Details of the Target
General Information of Target
| Target ID | LDTP14126 | |||||
|---|---|---|---|---|---|---|
| Target Name | Mitochondrial import inner membrane translocase subunit Tim9 (TIMM9) | |||||
| Gene Name | TIMM9 | |||||
| Gene ID | 26520 | |||||
| Synonyms |
TIM9; TIM9A; TIMM9A; Mitochondrial import inner membrane translocase subunit Tim9 |
|||||
| 3D Structure | ||||||
| Sequence |
MERQQQQQQQLRNLRDFLLVYNRMTELCFQRCVPSLHHRALDAEEEACLHSCAGKLIHSN
HRLMAAYVQLMPALVQRRIADYEAASAVPGVAAEQPGVSPSGS |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
Small Tim family
|
|||||
| Subcellular location |
Mitochondrion inner membrane
|
|||||
| Function |
Mitochondrial intermembrane chaperone that participates in the import and insertion of multi-pass transmembrane proteins into the mitochondrial inner membrane. May also be required for the transfer of beta-barrel precursors from the TOM complex to the sorting and assembly machinery (SAM complex) of the outer membrane. Acts as a chaperone-like protein that protects the hydrophobic precursors from aggregation and guide them through the mitochondrial intermembrane space.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K81(20.00) | LDD2218 | [1] | |
|
Probe 1 Probe Info |
 |
Y21(57.17); Y70(13.09) | LDD3495 | [2] | |
|
BTD Probe Info |
 |
C48(0.74) | LDD2100 | [3] | |
|
DBIA Probe Info |
 |
