Details of the Target
General Information of Target
| Target ID | LDTP06255 | |||||
|---|---|---|---|---|---|---|
| Target Name | GTPase-activating protein and VPS9 domain-containing protein 1 (GAPVD1) | |||||
| Gene Name | GAPVD1 | |||||
| Gene ID | 26130 | |||||
| Synonyms |
GAPEX5; KIAA1521; RAP6; GTPase-activating protein and VPS9 domain-containing protein 1; GAPex-5; Rab5-activating protein 6 |
|||||
| 3D Structure | ||||||
| Sequence |
MVKLDIHTLAHHLKQERLYVNSEKQLIQRLNADVLKTAEKLYRTAWIAKQQRINLDRLII
TSAEASPAECCQHAKILEDTQFVDGYKQLGFQETAYGEFLSRLRENPRLIASSLVAGEKL NQENTQSVIYTVFTSLYGNCIMQEDESYLLQVLRYLIEFELKESDNPRRLLRRGTCAFSI LFKLFSEGLFSAKLFLTATLHEPIMQLLVEDEDHLETDPNKLIERFSPSQQEKLFGEKGS DRFRQKVQEMVESNEAKLVALVNKFIGYLKQNTYCFPHSLRWIVSQMYKTLSCVDRLEVG EVRAMCTDLLLACFICPAVVNPEQYGIISDAPINEVARFNLMQVGRLLQQLAMTGSEEGD PRTKSSLGKFDKSCVAAFLDVVIGGRAVETPPLSSVNLLEGLSRTVVYITYSQLITLVNF MKSVMSGDQLREDRMALDNLLANLPPAKPGKSSSLEMTPYNTPQLSPATTPANKKNRLPI ATRSRSRTNMLMDLHMDHEGSSQETIQEVQPEEVLVISLGTGPQLTPGMMSENEVLNMQL SDGGQGDVPVDENKLHGKPDKTLRFSLCSDNLEGISEGPSNRSNSVSSLDLEGESVSELG AGPSGSNGVEALQLLEHEQATTQDNLDDKLRKFEIRDMMGLTDDRDISETVSETWSTDVL GSDFDPNIDEDRLQEIAGAAAENMLGSLLCLPGSGSVLLDPCTGSTISETTSEAWSVEVL PSDSEAPDLKQEERLQELESCSGLGSTSDDTDVREVSSRPSTPGLSVVSGISATSEDIPN KIEDLRSECSSDFGGKDSVTSPDMDEITHGAHQLTSPPSQSESLLAMFDPLSSHEGASAV VRPKVHYARPSHPPPDPPILEGAVGGNEARLPNFGSHVLTPAEMEAFKQRHSYPERLVRS RSSDIVSSVRRPMSDPSWNRRPGNEERELPPAAAIGATSLVAAPHSSSSSPSKDSSRGET EERKDSDDEKSDRNRPWWRKRFVSAMPKAPIPFRKKEKQEKDKDDLGPDRFSTLTDDPSP RLSAQAQVAEDILDKYRNAIKRTSPSDGAMANYESTGDNHDRDLSSKLLYHSDKEVMGDG ESAHDSPRDEALQNISADDLPDSASQAAHPQDSAFSYRDAKKKLRLALCSADSVAFPVLT HSTRNGLPDHTDPEDNEIVCFLKVQIAEAINLQDKNLMAQLQETMRCVCRFDNRTCRKLL ASIAEDYRKRAPYIAYLTRCRQGLQTTQAHLERLLQRVLRDKEVANRYFTTVCVRLLLES KEKKIREFIQDFQKLTAADDKTAQVEDFLQFLYGAMAQDVIWQNASEEQLQDAQLAIERS VMNRIFKLAFYPNQDGDILRDQVLHEHIQRLSKVVTANHRALQIPEVYLREAPWPSAQSE IRTISAYKTPRDKVQCILRMCSTIMNLLSLANEDSVPGADDFVPVLVFVLIKANPPCLLS TVQYISSFYASCLSGEESYWWMQFTAAVEFIKTIDDRK |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
GAPVD1 family
|
|||||
| Subcellular location |
Membrane
|
|||||
| Function |
Acts both as a GTPase-activating protein (GAP) and a guanine nucleotide exchange factor (GEF), and participates in various processes such as endocytosis, insulin receptor internalization or LC2A4/GLUT4 trafficking. Acts as a GEF for the Ras-related protein RAB31 by exchanging bound GDP for free GTP, leading to regulate LC2A4/GLUT4 trafficking. In the absence of insulin, it maintains RAB31 in an active state and promotes a futile cycle between LC2A4/GLUT4 storage vesicles and early endosomes, retaining LC2A4/GLUT4 inside the cells. Upon insulin stimulation, it is translocated to the plasma membrane, releasing LC2A4/GLUT4 from intracellular storage vesicles. Also involved in EGFR trafficking and degradation, possibly by promoting EGFR ubiquitination and subsequent degradation by the proteasome. Has GEF activity for Rab5 and GAP activity for Ras.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
14.40 | LDD0402 | [1] | |
|
A-EBA Probe Info |
 |
2.72 | LDD0215 | [2] | |
|
CY-1 Probe Info |
 |
100.00 | LDD0243 | [3] | |
|
CY4 Probe Info |
 |
100.00 | LDD0244 | [3] | |
|
N1 Probe Info |
 |
100.00 | LDD0242 | [3] | |
|
TH211 Probe Info |
 |
Y1331(20.00); Y1368(13.97); Y1207(9.09); Y1387(5.12) | LDD0257 | [4] | |
|
TH216 Probe Info |
 |
Y1207(20.00) | LDD0259 | [4] | |
|
ONAyne Probe Info |
 |
K1035(1.59) | LDD0274 | [5] | |
|
Probe 1 Probe Info |
 |
Y19(44.86); Y86(19.12); Y96(5.41); Y1216(3.73) | LDD3495 | [6] | |
|
BTD Probe Info |
 |
C1396(1.08) | LDD2098 | [7] | |
|
DA-P3 Probe Info |
 |
5.57 | LDD0182 | [8] | |
|
AHL-Pu-1 Probe Info |
 |
C789(4.03); C374(2.31); C568(2.12) | LDD0169 | [9] | |
|
DBIA Probe Info |
 |
C176(4.71); C293(12.72); C176(92.05); C1396(26.86) | LDD0209 | [10] | |
