Details of the Target
General Information of Target
| Target ID | LDTP10684 | |||||
|---|---|---|---|---|---|---|
| Target Name | RNA-binding protein 14 (RBM14) | |||||
| Gene Name | RBM14 | |||||
| Gene ID | 100526737 | |||||
| Synonyms |
SIP; RNA-binding protein 14; Paraspeckle protein 2; PSP2; RNA-binding motif protein 14; RRM-containing coactivator activator/modulator; Synaptotagmin-interacting protein; SYT-interacting protein |
|||||
| 3D Structure | ||||||
| Sequence |
MLLWASLLAFAPVCGQSAAAHKPVISVHPPWTTFFKGERVTLTCNGFQFYATEKTTWYHR
HYWGEKLTLTPGNTLEVRESGLYRCQARGSPRSNPVRLLFSSDSLILQAPYSVFEGDTLV LRCHRRRKEKLTAVKYTWNGNILSISNKSWDLLIPQASSNNNGNYRCIGYGDENDVFRSN FKIIKIQELFPHPELKATDSQPTEGNSVNLSCETQLPPERSDTPLHFNFFRDGEVILSDW STYPELQLPTVWRENSGSYWCGAETVRGNIHKHSPSLQIHVQRIPVSGVLLETQPSGGQA VEGEMLVLVCSVAEGTGDTTFSWHREDMQESLGRKTQRSLRAELELPAIRQSHAGGYYCT