(1.01) | LDD0696 | [12] |
| LDCM0380 | CL110 | PaTu 8988t | C1479(0.87); C1475(0.87) | LDD1259 | [12] |
| LDCM0381 | CL111 | PaTu 8988t | C1479(0.62); C1475(0.62) | LDD1260 | [12] |
| LDCM0382 | CL112 | HEK-293T | C1479(1.40) | LDD0980 | [12] |
| LDCM0383 | CL113 | HEK-293T | C1479(1.42) | LDD0981 | [12] |
| LDCM0384 | CL114 | HEK-293T | C1479(1.87) | LDD0982 | [12] |
| LDCM0385 | CL115 | HEK-293T | C1479(1.97) | LDD0983 | [12] |
| LDCM0386 | CL116 | HEK-293T | C1479(2.07) | LDD0984 | [12] |
| LDCM0387 | CL117 | PaTu 8988t | C1479(1.01); C1475(1.01) | LDD1266 | [12] |
| LDCM0388 | CL118 | PaTu 8988t | C1479(1.34); C1475(1.34) | LDD1267 | [12] |
| LDCM0389 | CL119 | PaTu 8988t | C1479(1.22); C1475(1.22) | LDD1268 | [12] |
| LDCM0390 | CL12 | HCT 116 | C1475(1.18) | LDD0707 | [12] |
| LDCM0391 | CL120 | PaTu 8988t | C1479(1.24); C1475(1.24) | LDD1270 | [12] |
| LDCM0392 | CL121 | HCT 116 | C1386(1.02) | LDD0709 | [12] |
| LDCM0393 | CL122 | HCT 116 | C1386(1.01) | LDD0710 | [12] |
| LDCM0394 | CL123 | HCT 116 | C1386(0.87) | LDD0711 | [12] |
| LDCM0395 | CL124 | HCT 116 | C1386(0.68) | LDD0712 | [12] |
| LDCM0396 | CL125 | HCT 116 | C1386(0.83) | LDD0713 | [12] |
| LDCM0397 | CL126 | HCT 116 | C1386(1.08) | LDD0714 | [12] |
| LDCM0398 | CL127 | HCT 116 | C1386(1.22) | LDD0715 | [12] |
| LDCM0399 | CL128 | HCT 116 | C1386(0.88) | LDD0716 | [12] |
| LDCM0400 | CL13 | HCT 116 | C1475(1.08) | LDD0717 | [12] |
| LDCM0401 | CL14 | HCT 116 | C1475(1.22) | LDD0718 | [12] |
| LDCM0402 | CL15 | HCT 116 | C1475(1.05) | LDD0719 | [12] |
| LDCM0403 | CL16 | HCT 116 | C1475(0.88) | LDD0720 | [12] |
| LDCM0404 | CL17 | HCT 116 | C1475(0.60); C1386(0.77) | LDD0721 | [12] |
| LDCM0405 | CL18 | HCT 116 | C1386(0.78); C1475(0.91) | LDD0722 | [12] |
| LDCM0406 | CL19 | HCT 116 | C1475(0.61); C1386(1.13) | LDD0723 | [12] |
| LDCM0407 | CL2 | HCT 116 | C1475(1.12) | LDD0724 | [12] |
| LDCM0408 | CL20 | HCT 116 | C1475(0.76); C1386(0.87) | LDD0725 | [12] |
| LDCM0409 | CL21 | HCT 116 | C1475(0.40); C1386(0.84) | LDD0726 | [12] |
| LDCM0410 | CL22 | HCT 116 | C1475(0.21); C1386(1.45) | LDD0727 | [12] |
| LDCM0411 | CL23 | HCT 116 | C1386(0.74); C1475(1.67) | LDD0728 | [12] |
| LDCM0412 | CL24 | HCT 116 | C1386(0.89); C1475(0.55) | LDD0729 | [12] |
| LDCM0413 | CL25 | HCT 116 | C1386(0.86); C1475(0.46) | LDD0730 | [12] |
| LDCM0414 | CL26 | HCT 116 | C1386(0.88); C1475(0.80) | LDD0731 | [12] |
| LDCM0415 | CL27 | HCT 116 | C1386(1.09); C1475(0.85) | LDD0732 | [12] |
| LDCM0416 | CL28 | HCT 116 | C1386(0.86); C1475(0.68) | LDD0733 | [12] |
| LDCM0417 | CL29 | HCT 116 | C1386(1.13); C1475(0.59) | LDD0734 | [12] |