|
HHS-482 Probe Info |
 |
Y1207(1.05) | LDD0285 | [11] | |
|
HHS-475 Probe Info |
 |
Y1036(0.80); Y460(0.89); Y86(0.95); Y1207(1.17) | LDD0264 | [12] | |
|
HHS-465 Probe Info |
 |
Y1036(10.00); Y460(2.92); Y893(9.41) | LDD2237 | [13] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [14] | |
|
ATP probe Probe Info |
 |
K1198(0.00); K1035(0.00) | LDD0199 | [15] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C1396(0.00); C568(0.00); C1129(0.00); C275(0.00) | LDD0038 | [16] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0032 | [17] | |
|
Lodoacetamide azide Probe Info |
 |
C1396(0.00); C568(0.00); C1129(0.00); C275(0.00) | LDD0037 | [16] | |
|
JW-RF-010 Probe Info |
 |
C71(0.00); C1129(0.00); C70(0.00) | LDD0026 | [18] | |
|
NAIA_4 Probe Info |
 |
C374(0.00); C741(0.00) | LDD2226 | [19] | |
|
TFBX Probe Info |
 |
C1253(0.00); C71(0.00); C1220(0.00); C1129(0.00) | LDD0027 | [18] | |
|
Compound 10 Probe Info |
 |
C1129(0.00); C568(0.00) | LDD2216 | [20] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [20] | |
|
ENE Probe Info |
 |
C741(0.00); C789(0.00) | LDD0006 | [21] | |
|
IPM Probe Info |
 |
C568(0.00); C1396(0.00); C293(0.00); C275(0.00) | LDD0005 | [21] | |
|
SF Probe Info |
 |
Y893(0.00); Y1036(0.00) | LDD0028 | [22] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [21] | |
|
VSF Probe Info |
 |
C1129(0.00); C568(0.00); C789(0.00); C741(0.00) | LDD0007 | [21] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [23] | |
|
Acrolein Probe Info |
 |
C741(0.00); C568(0.00); C1129(0.00); C789(0.00) | LDD0217 | [24] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [24] | |
|
Methacrolein Probe Info |
 |
C741(0.00); C568(0.00); C1129(0.00); C70(0.00) | LDD0218 | [24] | |
|
W1 Probe Info |
 |
C1253(0.00); C741(0.00) | LDD0236 | [25] | |
|
AOyne Probe Info |
 |
14.90 | LDD0443 | [26] | |
|
NAIA_5 Probe Info |
 |
C1220(0.00); C1396(0.00); C176(0.00); C1129(0.00) | LDD2223 | [19] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C317 Probe Info |
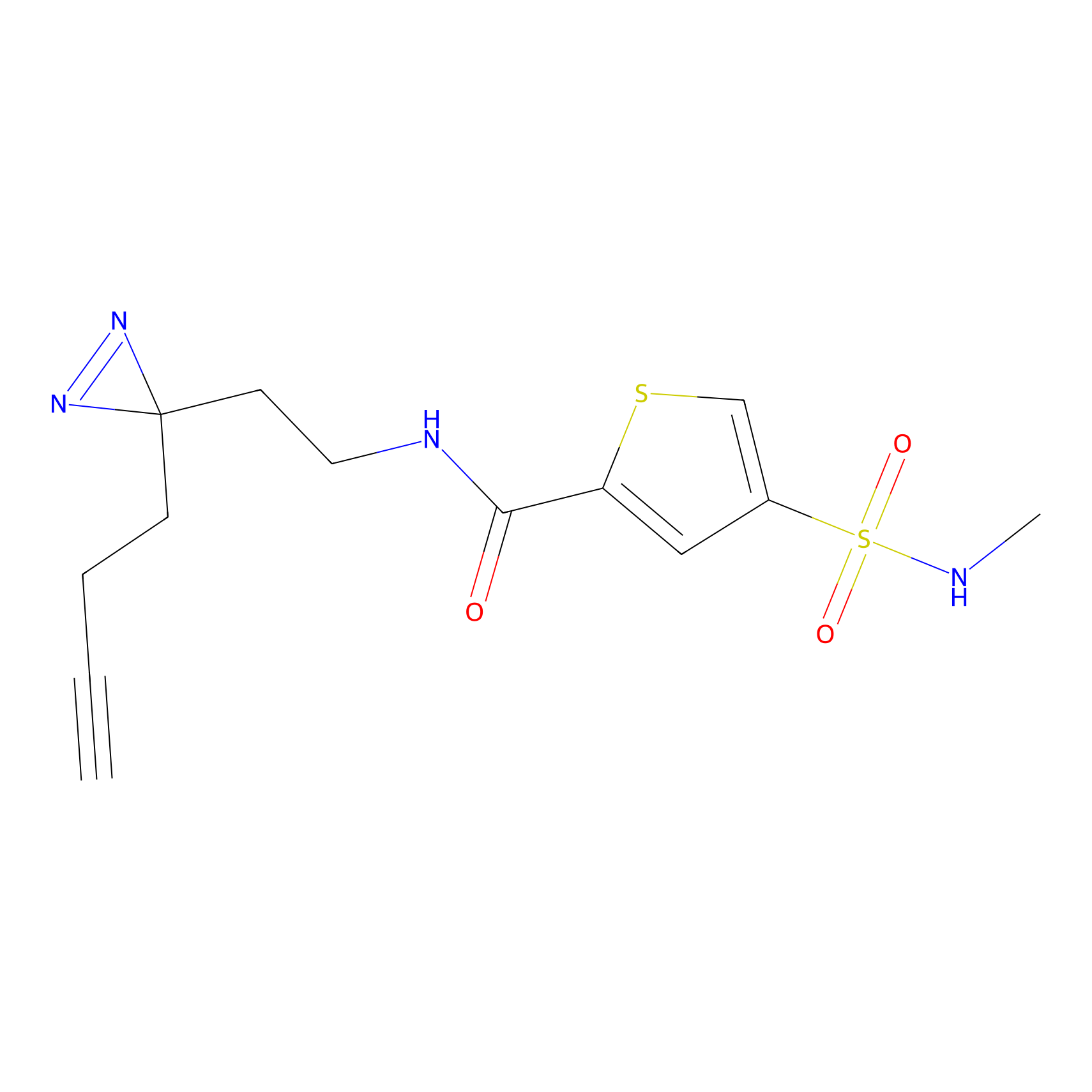 |
6.11 | LDD1983 | [27] | |
|
C320 Probe Info |
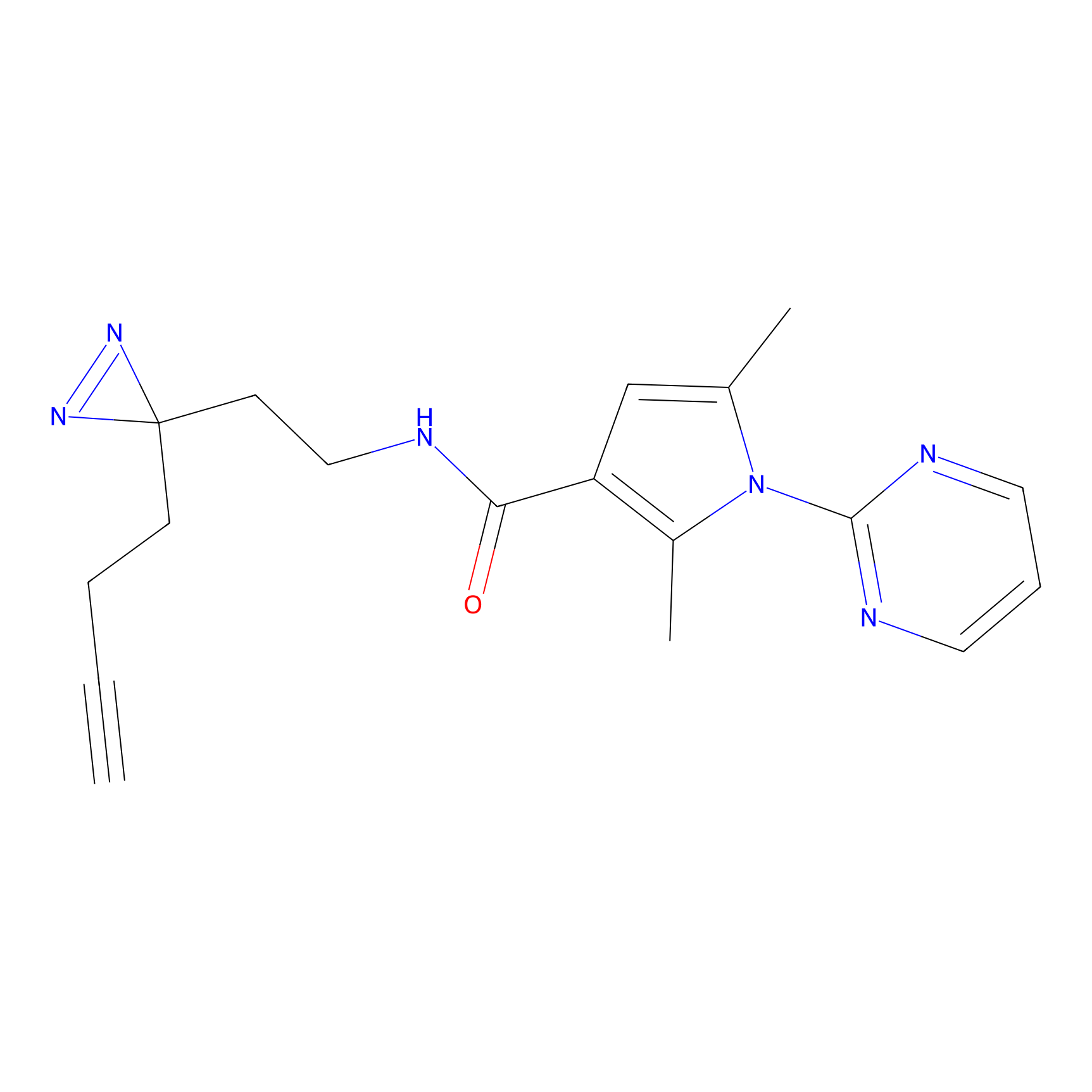 |
5.62 | LDD1986 | [27] | |
|
C433 Probe Info |
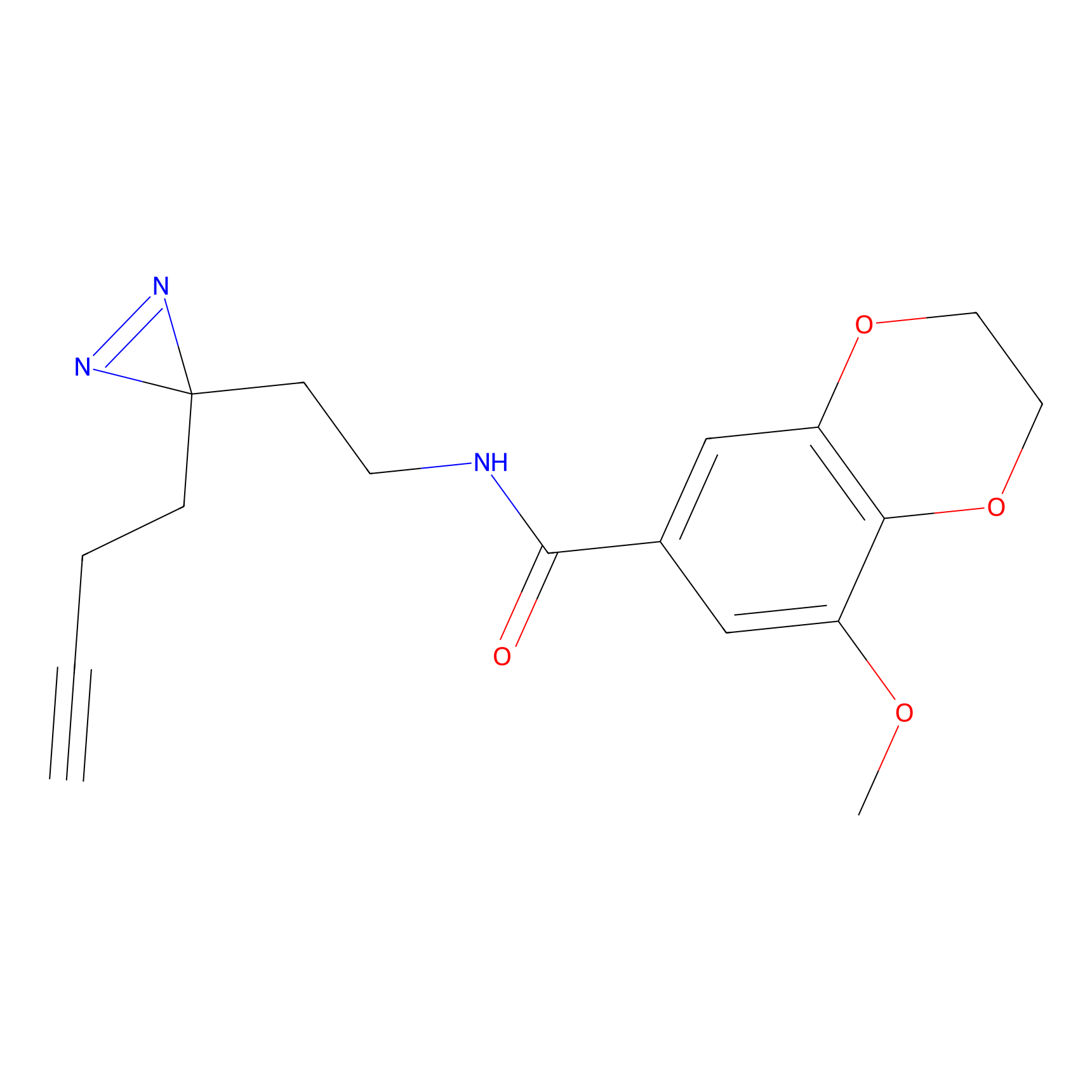 |
5.94 | LDD2088 | [27] | |
|
FFF probe3 Probe Info |
 |
7.06 | LDD0465 | [28] | |
|
Alk-rapa Probe Info |
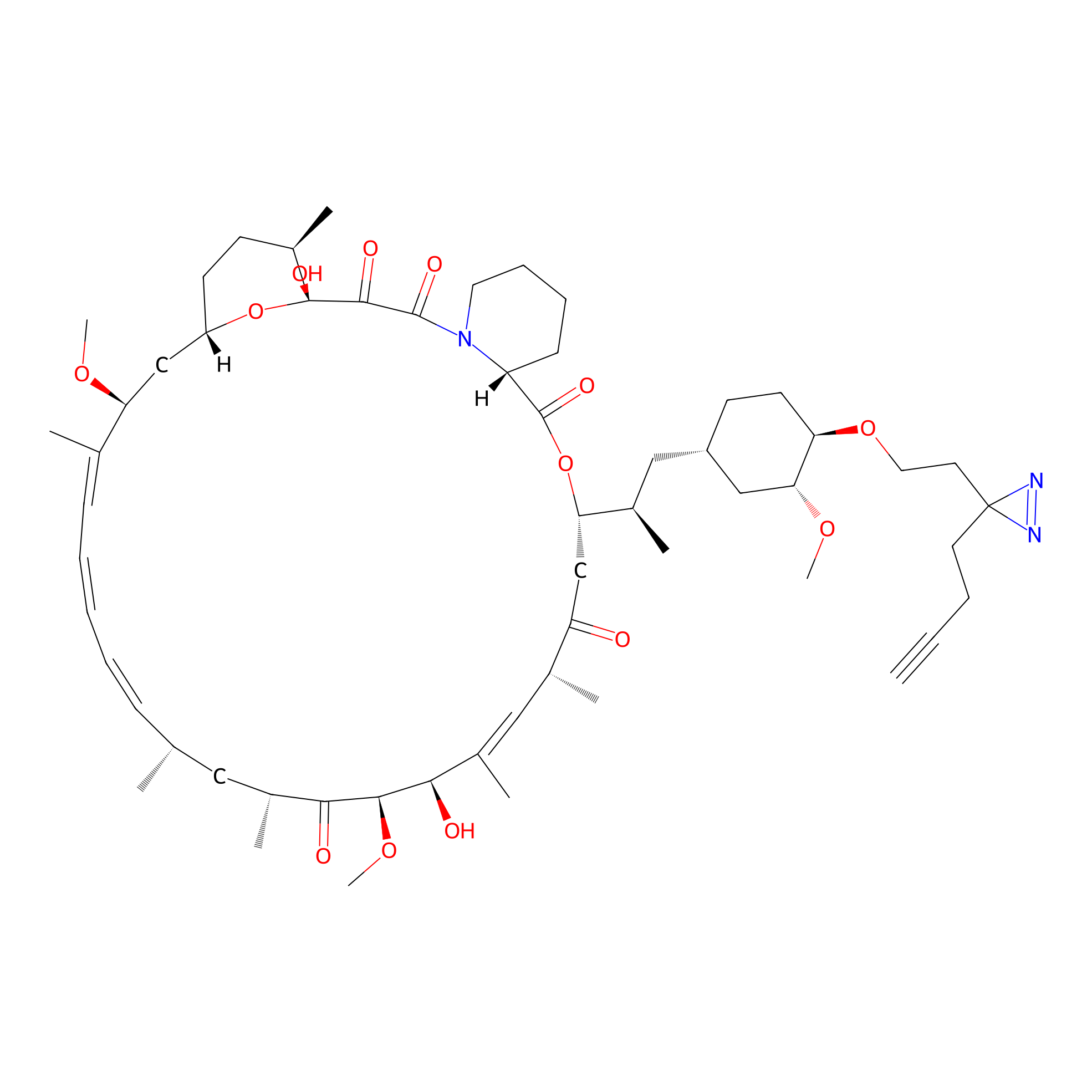 |
3.44 | LDD0213 | [29] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | DM93 | C70(20.00) | LDD0170 | [9] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C789(4.03); C374(2.31); C568(2.12) | LDD0169 | [9] |
| LDCM0561 | Abegg_cp(-)-10 | HeLa | C1396(2.12) | LDD0312 | [18] |
| LDCM0214 | AC1 | HEK-293T | C293(0.89); C789(0.99); C1253(1.04); C568(0.99) | LDD1507 | [30] |
| LDCM0215 | AC10 | HEK-293T | C293(0.95); C789(0.98); C1253(1.08); C568(1.03) | LDD1508 | [30] |
| LDCM0226 | AC11 | HEK-293T | C293(1.01); C789(1.05); C1253(0.90); C568(1.00) | LDD1509 | [30] |
| LDCM0237 | AC12 | HEK-293T | C293(0.98); C789(0.95); C1253(0.97); C568(1.05) | LDD1510 | [30] |
| LDCM0259 | AC14 | HEK-293T | C293(0.98); C789(0.95); C1253(0.96); C568(0.94) | LDD1512 | [30] |
| LDCM0270 | AC15 | HEK-293T | C293(1.00); C789(0.97); C1253(1.00); C568(1.00) | LDD1513 | [30] |
| LDCM0276 | AC17 | HEK-293T | C293(0.95); C789(1.00); C1253(1.01); C568(1.03) | LDD1515 | [30] |
| LDCM0277 | AC18 | HEK-293T | C293(0.95); C789(1.01); C1253(1.06); C568(0.97) | LDD1516 | [30] |
| LDCM0278 | AC19 | HEK-293T | C293(0.98); C789(1.13); C1253(1.03); C568(1.05) | LDD1517 | [30] |
| LDCM0279 | AC2 | HEK-293T | C293(0.98); C789(0.98); C1253(0.95); C568(1.12) | LDD1518 | [30] |
| LDCM0280 | AC20 | HEK-293T | C293(0.93); C789(0.96); C1253(0.98); C568(1.00) | LDD1519 | [30] |
| LDCM0281 | AC21 | HEK-293T | C293(1.00); C789(0.96); C1253(0.95); C568(1.03) | LDD1520 | [30] |