ADNSYGPVQSMVLNVTVRETPGNRDGLVAAGATGGLLSALLLAVALLFHCWRRRKSGVGF LGDETRLPPAPGPGESSHSICPAQVELQSLYVDVHPKKGDLVYSEIQTTQLGEEEEANTS RTLLEDKDVSVVYSEVKTQHPDNSAGKISSKDEES |
|||||
| Target Bioclass |
Other
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
Isoform 1 may function as a nuclear receptor coactivator, enhancing transcription through other coactivators such as NCOA6 and CITED1. Isoform 2, functions as a transcriptional repressor, modulating transcriptional activities of coactivators including isoform 1, NCOA6 and CITED1. Regulates centriole biogenesis by suppressing the formation of aberrant centriolar protein complexes in the cytoplasm and thus preserving mitotic spindle integrity. Prevents the formation of the STIL-CENPJ complex (which can induce the formation of aberrant centriolar protein complexes) by interfering with the interaction of STIL with CENPJ. Plays a role in the regulation of DNA virus-mediated innate immune response by assembling into the HDP-RNP complex, a complex that serves as a platform for IRF3 phosphorylation and subsequent innate immune response activation through the cGAS-STING pathway. Also involved in the regulation of pre-mRNA alternative splicing.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| 22RV1 | SNV: p.A268T | DBIA Probe Info | |||
| EFO27 | Deletion: p.A441del | DBIA Probe Info | |||
| G361 | SNV: p.A513E | DBIA Probe Info | |||
| HCT15 | SNV: p.G307E | DBIA Probe Info | |||
| JURKAT | SNV: p.A418V | Compound 10 Probe Info | |||
| LOXIMVI | SNV: p.D589E | DBIA Probe Info | |||
| MCF7 | SNV: p.T454S | . | |||
| NCIH146 | SNV: p.T206I | DBIA Probe Info | |||
| RKO | SNV: p.G287C | DBIA Probe Info | |||