C48(1.28) | LDD1571 | [4] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [5] | |
|
m-APA Probe Info |
 |
N.A. | LDD2231 | [5] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0175 | [6] | |
|
1d-yne Probe Info |
 |
N.A. | LDD0356 | [7] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [7] | |
|
Acrolein Probe Info |
 |
H71(0.00); H51(0.00) | LDD0217 | [8] | |
|
Methacrolein Probe Info |
 |
C48(0.00); C28(0.00); C52(0.00) | LDD0218 | [8] | |
|
NAIA_5 Probe Info |
 |
C32(0.00); C48(0.00); C28(0.00) | LDD2223 | [9] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C056 Probe Info |
 |
37.79 | LDD1753 | [10] | |
|
C112 Probe Info |
 |
47.84 | LDD1799 | [10] | |
|
C159 Probe Info |
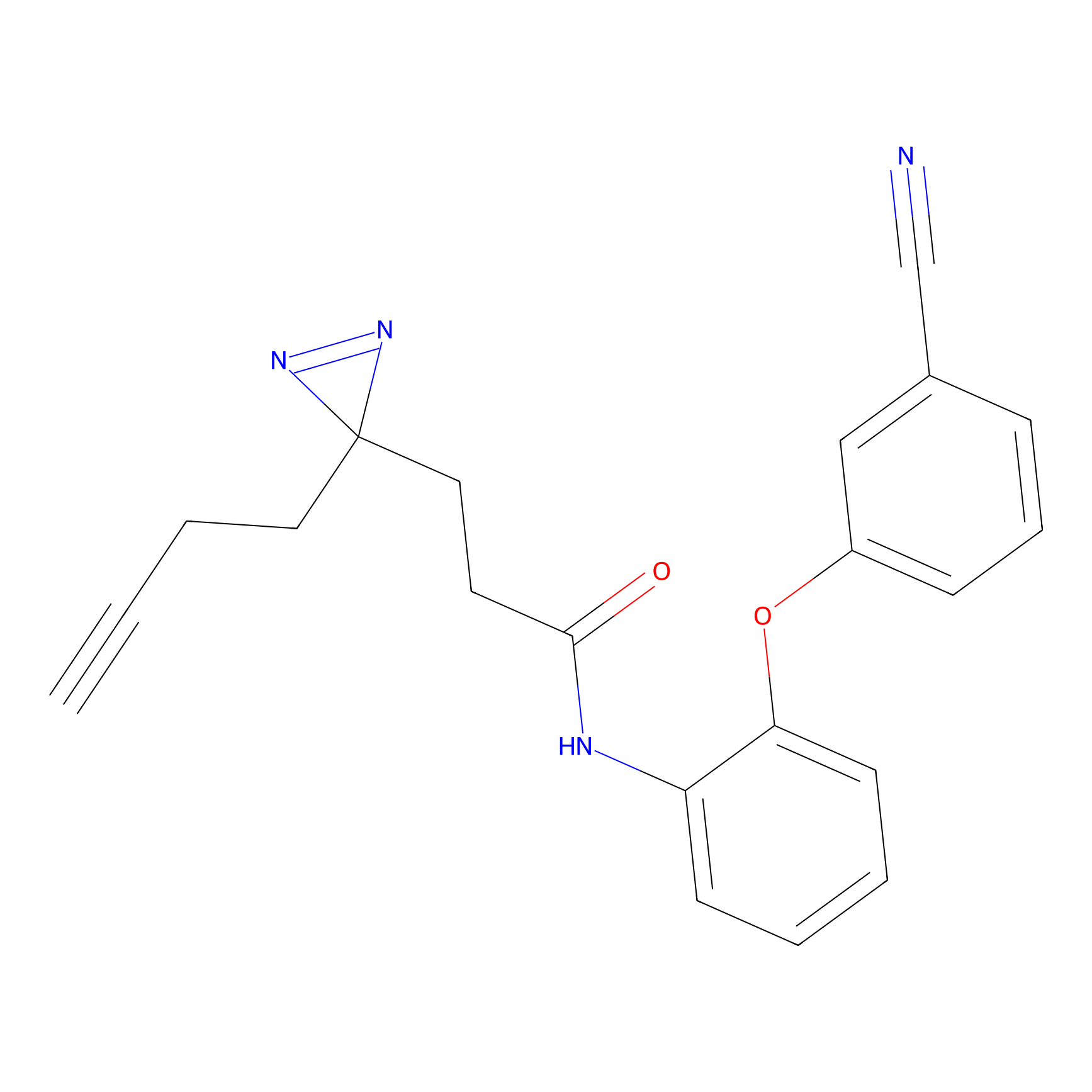 |
8.22 | LDD1839 | [10] | |
|
C160 Probe Info |
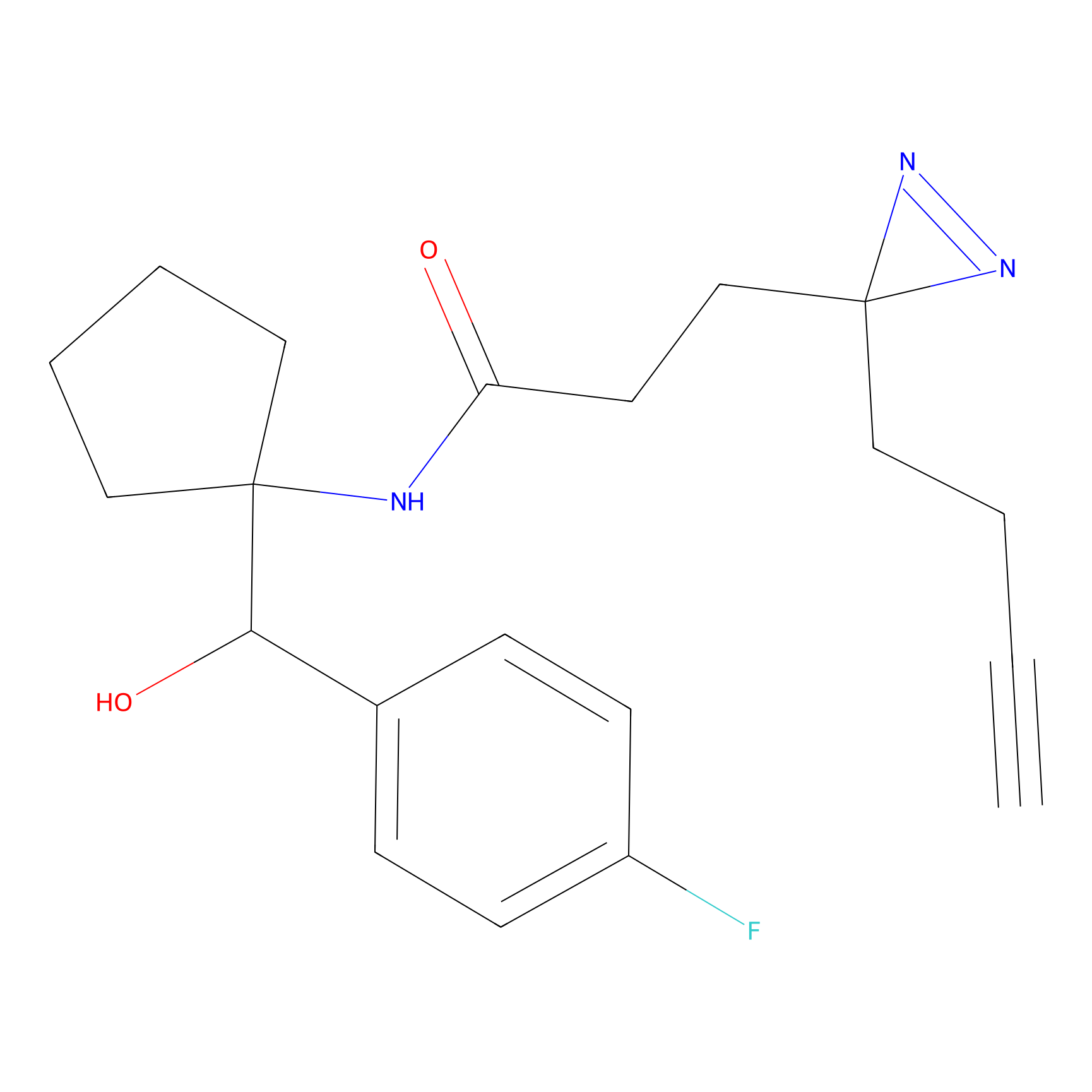 |
7.62 | LDD1840 | [10] | |
|
C161 Probe Info |
 |
22.94 | LDD1841 | [10] | |
|
C218 Probe Info |
 |
15.89 | LDD1892 | [10] | |
|
C220 Probe Info |
 |
38.05 | LDD1894 | [10] | |
|
C228 Probe Info |
 |
15.24 | LDD1901 | [10] | |
|
C243 Probe Info |
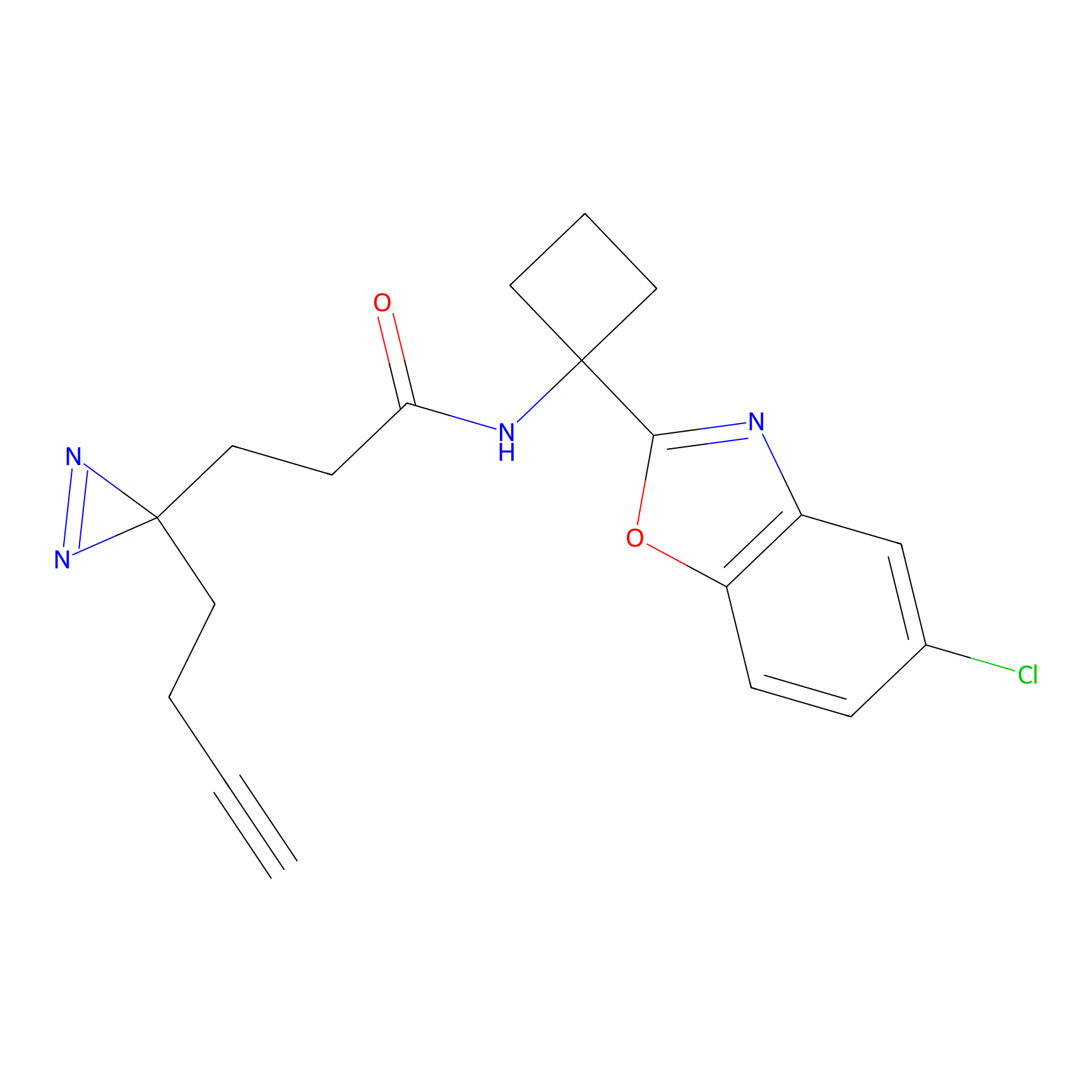 |
12.30 | LDD1916 | [10] | |
|
C244 Probe Info |
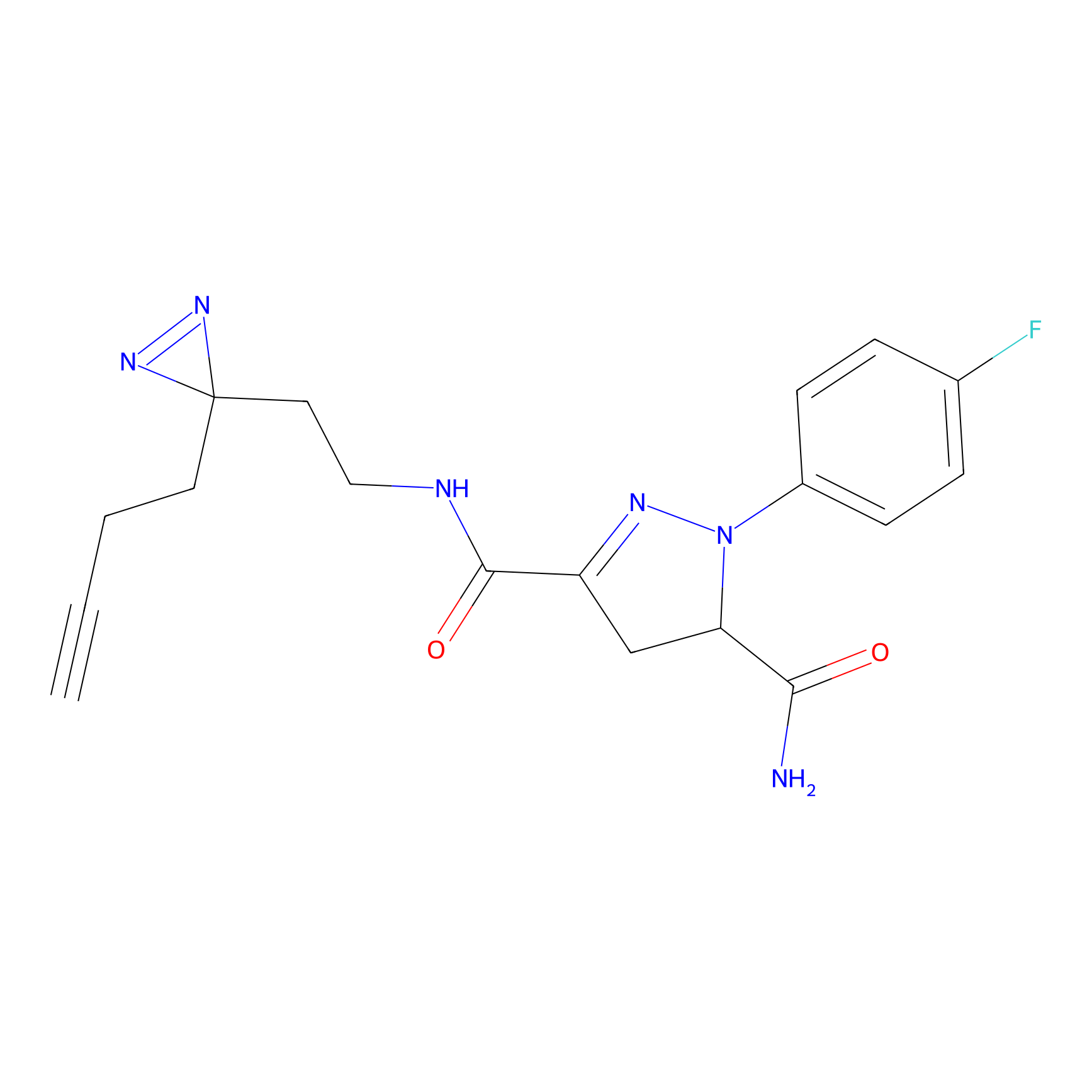 |
17.75 | LDD1917 | [10] | |
|
C246 Probe Info |
 |
15.03 | LDD1919 | [10] | |
|
C313 Probe Info |
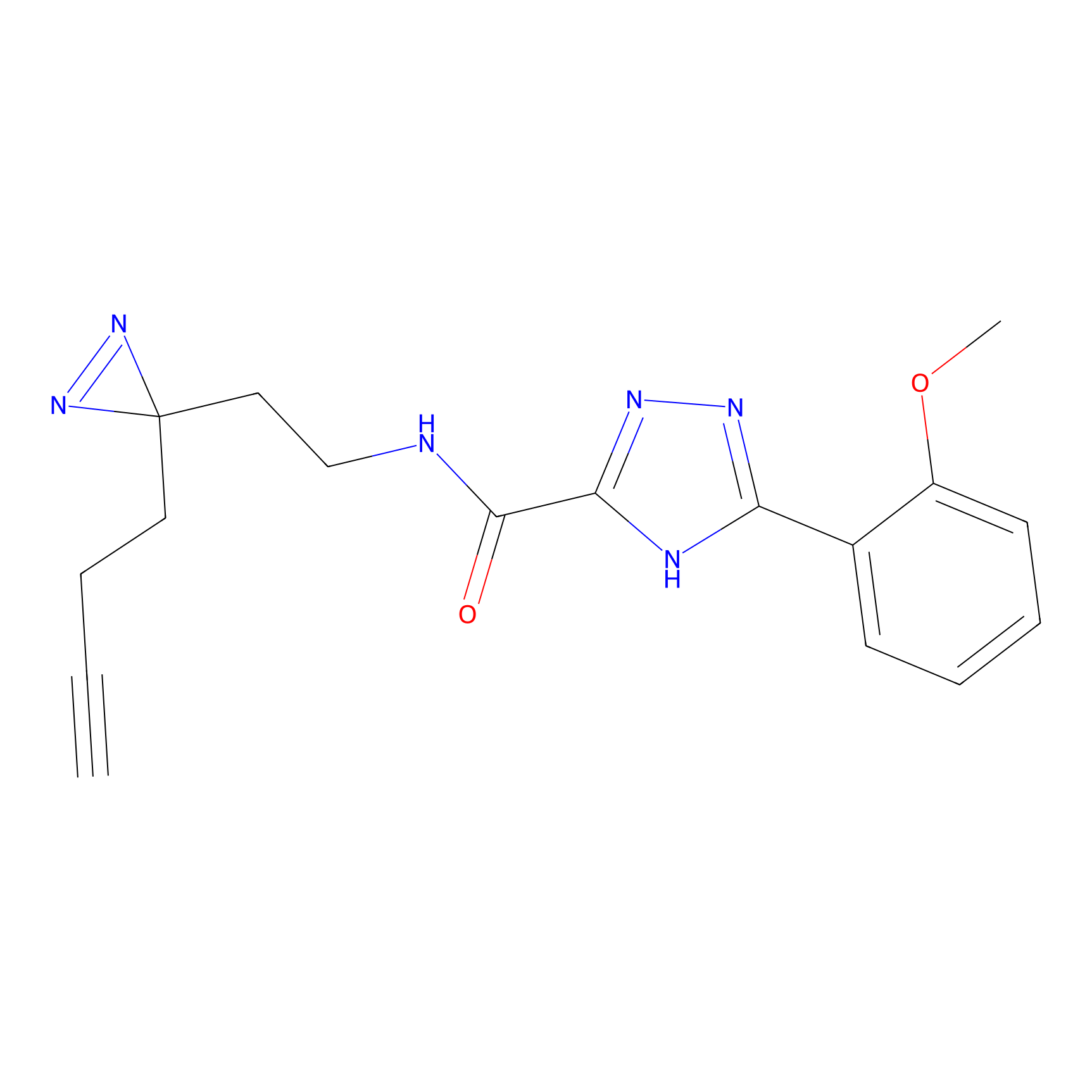 |
15.78 | LDD1980 | [10] | |
|
C343 Probe Info |