| LDCM0282 | AC22 | HEK-293T | C293(0.95); C789(0.94); C1253(0.96); C568(1.03) | LDD1521 | [30] |
| LDCM0283 | AC23 | HEK-293T | C293(1.02); C789(0.99); C1253(0.93); C568(1.05) | LDD1522 | [30] |
| LDCM0284 | AC24 | HEK-293T | C293(0.98); C789(0.95); C1253(0.90); C568(0.99) | LDD1523 | [30] |
| LDCM0285 | AC25 | HEK-293T | C293(0.93); C789(0.96); C1253(0.98); C568(1.01) | LDD1524 | [30] |
| LDCM0286 | AC26 | HEK-293T | C293(0.94); C789(1.03); C1253(1.03); C568(0.95) | LDD1525 | [30] |
| LDCM0287 | AC27 | HEK-293T | C293(0.98); C789(0.96); C1253(1.00); C568(1.12) | LDD1526 | [30] |
| LDCM0288 | AC28 | HEK-293T | C293(0.97); C789(1.05); C1253(1.00); C568(1.07) | LDD1527 | [30] |
| LDCM0289 | AC29 | HEK-293T | C293(0.98); C789(1.04); C1253(0.90); C568(1.04) | LDD1528 | [30] |
| LDCM0290 | AC3 | HEK-293T | C293(1.01); C789(0.99); C1253(1.06); C568(0.99) | LDD1529 | [30] |
| LDCM0291 | AC30 | HEK-293T | C293(0.94); C789(0.95); C1253(0.96); C568(0.97) | LDD1530 | [30] |
| LDCM0292 | AC31 | HEK-293T | C293(1.01); C789(0.96); C1253(0.95); C568(1.06) | LDD1531 | [30] |
| LDCM0293 | AC32 | HEK-293T | C293(0.99); C789(0.97); C1253(0.98); C568(0.99) | LDD1532 | [30] |
| LDCM0294 | AC33 | HEK-293T | C293(0.95); C789(0.99); C1253(1.00); C568(0.91) | LDD1533 | [30] |
| LDCM0295 | AC34 | HEK-293T | C293(0.95); C789(1.03); C1253(1.13); C568(0.92) | LDD1534 | [30] |
| LDCM0296 | AC35 | HEK-293T | C293(0.92); C789(1.00); C1253(0.98); C568(1.04) | LDD1535 | [30] |
| LDCM0297 | AC36 | HEK-293T | C293(0.91); C789(0.98); C1253(1.00); C568(0.98) | LDD1536 | [30] |
| LDCM0298 | AC37 | HEK-293T | C293(0.97); C789(0.97); C1253(0.86); C568(1.08) | LDD1537 | [30] |
| LDCM0299 | AC38 | HEK-293T | C293(0.91); C789(0.92); C1253(0.96); C568(0.93) | LDD1538 | [30] |
| LDCM0300 | AC39 | HEK-293T | C293(0.91); C789(0.98); C1253(0.99); C568(0.92) | LDD1539 | [30] |
| LDCM0301 | AC4 | HEK-293T | C293(0.93); C789(1.03); C1253(1.02); C568(0.99) | LDD1540 | [30] |
| LDCM0302 | AC40 | HEK-293T | C293(0.97); C789(0.91); C1253(0.96); C568(1.10) | LDD1541 | [30] |
| LDCM0303 | AC41 | HEK-293T | C293(0.94); C789(0.99); C1253(0.99); C568(0.99) | LDD1542 | [30] |
| LDCM0304 | AC42 | HEK-293T | C293(0.93); C789(0.96); C1253(0.99); C568(1.13) | LDD1543 | [30] |
| LDCM0305 | AC43 | HEK-293T | C293(0.97); C789(0.99); C1253(0.98); C568(1.13) | LDD1544 | [30] |
| LDCM0306 | AC44 | HEK-293T | C293(0.92); C789(1.01); C1253(1.02); C568(1.03) | LDD1545 | [30] |
| LDCM0307 | AC45 | HEK-293T | C293(1.01); C789(1.01); C1253(0.92); C568(1.04) | LDD1546 | [30] |
| LDCM0308 | AC46 | HEK-293T | C293(0.92); C789(0.91); C1253(0.95); C568(0.86) | LDD1547 | [30] |
| LDCM0309 | AC47 | HEK-293T | C293(0.92); C789(1.02); C1253(0.96); C568(1.00) | LDD1548 | [30] |
| LDCM0310 | AC48 | HEK-293T | C293(0.96); C789(0.97); C1253(0.94); C568(1.13) | LDD1549 | [30] |
| LDCM0311 | AC49 | HEK-293T | C293(0.94); C789(0.96); C1253(0.93); C568(0.99) | LDD1550 | [30] |
| LDCM0312 | AC5 | HEK-293T | C293(1.01); C789(0.94); C1253(0.93); C568(1.09) | LDD1551 | [30] |
| LDCM0313 | AC50 | HEK-293T | C293(0.93); C789(1.00); C1253(1.07); C568(1.23) | LDD1552 | [30] |
| LDCM0314 | AC51 | HEK-293T | C293(1.02); C789(1.00); C1253(1.00); C568(1.07) | LDD1553 | [30] |
| LDCM0315 | AC52 | HEK-293T | C293(0.94); C789(1.01); C1253(0.98); C568(0.96) | LDD1554 | [30] |
| LDCM0316 | AC53 | HEK-293T | C293(0.94); C789(0.99); C1253(0.89); C568(1.06) | LDD1555 | [30] |
| LDCM0317 | AC54 | HEK-293T | C293(0.94); C789(0.93); C1253(1.03); C568(1.07) | LDD1556 | [30] |
| LDCM0318 | AC55 | HEK-293T | C293(0.95); C789(0.97); C1253(0.99); C568(1.14) | LDD1557 | [30] |
| LDCM0319 | AC56 | HEK-293T | C293(0.99); C789(0.95); C1253(0.97); C568(0.95) | LDD1558 | [30] |