| TOV21G | SNV: p.P304L | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
A-EBA Probe Info |
 |
2.90 | LDD0215 | [1] | |
|
CY-1 Probe Info |
 |
100.00 | LDD0243 | [2] | |
|
C-Sul Probe Info |
 |
4.73 | LDD0066 | [3] | |
|
TH211 Probe Info |
 |
Y249(20.00); Y285(20.00); Y540(20.00); Y645(20.00) | LDD0257 | [4] | |
|
TH216 Probe Info |
 |
Y114(20.00); Y665(20.00); Y249(17.17); Y614(11.07) | LDD0259 | [4] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [5] | |
|
Probe 1 Probe Info |
 |
Y237(19.60); Y249(43.68); Y261(48.51); Y273(7.88) | LDD3495 | [6] | |
|
BTD Probe Info |
 |
C108(0.90) | LDD2090 | [7] | |
|
HPAP Probe Info |
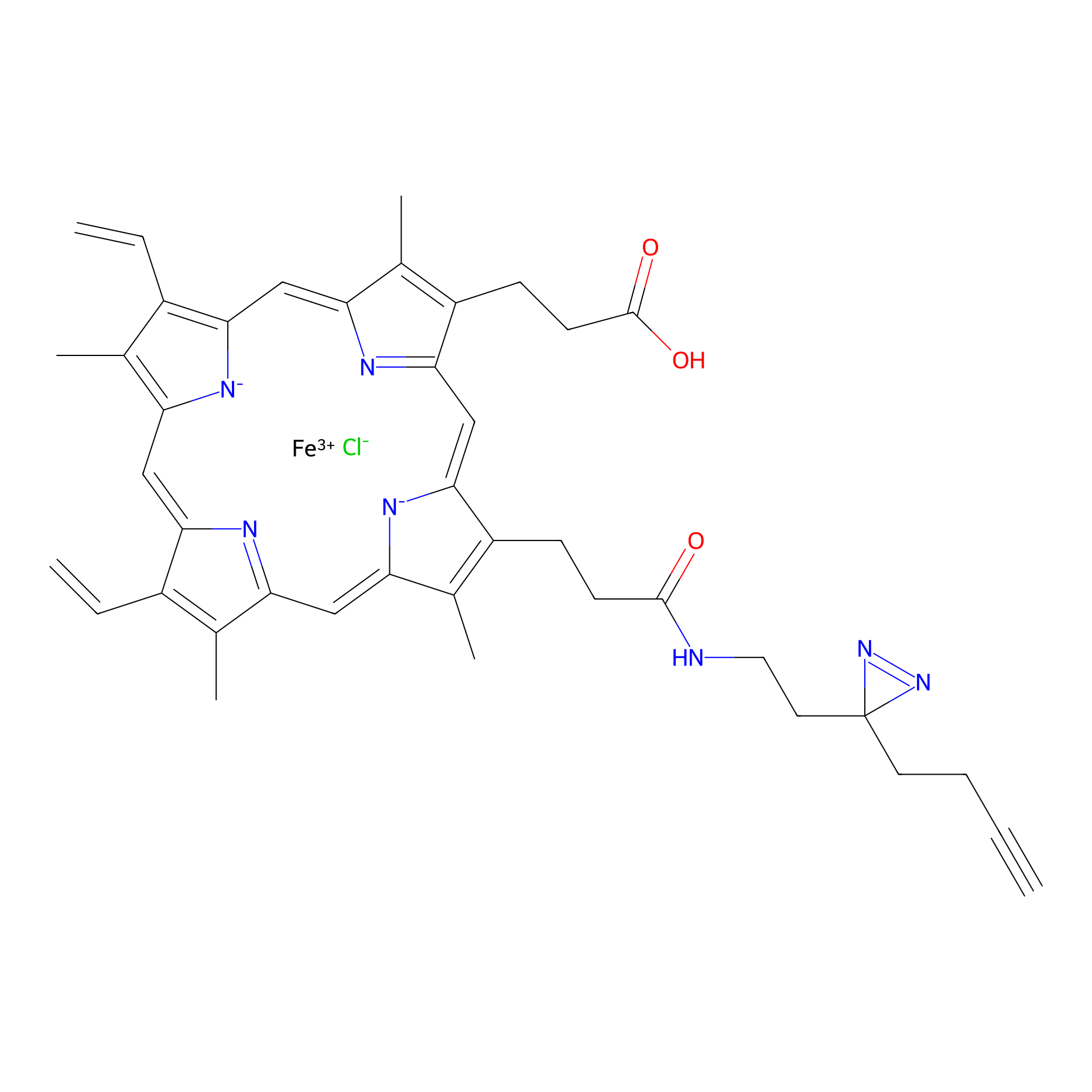 |
3.37 | LDD0063 | [8] | |
|
HHS-475 Probe Info |
 |
Y540(0.13); Y665(0.53); Y249(1.15); Y226(1.90) | LDD0264 | [9] | |
|
HHS-465 Probe Info |
 |
Y665(10.00) | LDD2237 | [10] | |
|