| LDCM0418 | CL3 | HCT 116 | C1475(1.13) | LDD0735 | [12] |
| LDCM0419 | CL30 | HCT 116 | C1386(0.75); C1475(0.67) | LDD0736 | [12] |
| LDCM0420 | CL31 | HCT 116 | C1386(0.93); C1475(1.06) | LDD0737 | [12] |
| LDCM0421 | CL32 | HCT 116 | C1475(0.94) | LDD0738 | [12] |
| LDCM0422 | CL33 | HCT 116 | C1475(0.99) | LDD0739 | [12] |
| LDCM0423 | CL34 | HCT 116 | C1475(0.91) | LDD0740 | [12] |
| LDCM0424 | CL35 | HCT 116 | C1475(1.15) | LDD0741 | [12] |
| LDCM0425 | CL36 | HCT 116 | C1475(1.18) | LDD0742 | [12] |
| LDCM0426 | CL37 | HCT 116 | C1475(0.98) | LDD0743 | [12] |
| LDCM0428 | CL39 | HCT 116 | C1475(1.07) | LDD0745 | [12] |
| LDCM0429 | CL4 | HCT 116 | C1475(1.10) | LDD0746 | [12] |
| LDCM0430 | CL40 | HCT 116 | C1475(1.13) | LDD0747 | [12] |
| LDCM0431 | CL41 | HCT 116 | C1475(1.50) | LDD0748 | [12] |
| LDCM0432 | CL42 | HCT 116 | C1475(0.50) | LDD0749 | [12] |
| LDCM0433 | CL43 | HCT 116 | C1475(0.81) | LDD0750 | [12] |
| LDCM0434 | CL44 | HCT 116 | C1475(0.89) | LDD0751 | [12] |
| LDCM0435 | CL45 | HCT 116 | C1475(0.91) | LDD0752 | [12] |
| LDCM0436 | CL46 | HEK-293T | C1479(1.28) | LDD1034 | [12] |
| LDCM0437 | CL47 | HEK-293T | C1479(1.09) | LDD1035 | [12] |
| LDCM0438 | CL48 | HEK-293T | C1479(1.11) | LDD1036 | [12] |
| LDCM0439 | CL49 | HEK-293T | C1479(1.27) | LDD1037 | [12] |
| LDCM0440 | CL5 | HCT 116 | C1475(1.17) | LDD0757 | [12] |
| LDCM0441 | CL50 | HEK-293T | C1479(1.36) | LDD1039 | [12] |
| LDCM0442 | CL51 | HEK-293T | C1479(1.15) | LDD1040 | [12] |
| LDCM0443 | CL52 | HEK-293T | C1479(0.94) | LDD1041 | [12] |
| LDCM0444 | CL53 | HEK-293T | C1479(1.03) | LDD1042 | [12] |
| LDCM0445 | CL54 | HEK-293T | C1479(1.15) | LDD1043 | [12] |
| LDCM0446 | CL55 | HEK-293T | C1479(1.41) | LDD1044 | [12] |
| LDCM0447 | CL56 | HEK-293T | C1479(0.96) | LDD1045 | [12] |
| LDCM0448 | CL57 | HEK-293T | C1479(1.18) | LDD1046 | [12] |
| LDCM0449 | CL58 | HEK-293T | C1479(1.22) | LDD1047 | [12] |
| LDCM0450 | CL59 | HEK-293T | C1479(1.36) | LDD1048 | [12] |
| LDCM0451 | CL6 | HCT 116 | C1475(1.06) | LDD0768 | [12] |
| LDCM0452 | CL60 | HEK-293T | C1479(1.55) | LDD1050 | [12] |
| LDCM0453 | CL61 | HCT 116 | C1386(1.17); C1475(0.83) | LDD0770 | [12] |
| LDCM0454 | CL62 | HCT 116 | C1386(1.05); C1475(0.79) | LDD0771 | [12] |
| LDCM0455 | CL63 | HCT 116 | C1386(1.79); C1475(0.98) | LDD0772 | [12] |
| LDCM0456 | CL64 | HCT 116 | C1386(1.18); C1475(0.83) | LDD0773 | [12] |
| LDCM0457 | CL65 | HCT 116 | C1386(1.37); C1475(0.98) | LDD0774 | [12] |
| LDCM0458 | CL66 | HCT 116 | C1386(1.16); C1475(0.93) | LDD0775 | [12] |