| LDCM0320 | AC57 | HEK-293T | C293(0.95); C789(1.02); C1253(0.96); C568(0.95) | LDD1559 | [30] |
| LDCM0321 | AC58 | HEK-293T | C293(0.97); C789(0.97); C1253(1.00); C568(0.93) | LDD1560 | [30] |
| LDCM0322 | AC59 | HEK-293T | C293(0.96); C789(1.00); C1253(1.00); C568(1.05) | LDD1561 | [30] |
| LDCM0323 | AC6 | HEK-293T | C293(0.91); C789(0.93); C1253(0.95); C568(1.12) | LDD1562 | [30] |
| LDCM0324 | AC60 | HEK-293T | C293(0.99); C789(0.99); C1253(1.01); C568(0.97) | LDD1563 | [30] |
| LDCM0325 | AC61 | HEK-293T | C293(0.95); C789(0.98); C1253(1.01); C568(1.04) | LDD1564 | [30] |
| LDCM0326 | AC62 | HEK-293T | C293(0.97); C789(0.96); C1253(1.01); C568(0.88) | LDD1565 | [30] |
| LDCM0327 | AC63 | HEK-293T | C293(1.02); C789(1.03); C1253(1.04); C568(1.08) | LDD1566 | [30] |
| LDCM0328 | AC64 | HEK-293T | C293(1.01); C789(0.95); C1253(0.94); C568(0.87) | LDD1567 | [30] |
| LDCM0334 | AC7 | HEK-293T | C293(0.96); C789(1.01); C1253(0.94); C568(0.95) | LDD1568 | [30] |
| LDCM0345 | AC8 | HEK-293T | C293(0.98); C789(0.96); C1253(1.02); C568(1.03) | LDD1569 | [30] |
| LDCM0248 | AKOS034007472 | HEK-293T | C293(0.93); C789(0.95); C1253(0.90); C568(0.98) | LDD1511 | [30] |
| LDCM0356 | AKOS034007680 | HEK-293T | C293(0.94); C789(1.03); C1253(1.00); C568(1.07) | LDD1570 | [30] |
| LDCM0275 | AKOS034007705 | HEK-293T | C293(0.96); C789(0.92); C1253(0.98); C568(1.17) | LDD1514 | [30] |
| LDCM0156 | Aniline | NCI-H1299 | 11.70 | LDD0403 | [1] |
| LDCM0087 | Capsaicin | HEK-293T | 8.96 | LDD0185 | [8] |
| LDCM0108 | Chloroacetamide | HeLa | C741(0.00); C568(0.00); C1129(0.00); C789(0.00) | LDD0222 | [24] |
| LDCM0632 | CL-Sc | Hep-G2 | C1129(1.83); C293(1.48); C1160(0.91); C70(0.84) | LDD2227 | [19] |
| LDCM0367 | CL1 | HEK-293T | C293(0.87); C789(0.83); C1253(1.08); C568(0.97) | LDD1571 | [30] |
| LDCM0368 | CL10 | HEK-293T | C293(1.20); C789(0.78); C1253(1.16); C568(1.02) | LDD1572 | [30] |
| LDCM0369 | CL100 | HEK-293T | C293(0.90); C789(0.97); C1253(0.96); C568(1.00) | LDD1573 | [30] |
| LDCM0370 | CL101 | HEK-293T | C293(0.97); C789(1.01); C1253(0.98); C568(1.01) | LDD1574 | [30] |
| LDCM0371 | CL102 | HEK-293T | C293(0.91); C1253(0.87); C568(1.04); C1160(0.84) | LDD1575 | [30] |
| LDCM0372 | CL103 | HEK-293T | C293(0.98); C789(1.01); C1253(1.04); C568(0.93) | LDD1576 | [30] |
| LDCM0373 | CL104 | HEK-293T | C293(0.96); C789(0.94); C1253(1.01); C568(0.90) | LDD1577 | [30] |
| LDCM0374 | CL105 | HEK-293T | C293(0.93); C789(0.99); C1253(0.94); C568(1.03) | LDD1578 | [30] |
| LDCM0375 | CL106 | HEK-293T | C293(0.92); C1253(0.98); C568(0.93); C1160(1.01) | LDD1579 | [30] |
| LDCM0376 | CL107 | HEK-293T | C293(0.97); C789(0.99); C1253(1.05); C568(1.08) | LDD1580 | [30] |
| LDCM0377 | CL108 | HEK-293T | C293(0.95); C789(0.94); C1253(0.98); C568(1.06) | LDD1581 | [30] |
| LDCM0378 | CL109 | HEK-293T | C293(0.88); C789(0.97); C1253(1.00); C568(0.95) | LDD1582 | [30] |
| LDCM0379 | CL11 | HEK-293T | C293(1.10); C789(0.95); C1253(1.02); C568(1.00) | LDD1583 | [30] |
| LDCM0380 | CL110 | HEK-293T | C293(1.00); C1253(0.92); C568(1.06); C1160(1.00) | LDD1584 | [30] |
| LDCM0381 | CL111 | HEK-293T | C293(1.03); C789(0.97); C1253(1.08); C568(1.01) | LDD1585 | [30] |
| LDCM0382 | CL112 | HEK-293T | C293(0.99); C789(0.97); C1253(0.93); C568(1.04) | LDD1586 | [30] |
| LDCM0383 | CL113 | HEK-293T | C293(0.97); C789(0.95); C1253(1.00); C568(0.99) | LDD1587 | [30] |
| LDCM0384 | CL114 | HEK-293T | C293(0.99); C1253(0.98); C568(0.97); C1160(0.61) | LDD1588 | [30] |
| LDCM0385 | CL115 | HEK-293T | C293(0.96); C789(1.00); C1253(1.02); C568(0.98) | LDD1589 | [30] |
| LDCM0386 | CL116 | HEK-293T | C293(0.94); C789(0.95); C1253(0.90); C568(0.99) | LDD1590 | [30] |
| LDCM0387 | CL117 | HEK-293T | C293(0.95); C789(1.02); C1253(0.90); C568(1.01) | LDD1591 | [30] |