DBIA Probe Info |
 |
C90(0.88) | LDD0078 | [11] | |
|
5E-2FA Probe Info |
 |
H662(0.00); H56(0.00); H642(0.00); H533(0.00) | LDD2235 | [12] | |
|
ATP probe Probe Info |
 |
K149(0.00); K152(0.00); K121(0.00); K126(0.00) | LDD0199 | [13] | |
|
m-APA Probe Info |
 |
H662(0.00); H56(0.00); H642(0.00); H533(0.00) | LDD2231 | [12] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C31(0.00); C90(0.00); C108(0.00) | LDD0038 | [14] | |
|
IA-alkyne Probe Info |
 |
C90(0.00); C108(0.00) | LDD0032 | [15] | |
|
IPIAA_L Probe Info |
 |
C108(0.00); C90(0.00) | LDD0031 | [16] | |
|
Lodoacetamide azide Probe Info |
 |
C31(0.00); C90(0.00); C108(0.00) | LDD0037 | [14] | |
|
IPM Probe Info |
 |
N.A. | LDD0025 | [17] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [17] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [17] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [18] | |
|
Compound 10 Probe Info |
 |
C108(0.00); C90(0.00) | LDD2216 | [19] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [19] | |
|
ENE Probe Info |
 |
N.A. | LDD0006 | [18] | |
|
NHS Probe Info |
 |
K149(0.00); K153(0.00); K135(0.00); K126(0.00) | LDD0010 | [18] | |
|
PF-06672131 Probe Info |
 |
C90(0.00); C108(0.00) | LDD0017 | [20] | |
|
SF Probe Info |
 |
Y285(0.00); Y645(0.00); Y261(0.00); Y588(0.00) | LDD0028 | [21] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [18] | |
|
Acrolein Probe Info |
 |
C90(0.00); C108(0.00); H642(0.00); H58(0.00) | LDD0217 | [22] | |
|
Cinnamaldehyde Probe Info |
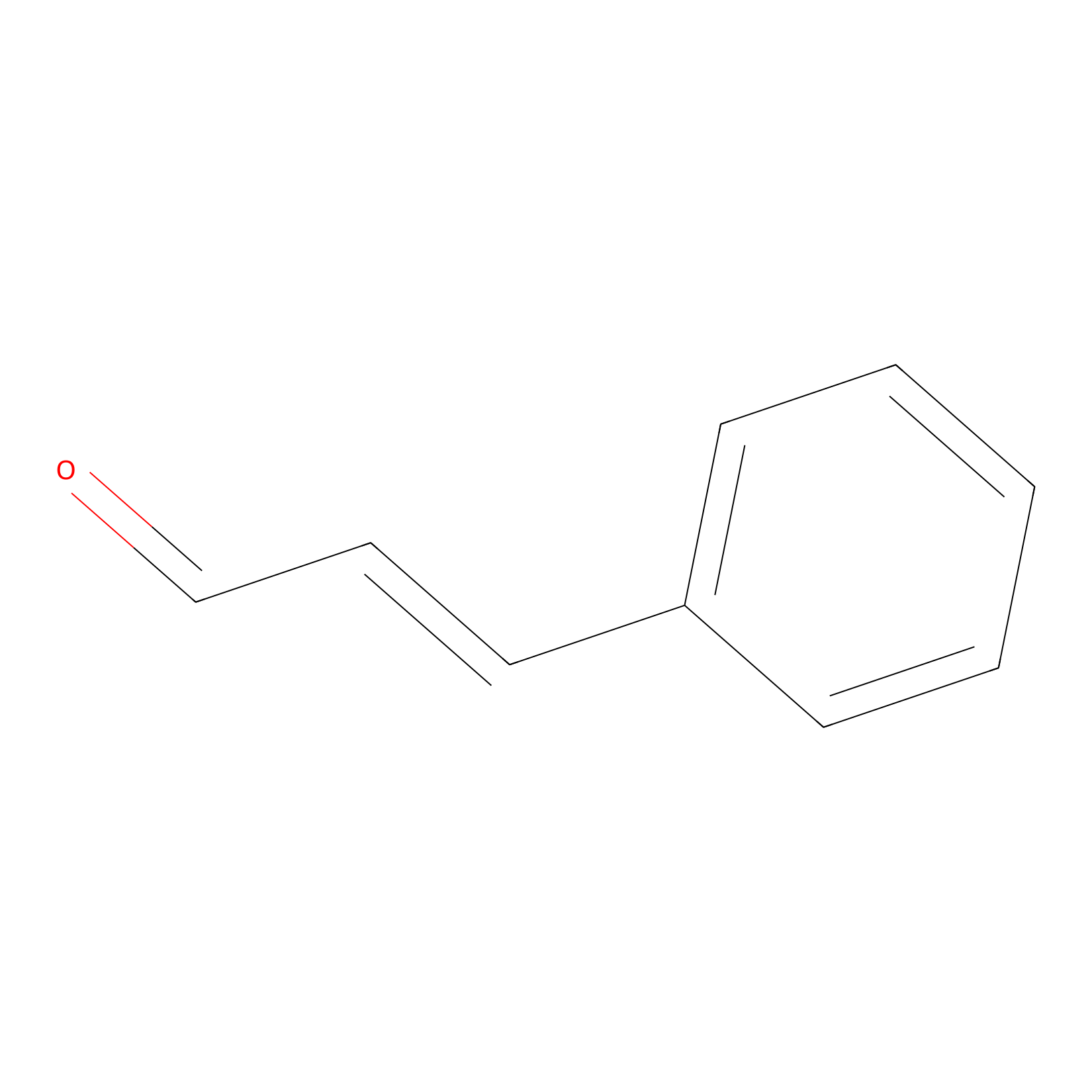 |
N.A. | LDD0220 | [22] | |
|
Crotonaldehyde Probe Info |
 |
H533(0.00); C90(0.00); H58(0.00); C108(0.00) | LDD0219 | [22] | |
|
Methacrolein Probe Info |
 |