 |
13.00 | LDD2005 | [10] | |
|
C348 Probe Info |
 |
13.09 | LDD2009 | [10] | |
|
C349 Probe Info |
 |
14.22 | LDD2010 | [10] | |
|
C350 Probe Info |
 |
51.63 | LDD2011 | [10] | |
|
C353 Probe Info |
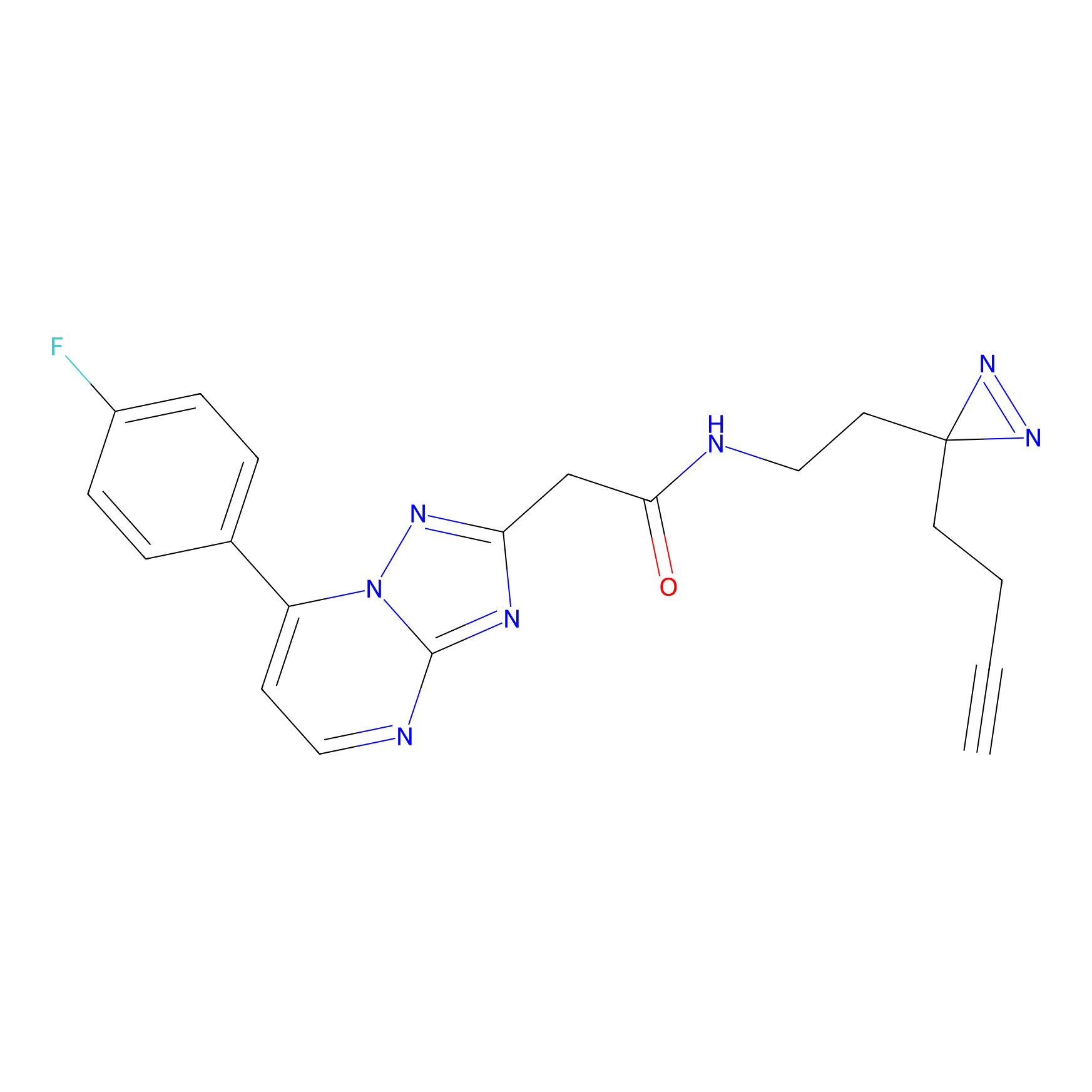 |
6.28 | LDD2014 | [10] | |
|
C362 Probe Info |
 |
56.10 | LDD2023 | [10] | |
|
C364 Probe Info |
 |
21.41 | LDD2025 | [10] | |
|
C383 Probe Info |
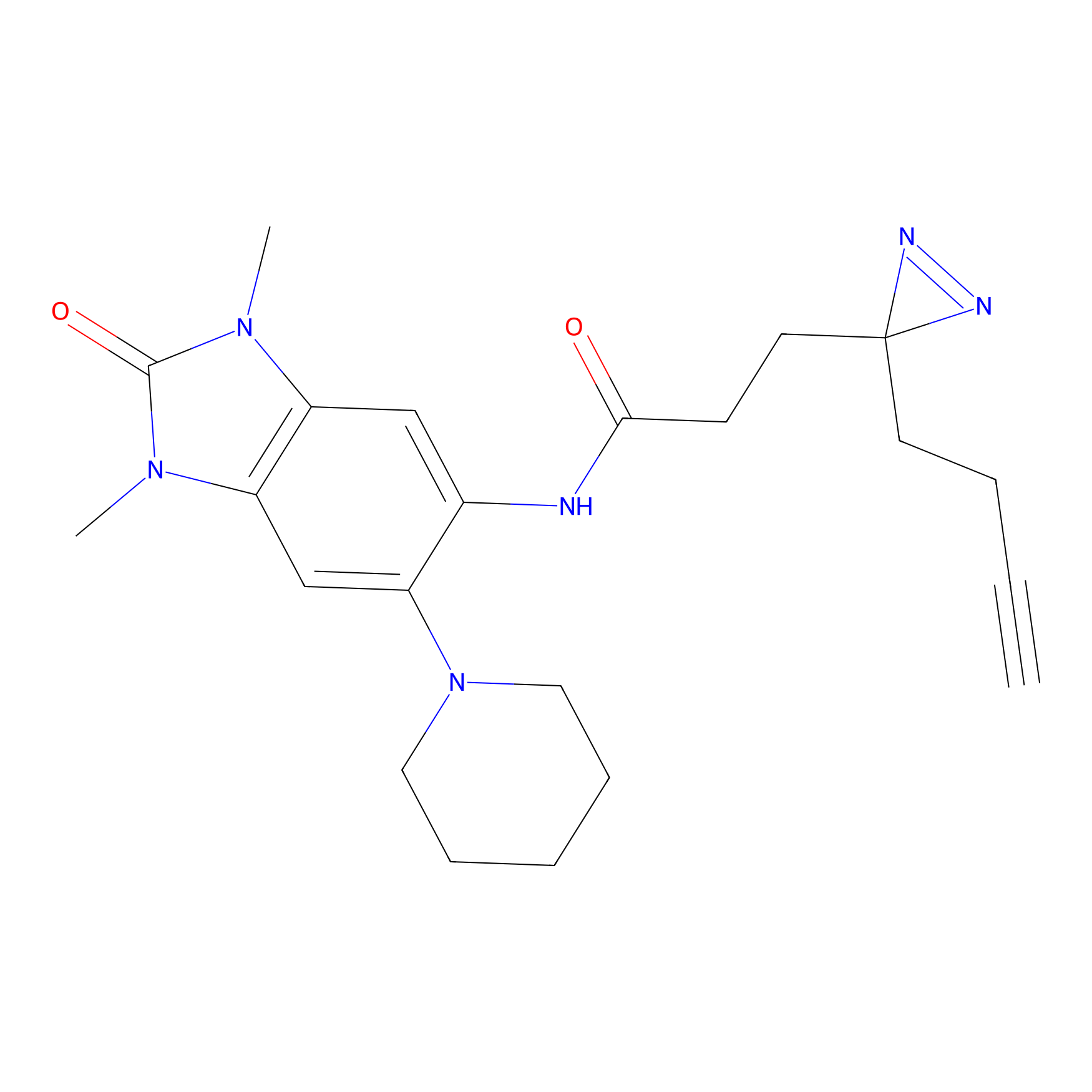 |
14.42 | LDD2042 | [10] | |
|
C386 Probe Info |
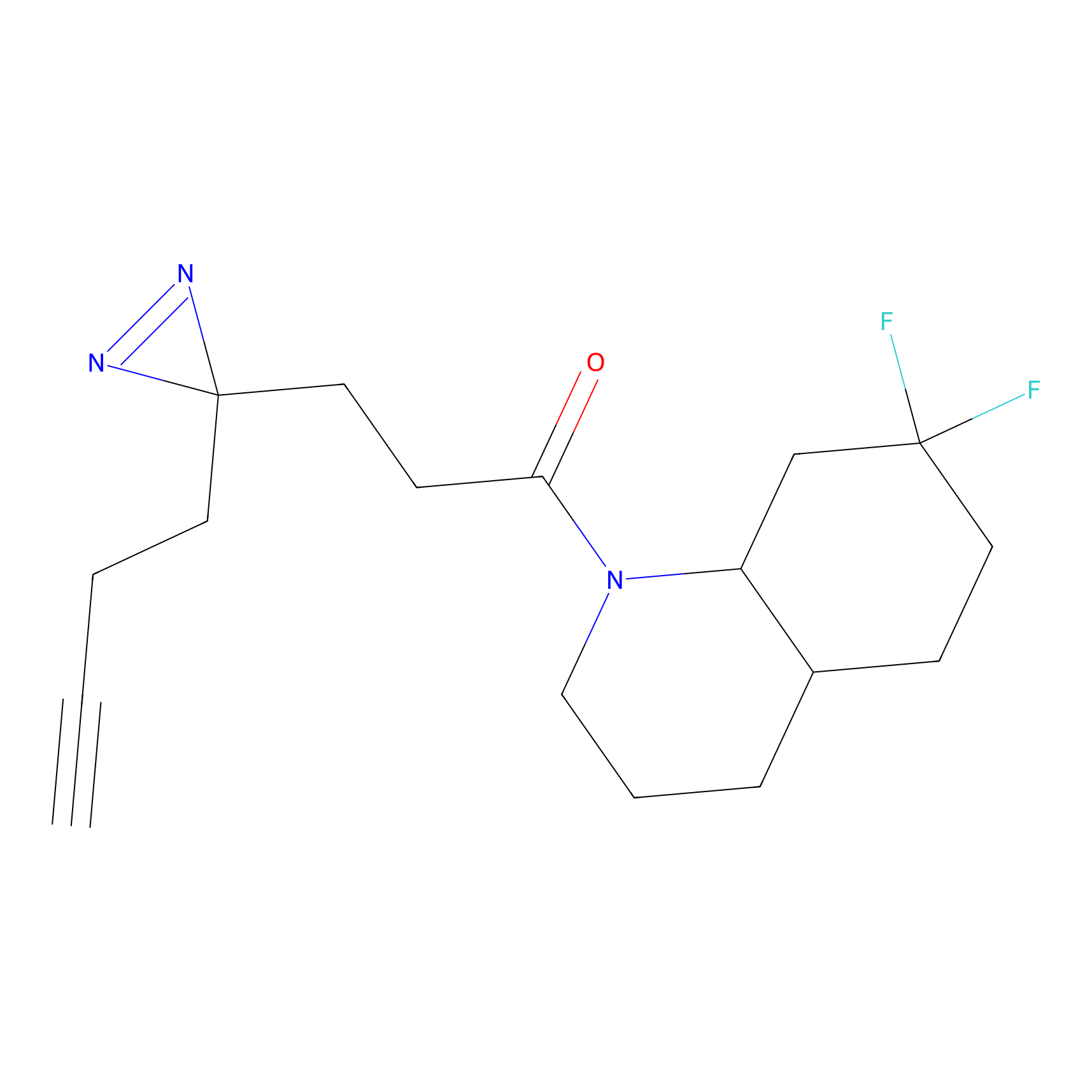 |
6.82 | LDD2045 | [10] | |
|
C388 Probe Info |
 |
84.45 | LDD2047 | [10] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C52(0.73); C48(0.81); C32(0.81) | LDD2112 | [3] |