| LDCM0388 | CL118 | HEK-293T | C293(0.98); C1253(0.92); C568(1.00); C1160(0.98) | LDD1592 | [30] |
| LDCM0389 | CL119 | HEK-293T | C293(1.00); C789(1.03); C1253(1.07); C568(0.89) | LDD1593 | [30] |
| LDCM0390 | CL12 | HEK-293T | C293(1.05); C789(0.93); C1253(0.94); C568(1.00) | LDD1594 | [30] |
| LDCM0391 | CL120 | HEK-293T | C293(0.96); C789(0.98); C1253(0.96); C568(1.00) | LDD1595 | [30] |
| LDCM0392 | CL121 | HEK-293T | C293(0.98); C789(0.96); C1253(0.98); C568(0.99) | LDD1596 | [30] |
| LDCM0393 | CL122 | HEK-293T | C293(0.94); C1253(0.94); C568(1.00); C1160(0.93) | LDD1597 | [30] |
| LDCM0394 | CL123 | HEK-293T | C293(1.04); C789(0.94); C1253(1.08); C568(0.87) | LDD1598 | [30] |
| LDCM0395 | CL124 | HEK-293T | C293(0.92); C789(0.95); C1253(0.99); C568(0.93) | LDD1599 | [30] |
| LDCM0396 | CL125 | HEK-293T | C293(0.92); C789(0.91); C1253(0.93); C568(1.00) | LDD1600 | [30] |
| LDCM0397 | CL126 | HEK-293T | C293(0.95); C1253(0.96); C568(0.96); C1160(0.80) | LDD1601 | [30] |
| LDCM0398 | CL127 | HEK-293T | C293(0.97); C789(1.04); C1253(1.04); C568(0.92) | LDD1602 | [30] |
| LDCM0399 | CL128 | HEK-293T | C293(0.96); C789(0.93); C1253(0.97); C568(1.00) | LDD1603 | [30] |
| LDCM0400 | CL13 | HEK-293T | C293(0.85); C789(0.95); C1253(0.95); C568(1.23) | LDD1604 | [30] |
| LDCM0401 | CL14 | HEK-293T | C293(0.91); C1253(0.95); C568(0.91); C1160(0.69) | LDD1605 | [30] |
| LDCM0402 | CL15 | HEK-293T | C293(0.88); C789(0.99); C1253(1.11); C568(0.89) | LDD1606 | [30] |
| LDCM0403 | CL16 | HEK-293T | C293(1.05); C789(0.96); C1253(1.01); C568(1.00) | LDD1607 | [30] |
| LDCM0404 | CL17 | HEK-293T | C293(0.86); C789(0.90); C1253(1.11); C568(1.12) | LDD1608 | [30] |
| LDCM0405 | CL18 | HEK-293T | C293(0.93); C789(1.02); C1253(0.99); C568(1.03) | LDD1609 | [30] |
| LDCM0406 | CL19 | HEK-293T | C293(1.00); C789(1.02); C1253(0.99); C568(1.08) | LDD1610 | [30] |
| LDCM0407 | CL2 | HEK-293T | C293(0.95); C1253(0.97); C568(0.93); C1160(0.98) | LDD1611 | [30] |
| LDCM0408 | CL20 | HEK-293T | C293(1.04); C789(0.97); C1253(1.03); C568(1.02) | LDD1612 | [30] |
| LDCM0409 | CL21 | HEK-293T | C293(0.89); C789(0.90); C1253(0.99); C568(0.93) | LDD1613 | [30] |
| LDCM0410 | CL22 | HEK-293T | C293(1.12); C789(0.93); C1253(1.02); C568(1.07) | LDD1614 | [30] |
| LDCM0411 | CL23 | HEK-293T | C293(1.15); C789(1.03); C1253(0.96); C568(0.96) | LDD1615 | [30] |
| LDCM0412 | CL24 | HEK-293T | C293(1.11); C789(1.01); C1253(0.99); C568(1.13) | LDD1616 | [30] |
| LDCM0413 | CL25 | HEK-293T | C293(0.99); C789(0.90); C1253(0.97); C568(0.94) | LDD1617 | [30] |
| LDCM0414 | CL26 | HEK-293T | C293(0.90); C1253(1.00); C568(0.92); C1160(1.00) | LDD1618 | [30] |
| LDCM0415 | CL27 | HEK-293T | C293(0.95); C789(0.99); C1253(1.02); C568(0.92) | LDD1619 | [30] |
| LDCM0416 | CL28 | HEK-293T | C293(0.95); C789(0.93); C1253(0.95); C568(0.93) | LDD1620 | [30] |
| LDCM0417 | CL29 | HEK-293T | C293(0.97); C789(0.98); C1253(1.05); C568(1.02) | LDD1621 | [30] |
| LDCM0418 | CL3 | HEK-293T | C293(0.97); C789(1.05); C1253(1.09); C568(0.98) | LDD1622 | [30] |
| LDCM0419 | CL30 | HEK-293T | C293(1.00); C789(0.99); C1253(1.04); C568(1.24) | LDD1623 | [30] |
| LDCM0420 | CL31 | HEK-293T | C293(1.08); C789(0.99); C1253(1.02); C568(0.91) | LDD1624 | [30] |
| LDCM0421 | CL32 | HEK-293T | C293(1.06); C789(1.05); C1253(0.99); C568(1.05) | LDD1625 | [30] |
| LDCM0422 | CL33 | HEK-293T | C293(0.96); C789(0.97); C1253(1.25); C568(1.02) | LDD1626 | [30] |
| LDCM0423 | CL34 | HEK-293T | C293(1.16); C789(0.95); C1253(1.02); C568(1.01) | LDD1627 | [30] |
| LDCM0424 | CL35 | HEK-293T | C293(1.22); C789(1.04); C1253(1.02); C568(1.06) | LDD1628 | [30] |
| LDCM0425 | CL36 | HEK-293T | C293(1.26); C789(1.00); C1253(1.03); C568(1.05) | LDD1629 | [30] |
| LDCM0426 | CL37 | HEK-293T | C293(0.89); C789(0.95); C1253(1.01); C568(0.94) | LDD1630 | [30] |