C90(0.00); C31(0.00); C108(0.00) | LDD0218 | [22] | |
|
W1 Probe Info |
 |
C108(0.00); C90(0.00) | LDD0236 | [23] | |
|
NAIA_5 Probe Info |
 |
C108(0.00); C31(0.00); C90(0.00) | LDD2223 | [24] | |
|
HHS-482 Probe Info |
 |
Y226(0.54); Y237(0.55); Y249(0.29); Y273(0.32) | LDD2239 | [10] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C165 Probe Info |
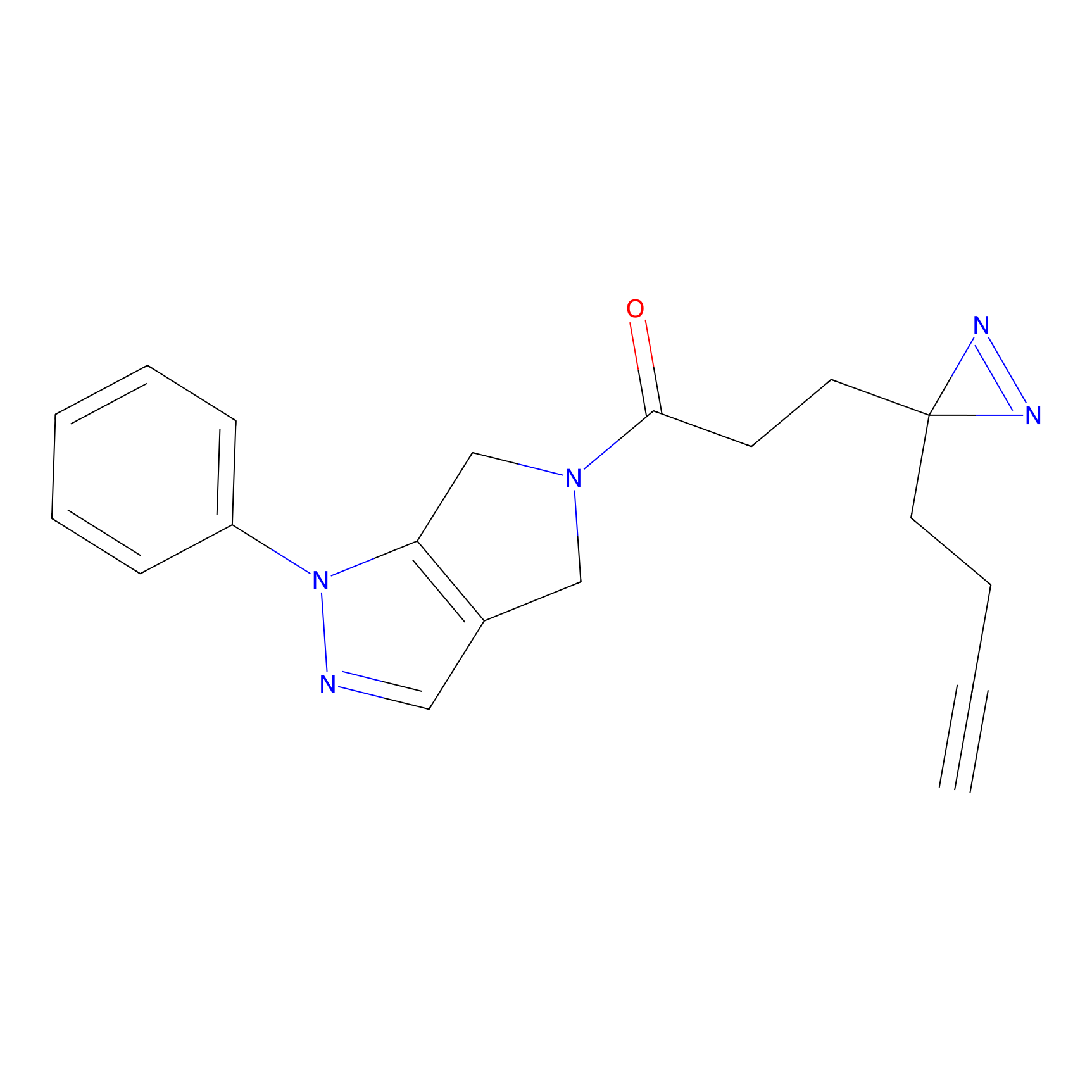 |
13.09 | LDD1845 | [25] | |
|
C187 Probe Info |
 |
25.99 | LDD1865 | [25] | |
|
C191 Probe Info |
 |
22.32 | LDD1868 | [25] | |
|
C193 Probe Info |
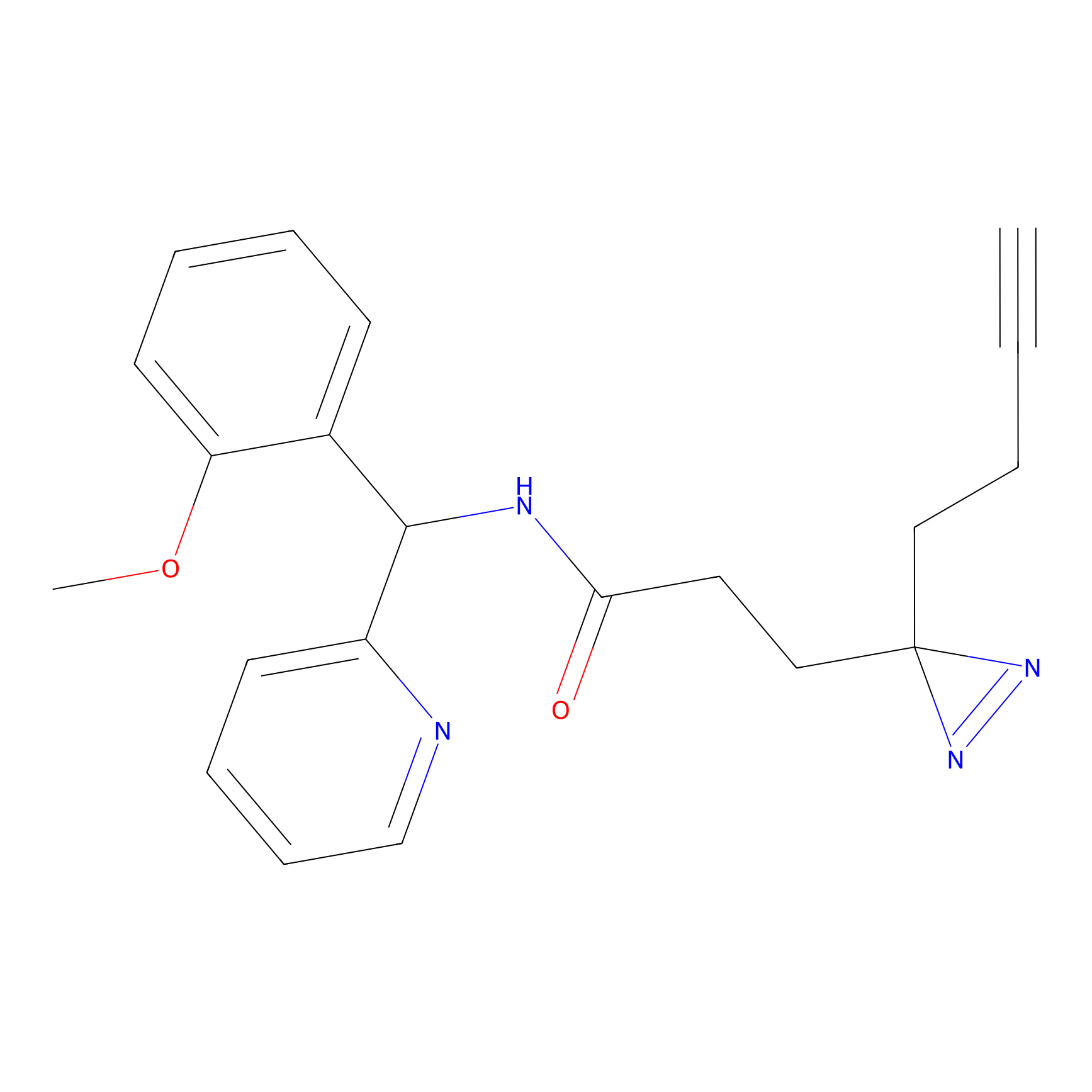 |
7.78 | LDD1869 | [25] | |
|
C232 Probe Info |
 |
33.82 | LDD1905 | [25] | |
|
C338 Probe Info |
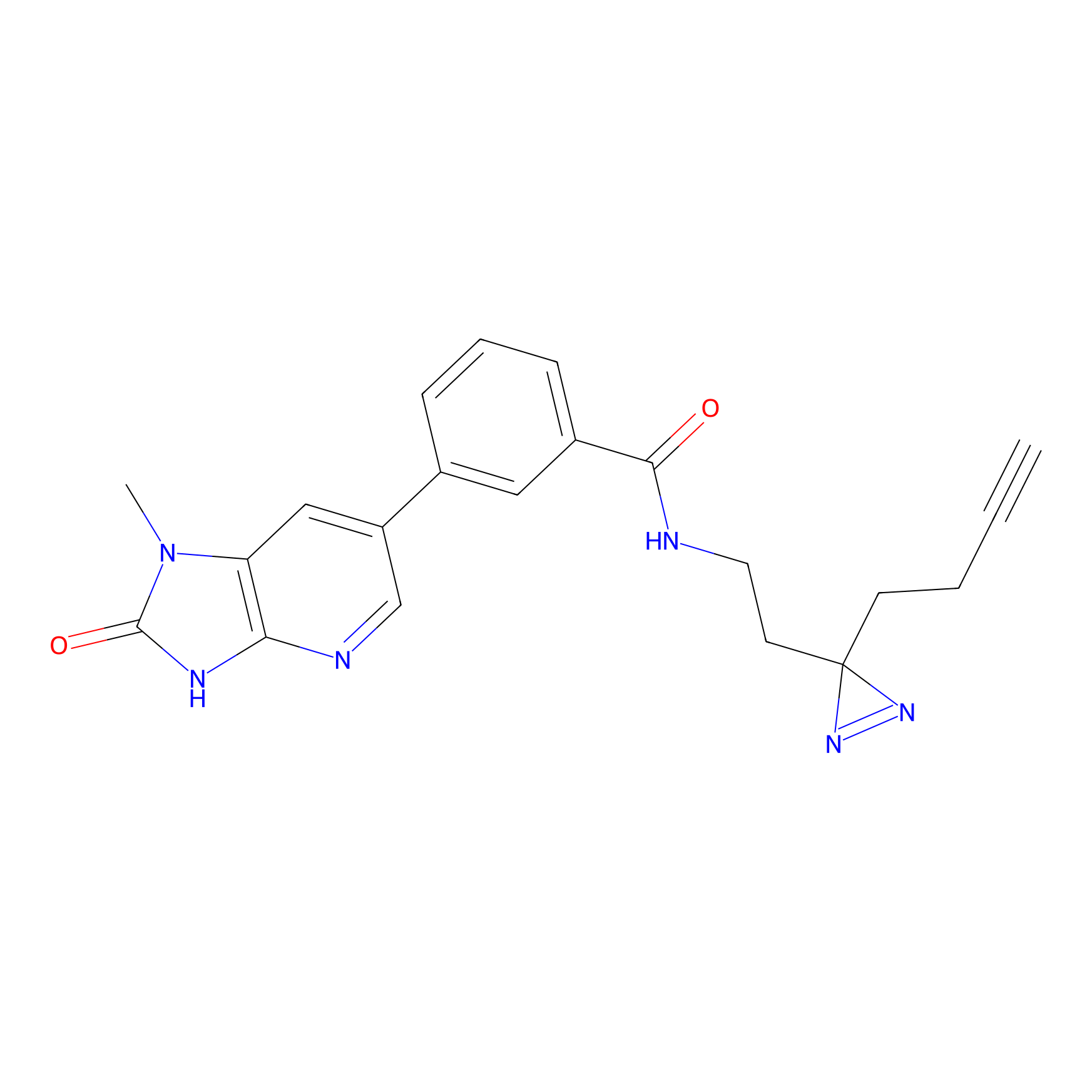 |
14.22 | LDD2001 | [25] | |
|
C348 Probe Info |
 |
10.27 | LDD2009 | [25] | |
|
FFF probe11 Probe Info |
 |
14.30 | LDD0471 | [26] | |