| LDCM0459 | CL67 | HCT 116 | C1386(1.81); C1475(0.98) | LDD0776 | [12] |
| LDCM0460 | CL68 | HCT 116 | C1386(3.04); C1475(0.92) | LDD0777 | [12] |
| LDCM0461 | CL69 | HCT 116 | C1386(1.30); C1475(0.92) | LDD0778 | [12] |
| LDCM0462 | CL7 | HCT 116 | C1475(1.10) | LDD0779 | [12] |
| LDCM0463 | CL70 | HCT 116 | C1386(1.16); C1475(0.92) | LDD0780 | [12] |
| LDCM0464 | CL71 | HCT 116 | C1386(1.38); C1475(0.88) | LDD0781 | [12] |
| LDCM0465 | CL72 | HCT 116 | C1386(1.50); C1475(0.88) | LDD0782 | [12] |
| LDCM0466 | CL73 | HCT 116 | C1386(1.63); C1475(0.76) | LDD0783 | [12] |
| LDCM0467 | CL74 | HCT 116 | C1386(1.82); C1475(1.07) | LDD0784 | [12] |
| LDCM0469 | CL76 | PaTu 8988t | C1475(0.89) | LDD1348 | [12] |
| LDCM0470 | CL77 | PaTu 8988t | C1475(0.81) | LDD1349 | [12] |
| LDCM0471 | CL78 | PaTu 8988t | C1475(1.16) | LDD1350 | [12] |
| LDCM0472 | CL79 | PaTu 8988t | C1475(1.08) | LDD1351 | [12] |
| LDCM0473 | CL8 | HCT 116 | C1475(0.99) | LDD0790 | [12] |
| LDCM0474 | CL80 | PaTu 8988t | C1475(1.05) | LDD1353 | [12] |
| LDCM0475 | CL81 | PaTu 8988t | C1475(0.94) | LDD1354 | [12] |
| LDCM0476 | CL82 | PaTu 8988t | C1475(0.96) | LDD1355 | [12] |
| LDCM0477 | CL83 | PaTu 8988t | C1475(1.07) | LDD1356 | [12] |
| LDCM0478 | CL84 | PaTu 8988t | C1475(0.98) | LDD1357 | [12] |
| LDCM0479 | CL85 | PaTu 8988t | C1475(1.01) | LDD1358 | [12] |
| LDCM0480 | CL86 | PaTu 8988t | C1475(0.95) | LDD1359 | [12] |
| LDCM0481 | CL87 | PaTu 8988t | C1475(0.95) | LDD1360 | [12] |
| LDCM0482 | CL88 | PaTu 8988t | C1475(0.92) | LDD1361 | [12] |
| LDCM0483 | CL89 | PaTu 8988t | C1475(0.91) | LDD1362 | [12] |
| LDCM0484 | CL9 | HCT 116 | C1475(1.34) | LDD0801 | [12] |
| LDCM0485 | CL90 | PaTu 8988t | C1475(0.97) | LDD1364 | [12] |
| LDCM0486 | CL91 | HCT 116 | C1386(1.11) | LDD0803 | [12] |
| LDCM0487 | CL92 | HCT 116 | C1386(1.12) | LDD0804 | [12] |
| LDCM0488 | CL93 | HCT 116 | C1386(0.87) | LDD0805 | [12] |
| LDCM0489 | CL94 | HCT 116 | C1386(1.05) | LDD0806 | [12] |
| LDCM0490 | CL95 | HCT 116 | C1386(1.34) | LDD0807 | [12] |
| LDCM0491 | CL96 | HCT 116 | C1386(1.13) | LDD0808 | [12] |
| LDCM0492 | CL97 | HCT 116 | C1386(1.05) | LDD0809 | [12] |
| LDCM0493 | CL98 | HCT 116 | C1386(0.95) | LDD0810 | [12] |
| LDCM0494 | CL99 | HCT 116 | C1386(0.95) | LDD0811 | [12] |
| LDCM0191 | Compound 21 | HEK-293T | 9.62 | LDD0508 | [20] |
| LDCM0192 | Compound 35 | HEK-293T | 6.54 | LDD0509 | [20] |
| LDCM0193 | Compound 36 | HEK-293T | 18.35 | LDD0511 | [20] |
| LDCM0495 | E2913 | HEK-293T | C1024(1.11); C880(1.11); C1475(2.11); C1479(0.84) | LDD1698 | [25] |
| LDCM0468 | Fragment33 | HCT 116 | C1386(1.17); C1475(0.85) | LDD0785 | [12] |