| LDCM0428 | CL39 | HEK-293T | C293(0.96); C789(1.00); C1253(1.08); C568(0.93) | LDD1632 | [30] |
| LDCM0429 | CL4 | HEK-293T | C293(0.95); C789(0.95); C1253(0.91); C568(1.04) | LDD1633 | [30] |
| LDCM0430 | CL40 | HEK-293T | C293(0.98); C789(1.02); C1253(0.96); C568(1.08) | LDD1634 | [30] |
| LDCM0431 | CL41 | HEK-293T | C293(0.94); C789(0.98); C1253(0.91); C568(0.98) | LDD1635 | [30] |
| LDCM0432 | CL42 | HEK-293T | C293(1.02); C789(1.08); C1253(1.10); C568(1.22) | LDD1636 | [30] |
| LDCM0433 | CL43 | HEK-293T | C293(1.17); C789(1.00); C1253(1.00); C568(0.93) | LDD1637 | [30] |
| LDCM0434 | CL44 | HEK-293T | C293(1.07); C789(1.03); C1253(1.00); C568(1.06) | LDD1638 | [30] |
| LDCM0435 | CL45 | HEK-293T | C293(0.93); C789(0.95); C1253(0.98); C568(1.17) | LDD1639 | [30] |
| LDCM0436 | CL46 | HEK-293T | C293(1.20); C789(1.10); C1253(1.02); C568(1.04) | LDD1640 | [30] |
| LDCM0437 | CL47 | HEK-293T | C293(1.24); C789(1.01); C1253(1.00); C568(1.01) | LDD1641 | [30] |
| LDCM0438 | CL48 | HEK-293T | C293(1.16); C789(0.99); C1253(1.08); C568(1.19) | LDD1642 | [30] |
| LDCM0439 | CL49 | HEK-293T | C293(0.90); C789(0.94); C1253(0.99); C568(1.05) | LDD1643 | [30] |
| LDCM0440 | CL5 | HEK-293T | C293(0.91); C789(0.95); C1253(0.93); C568(1.11) | LDD1644 | [30] |
| LDCM0441 | CL50 | HEK-293T | C293(0.90); C1253(0.95); C568(0.98); C1160(0.75) | LDD1645 | [30] |
| LDCM0443 | CL52 | HEK-293T | C293(0.95); C789(1.00); C1253(0.94); C568(0.96) | LDD1646 | [30] |
| LDCM0444 | CL53 | HEK-293T | C293(0.93); C789(0.94); C1253(0.99); C568(0.88) | LDD1647 | [30] |
| LDCM0445 | CL54 | HEK-293T | C293(1.00); C789(0.93); C1253(1.13); C568(1.10) | LDD1648 | [30] |
| LDCM0446 | CL55 | HEK-293T | C293(1.05); C789(0.99); C1253(1.01); C568(1.10) | LDD1649 | [30] |
| LDCM0447 | CL56 | HEK-293T | C293(1.05); C789(1.03); C1253(1.01); C568(1.01) | LDD1650 | [30] |
| LDCM0448 | CL57 | HEK-293T | C293(0.89); C789(0.87); C1253(1.02); C568(0.98) | LDD1651 | [30] |
| LDCM0449 | CL58 | HEK-293T | C293(1.08); C789(0.97); C1253(1.14); C568(0.84) | LDD1652 | [30] |
| LDCM0450 | CL59 | HEK-293T | C293(1.13); C789(1.07); C1253(1.01); C568(0.97) | LDD1653 | [30] |
| LDCM0451 | CL6 | HEK-293T | C293(1.00); C789(0.97); C1253(1.08); C568(1.03) | LDD1654 | [30] |
| LDCM0452 | CL60 | HEK-293T | C293(1.13); C789(0.96); C1253(0.99); C568(0.93) | LDD1655 | [30] |
| LDCM0453 | CL61 | HEK-293T | C293(0.88); C789(0.96); C1253(1.03); C568(0.96) | LDD1656 | [30] |
| LDCM0454 | CL62 | HEK-293T | C293(0.88); C1253(0.96); C568(1.04); C1160(0.72) | LDD1657 | [30] |
| LDCM0455 | CL63 | HEK-293T | C293(0.96); C789(1.07); C1253(1.06); C568(0.97) | LDD1658 | [30] |
| LDCM0456 | CL64 | HEK-293T | C293(0.94); C789(0.94); C1253(0.96); C568(0.97) | LDD1659 | [30] |
| LDCM0457 | CL65 | HEK-293T | C293(0.96); C789(1.02); C1253(1.08); C568(0.97) | LDD1660 | [30] |
| LDCM0458 | CL66 | HEK-293T | C293(0.92); C789(1.02); C1253(1.09); C568(0.99) | LDD1661 | [30] |
| LDCM0459 | CL67 | HEK-293T | C293(1.00); C789(1.02); C1253(0.97); C568(0.98) | LDD1662 | [30] |
| LDCM0460 | CL68 | HEK-293T | C293(1.08); C789(1.01); C1253(1.00); C568(0.93) | LDD1663 | [30] |
| LDCM0461 | CL69 | HEK-293T | C293(0.99); C789(0.93); C1253(0.97); C568(0.97) | LDD1664 | [30] |
| LDCM0462 | CL7 | HEK-293T | C293(0.99); C789(0.99); C1253(1.01); C568(0.98) | LDD1665 | [30] |
| LDCM0463 | CL70 | HEK-293T | C293(1.19); C789(0.96); C1253(0.96); C568(1.05) | LDD1666 | [30] |
| LDCM0464 | CL71 | HEK-293T | C293(1.13); C789(1.08); C1253(0.95); C568(1.09) | LDD1667 | [30] |
| LDCM0465 | CL72 | HEK-293T | C293(1.12); C789(0.97); C1253(0.99); C568(1.11) | LDD1668 | [30] |
| LDCM0466 | CL73 | HEK-293T | C293(0.83); C789(0.94); C1253(0.92); C568(1.00) | LDD1669 | [30] |