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C48(0.77) | LDD2113 | [3] |
| LDCM0108 | Chloroacetamide | HeLa | C32(0.00); H71(0.00); C28(0.00); H51(0.00) | LDD0222 | [8] |
| LDCM0367 | CL1 | HEK-293T | C48(1.28) | LDD1571 | [4] |
| LDCM0370 | CL101 | HEK-293T | C48(1.38) | LDD1574 | [4] |
| LDCM0374 | CL105 | HEK-293T | C48(1.48) | LDD1578 | [4] |
| LDCM0378 | CL109 | HEK-293T | C48(1.03) | LDD1582 | [4] |
| LDCM0383 | CL113 | HEK-293T | C48(1.10) | LDD1587 | [4] |
| LDCM0387 | CL117 | HEK-293T | C48(1.17) | LDD1591 | [4] |
| LDCM0392 | CL121 | HEK-293T | C48(1.39) | LDD1596 | [4] |
| LDCM0396 | CL125 | HEK-293T | C48(1.14) | LDD1600 | [4] |
| LDCM0400 | CL13 | HEK-293T | C48(1.04) | LDD1604 | [4] |
| LDCM0413 | CL25 | HEK-293T | C48(1.16) | LDD1617 | [4] |
| LDCM0426 | CL37 | HEK-293T | C48(1.18) | LDD1630 | [4] |
| LDCM0439 | CL49 | HEK-293T | C48(1.24) | LDD1643 | [4] |
| LDCM0453 | CL61 | HEK-293T | C48(1.35) | LDD1656 | [4] |
| LDCM0466 | CL73 | HEK-293T | C48(1.12) | LDD1669 | [4] |
| LDCM0479 | CL85 | HEK-293T | C48(1.03) | LDD1682 | [4] |
| LDCM0492 | CL97 | HEK-293T | C48(1.16) | LDD1695 | [4] |
| LDCM0107 | IAA | HeLa | H71(0.00); H51(0.00) | LDD0221 | [8] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [8] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C48(0.74) | LDD2100 | [3] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C48(1.12) | LDD2101 | [3] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C48(0.48) | LDD2104 | [3] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C48(0.61); C32(0.54) | LDD2106 | [3] |
| LDCM0517 | Nucleophilic fragment 21b | MDA-MB-231 | C48(0.74); C32(0.67) | LDD2110 | [3] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C48(0.80) | LDD2114 | [3] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C52(0.98); C48(0.77) | LDD2115 | [3] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C52(0.46); C48(0.67) | LDD2134 | [3] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C52(0.44) | LDD2148 | [3] |
The Interaction Atlas With This Target
References