| LDCM0427 | Fragment51 | HCT 116 | C1475(0.63) | LDD0744 | [12] |
| LDCM0116 | HHS-0101 | DM93 | Y599(0.05) | LDD0264 | [9] |
| LDCM0118 | HHS-0301 | DM93 | Y599(2.36) | LDD0266 | [9] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [11] |
| LDCM0022 | KB02 | HCT 116 | C1493(1.10) | LDD0080 | [12] |
| LDCM0023 | KB03 | HCT 116 | C1493(0.99) | LDD0081 | [12] |
| LDCM0024 | KB05 | HCT 116 | C1493(1.06) | LDD0082 | [12] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [11] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C1024(1.08) | LDD2093 | [8] |
| LDCM0504 | Nucleophilic fragment 15a | MDA-MB-231 | C1024(0.70) | LDD2097 | [8] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C1024(1.20) | LDD2099 | [8] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C1024(1.06) | LDD2101 | [8] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C1024(1.04) | LDD2107 | [8] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C1024(0.50) | LDD2109 | [8] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C1024(1.25) | LDD2111 | [8] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C1024(0.50) | LDD2115 | [8] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C1024(1.83) | LDD2119 | [8] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C1024(1.03) | LDD2120 | [8] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C1024(0.92) | LDD2123 | [8] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C1024(0.83) | LDD2125 | [8] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C1024(0.92); C1493(0.78) | LDD2127 | [8] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C1024(1.23) | LDD2128 | [8] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C1024(1.25) | LDD2129 | [8] |
| LDCM0540 | Nucleophilic fragment 35 | MDA-MB-231 | C1024(0.66) | LDD2133 | [8] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C1024(0.54) | LDD2134 | [8] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C1024(2.13) | LDD2135 | [8] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C1024(1.30) | LDD2136 | [8] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C1024(1.00) | LDD2137 | [8] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C1024(3.38) | LDD1700 | [8] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C1024(0.76) | LDD2140 | [8] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C1024(1.39) | LDD2143 | [8] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C1024(2.29) | LDD2144 | [8] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C1024(0.97) | LDD2146 | [8] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C1024(0.47) | LDD2148 | [8] |
References