| LDCM0467 | CL74 | HEK-293T | C293(0.89); C1253(0.96); C568(0.98); C1160(0.84) | LDD1670 | [30] |
| LDCM0469 | CL76 | HEK-293T | C293(0.99); C789(0.96); C1253(1.00); C568(0.98) | LDD1672 | [30] |
| LDCM0470 | CL77 | HEK-293T | C293(0.93); C789(0.92); C1253(1.04); C568(1.01) | LDD1673 | [30] |
| LDCM0471 | CL78 | HEK-293T | C293(0.94); C789(1.01); C1253(1.10); C568(1.27) | LDD1674 | [30] |
| LDCM0472 | CL79 | HEK-293T | C293(1.05); C789(0.96); C1253(0.95); C568(1.10) | LDD1675 | [30] |
| LDCM0473 | CL8 | HEK-293T | C293(1.17); C789(0.74); C1253(1.23); C568(0.92) | LDD1676 | [30] |
| LDCM0474 | CL80 | HEK-293T | C293(0.96); C789(0.98); C1253(1.01); C568(0.99) | LDD1677 | [30] |
| LDCM0475 | CL81 | HEK-293T | C293(0.95); C789(0.99); C1253(0.94); C568(0.97) | LDD1678 | [30] |
| LDCM0476 | CL82 | HEK-293T | C293(1.04); C789(0.95); C1253(0.98); C568(1.01) | LDD1679 | [30] |
| LDCM0477 | CL83 | HEK-293T | C293(1.13); C789(0.96); C1253(1.07); C568(0.98) | LDD1680 | [30] |
| LDCM0478 | CL84 | HEK-293T | C293(1.06); C789(0.91); C1253(0.97); C568(0.96) | LDD1681 | [30] |
| LDCM0479 | CL85 | HEK-293T | C293(0.90); C789(0.99); C1253(0.95); C568(0.91) | LDD1682 | [30] |
| LDCM0480 | CL86 | HEK-293T | C293(0.85); C1253(0.92); C568(0.92); C1160(0.98) | LDD1683 | [30] |
| LDCM0481 | CL87 | HEK-293T | C293(0.97); C789(0.98); C1253(1.06); C568(0.89) | LDD1684 | [30] |
| LDCM0482 | CL88 | HEK-293T | C293(1.01); C789(0.96); C1253(0.88); C568(1.03) | LDD1685 | [30] |
| LDCM0483 | CL89 | HEK-293T | C293(0.96); C789(0.98); C1253(0.97); C568(0.93) | LDD1686 | [30] |
| LDCM0484 | CL9 | HEK-293T | C293(0.95); C789(0.94); C1253(0.97); C568(1.10) | LDD1687 | [30] |
| LDCM0485 | CL90 | HEK-293T | C293(1.06); C789(0.88); C1253(1.03); C568(0.96) | LDD1688 | [30] |
| LDCM0486 | CL91 | HEK-293T | C293(0.99); C789(1.00); C1253(0.93); C568(1.03) | LDD1689 | [30] |
| LDCM0487 | CL92 | HEK-293T | C293(1.00); C789(0.95); C1253(1.01); C568(0.95) | LDD1690 | [30] |
| LDCM0488 | CL93 | HEK-293T | C293(0.92); C789(0.93); C1253(1.01); C568(0.98) | LDD1691 | [30] |
| LDCM0489 | CL94 | HEK-293T | C293(1.02); C789(0.91); C1253(1.00); C568(0.99) | LDD1692 | [30] |
| LDCM0490 | CL95 | HEK-293T | C293(1.00); C789(0.90); C1253(0.95); C568(1.04) | LDD1693 | [30] |
| LDCM0491 | CL96 | HEK-293T | C293(1.01); C789(0.91); C1253(0.96); C568(1.05) | LDD1694 | [30] |
| LDCM0492 | CL97 | HEK-293T | C293(0.90); C789(0.91); C1253(0.95); C568(1.03) | LDD1695 | [30] |
| LDCM0493 | CL98 | HEK-293T | C293(0.91); C1253(0.98); C568(1.02); C1160(0.94) | LDD1696 | [30] |
| LDCM0494 | CL99 | HEK-293T | C293(0.96); C789(0.95); C1253(1.00); C568(0.94) | LDD1697 | [30] |
| LDCM0495 | E2913 | HEK-293T | C293(0.95); C789(1.02); C1253(1.12); C568(0.97) | LDD1698 | [30] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C1129(1.46) | LDD1702 | [7] |
| LDCM0625 | F8 | Ramos | C568(0.91); C1396(1.21); C1253(1.09); C1189(2.10) | LDD2187 | [31] |
| LDCM0572 | Fragment10 | Ramos | C568(1.01); C1396(2.67); C1253(1.22); 1.51 | LDD2189 | [31] |
| LDCM0573 | Fragment11 | Ramos | C568(0.55); C1253(4.35); 0.96; C1129(1.23) | LDD2190 | [31] |
| LDCM0574 | Fragment12 | Ramos | C568(0.95); C1396(3.43); C1253(2.02); 1.21 | LDD2191 | [31] |
| LDCM0575 | Fragment13 | Ramos | C568(0.67); C1396(1.01); C1253(1.08); C1189(0.71) | LDD2192 | [31] |
| LDCM0576 | Fragment14 | Ramos | C568(0.97); C1396(4.85); C1189(0.65); 1.32 | LDD2193 | [31] |
| LDCM0579 | Fragment20 | Ramos | C568(1.06); C1253(2.00); C1129(0.67) | LDD2194 | [31] |
| LDCM0580 | Fragment21 | Ramos | C568(0.89); C1396(1.22); C1253(1.02); C1189(0.81) | LDD2195 | [31] |
| LDCM0582 | Fragment23 | Ramos | C568(0.98); C1396(0.95); C1253(0.98); C1189(1.22) | LDD2196 | [31] |
| LDCM0578 | Fragment27 | Ramos | C568(0.93); C1396(0.86); C1253(0.96); C1189(1.04) | LDD2197 | [31] |
| LDCM0586 | Fragment28 | Ramos | C568(0.69); C1396(0.87); C1253(0.79); C1189(0.77) | LDD2198 | [31] |
| LDCM0588 | Fragment30 | Ramos | C568(1.07); C1396(1.33); C1253(1.03); C1189(1.15) | LDD2199 | [31] |
| LDCM0589 | Fragment31 | Ramos | C568(0.85); C1396(1.56); C1253(1.30); C1189(1.17) | LDD2200 | [31] |
| LDCM0590 | Fragment32 | Ramos | C568(1.10); C1396(2.30); C1253(1.47); C1189(1.79) | LDD2201 | [31] |
| LDCM0468 | Fragment33 | HEK-293T | C293(0.94); C789(1.01); C1253(1.04); C568(0.99) | LDD1671 | [30] |
| LDCM0596 | Fragment38 | Ramos | C568(0.94); C1396(1.41); C1253(0.85); C1189(1.44) | LDD2203 | [31] |
| LDCM0566 | Fragment4 | Ramos | C568(0.59); C1396(3.96); C1253(1.41); C1189(1.92) | LDD2184 | [31] |
| LDCM0427 | Fragment51 | HEK-293T | C293(0.93); C1253(0.95); C568(1.19); C1160(0.89) | LDD1631 | [30] |
| LDCM0610 | Fragment52 | Ramos | C568(0.95); C1396(1.27); C1253(1.16); C1189(1.74) | LDD2204 | [31] |
| LDCM0614 | Fragment56 | Ramos | C568(0.93); C1396(1.01); C1253(0.92); C1189(1.03) | LDD2205 | [31] |
| LDCM0569 | Fragment7 | Ramos | C568(0.85); C1396(2.99); C1253(1.23); C1189(1.45) | LDD2186 | [31] |
| LDCM0571 | Fragment9 | Ramos | C568(0.84); C1253(2.64); C1189(2.17); C1129(0.68) | LDD2188 | [31] |
| LDCM0116 | HHS-0101 | DM93 | Y1036(0.80); Y460(0.89); Y86(0.95); Y1207(1.17) | LDD0264 | [12] |
| LDCM0117 | HHS-0201 | DM93 | Y460(0.65); Y1036(0.83); Y1207(0.84); Y86(1.24) | LDD0265 | [12] |
| LDCM0118 | HHS-0301 | DM93 | Y1207(0.49); Y1036(0.79); Y86(0.83); Y460(3.15) | LDD0266 | [12] |
| LDCM0119 | HHS-0401 | DM93 | Y460(0.60); Y1036(0.72); Y86(0.90); Y1207(1.25) | LDD0267 | [12] |
| LDCM0120 | HHS-0701 | DM93 | Y1036(0.82); Y460(0.92); Y1207(1.07); Y86(1.21) | LDD0268 | [12] |
| LDCM0107 | IAA | HeLa | H73(0.00); H1141(0.00); C1253(0.00) | LDD0221 | [24] |
| LDCM0123 | JWB131 | DM93 | Y1207(1.05) | LDD0285 | [11] |
| LDCM0124 | JWB142 | DM93 | Y1207(1.40) | LDD0286 | [11] |
| LDCM0125 | JWB146 | DM93 | Y1036(2.52); Y1207(1.65) | LDD0287 | [11] |
| LDCM0126 | JWB150 | DM93 | Y1207(3.99) | LDD0288 | [11] |
| LDCM0127 | JWB152 | DM93 | Y1207(2.56) | LDD0289 | [11] |
| LDCM0128 | JWB198 | DM93 | Y1036(2.03); Y1207(0.96) | LDD0290 | [11] |
| LDCM0129 | JWB202 | DM93 | Y1207(0.90) | LDD0291 | [11] |
| LDCM0130 | JWB211 | DM93 | Y1207(1.08) | LDD0292 | [11] |
| LDCM0022 | KB02 | HEK-293T | C293(0.95); C789(0.97); C275(0.99); C1253(0.93) | LDD1492 | [30] |
| LDCM0023 | KB03 | Jurkat | C176(4.71); C293(12.72); C176(92.05); C1396(26.86) | LDD0209 | [10] |
| LDCM0024 | KB05 | COLO792 | C275(2.78); C789(1.18); C1262(2.52) | LDD3310 | [32] |
| LDCM0030 | Luteolin | HEK-293T | 5.57 | LDD0182 | [8] |
| LDCM0109 | NEM | HeLa | H1345(0.00); H1347(0.00); H877(0.00) | LDD0223 | [24] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C1396(1.08) | LDD2098 | [7] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C1396(0.63); C789(1.22) | LDD2099 | [7] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C1396(0.94) | LDD2107 | [7] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C275(0.92) | LDD2109 | [7] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C1396(0.86) | LDD2123 | [7] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C789(1.06) | LDD2125 | [7] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C1396(1.28) | LDD2136 | [7] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C1396(1.01); C1253(0.73) | LDD2137 | [7] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C1396(1.21) | LDD2146 | [7] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C374(1.25); C568(1.18); C176(0.24) | LDD2206 | [33] |
| LDCM0032 | Oleacein | HEK-293T | 5.43 | LDD0184 | [8] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C176(0.78); C71(0.62); C568(0.59) | LDD2207 | [33] |
| LDCM0090 | Rapamycin | JHH-7 | 3.44 | LDD0213 | [29] |
References