|
FFF probe12 Probe Info |
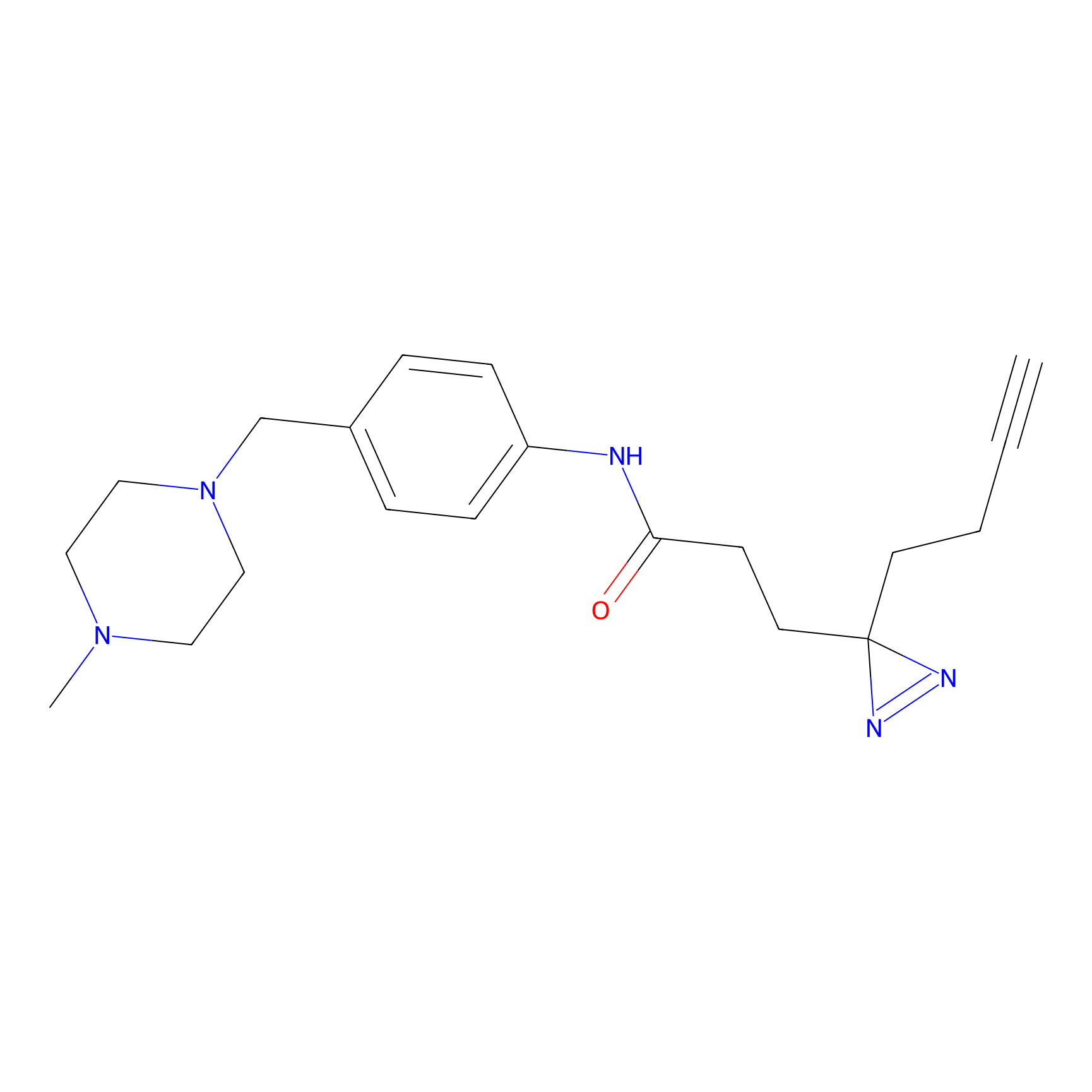 |
5.67 | LDD0473 | [26] | |
|
FFF probe13 Probe Info |
 |
19.98 | LDD0475 | [26] | |
|
FFF probe14 Probe Info |
 |
12.54 | LDD0477 | [26] | |
|
FFF probe2 Probe Info |
 |
20.00 | LDD0463 | [26] | |
|
FFF probe3 Probe Info |
 |
14.25 | LDD0464 | [26] | |
|
FFF probe4 Probe Info |
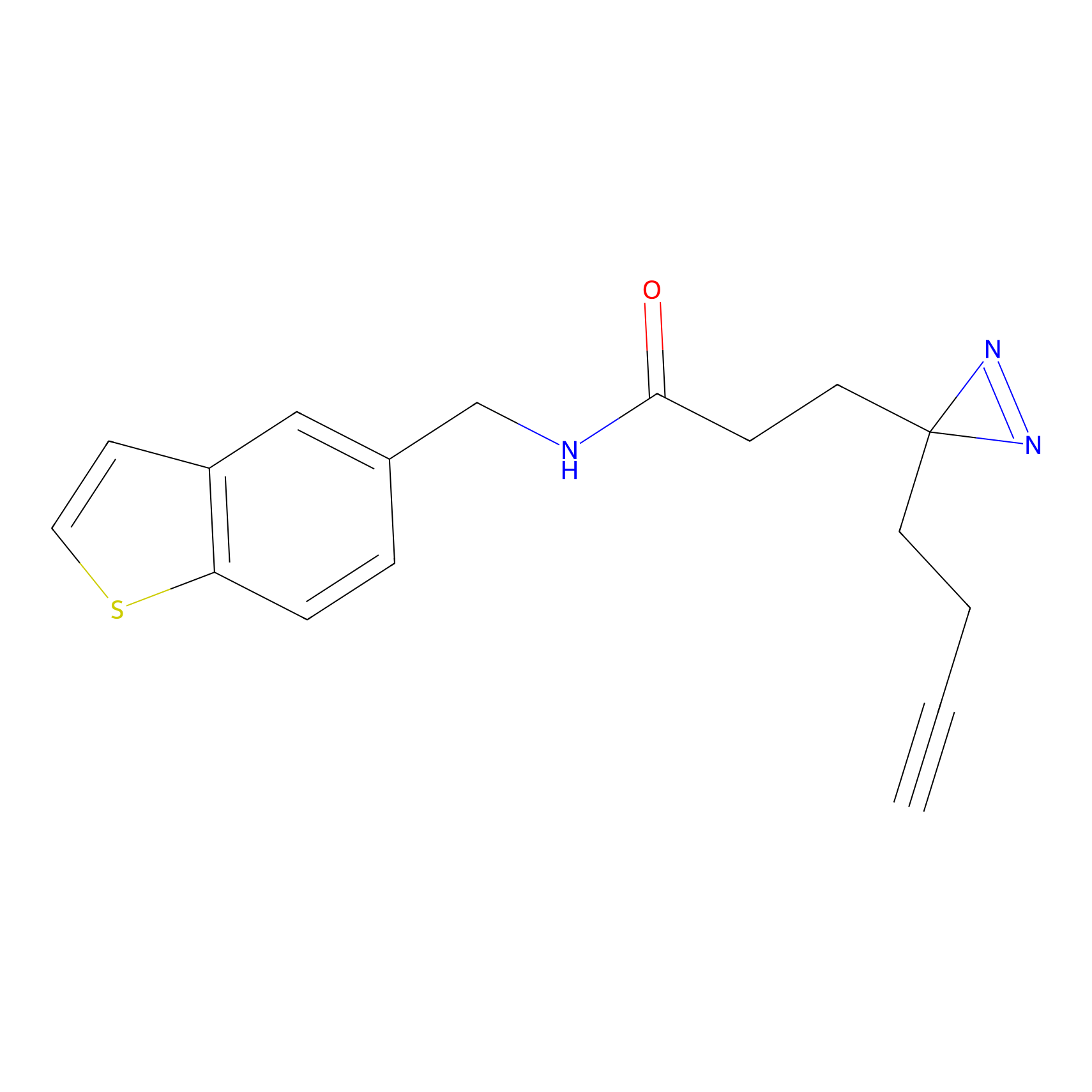 |
12.18 | LDD0466 | [26] | |
|
FFF probe6 Probe Info |
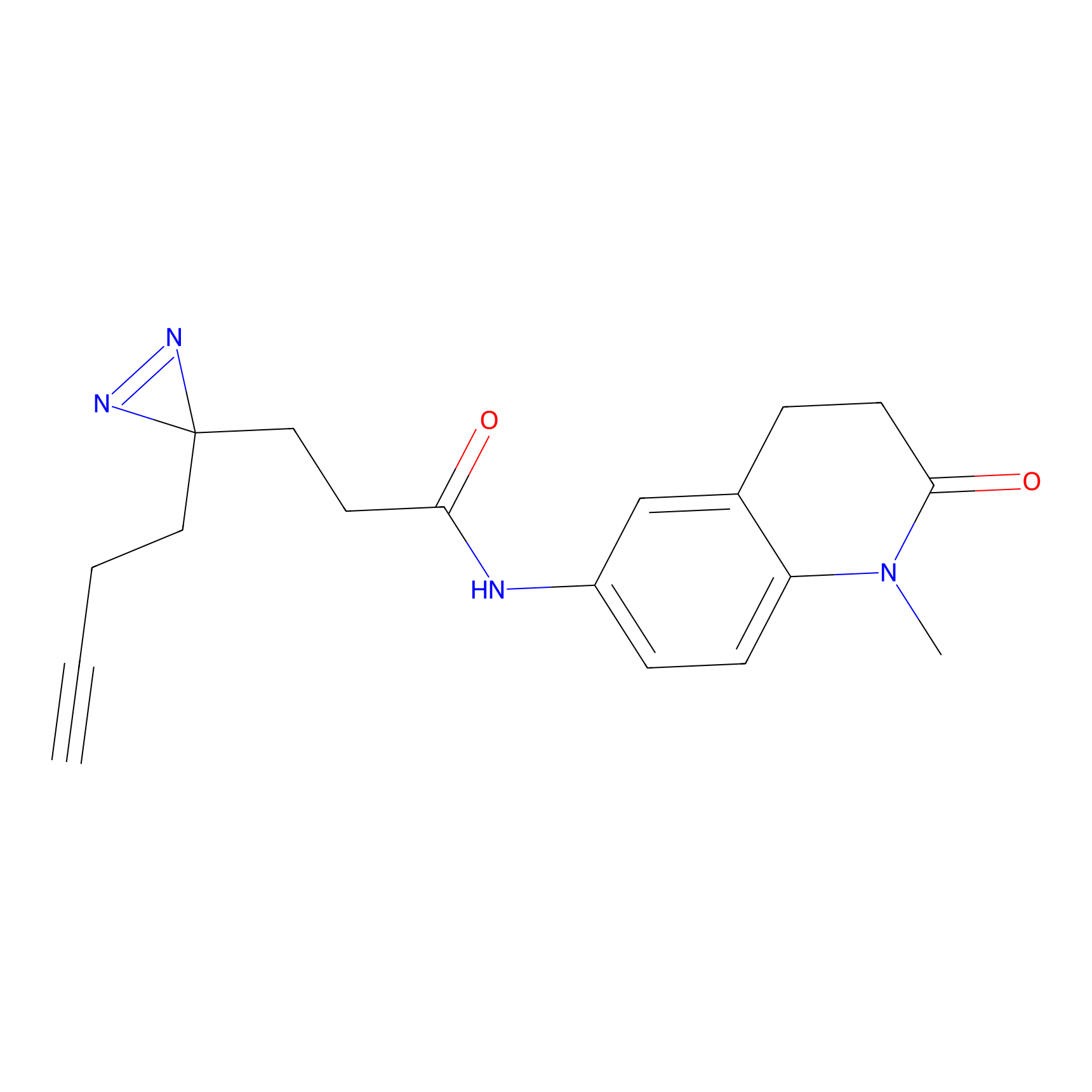 |
13.32 | LDD0467 | [26] | |
|
JN0003 Probe Info |
 |
20.00 | LDD0469 | [26] | |
|
OEA-DA Probe Info |
 |
8.52 | LDD0046 | [27] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C108(0.80) | LDD2117 | [7] |
| LDCM0214 | AC1 | HEK-293T | C108(0.90); C90(0.83) | LDD1507 | [28] |
| LDCM0215 | AC10 | HEK-293T | C108(0.90); C90(0.85); C31(1.07) | LDD1508 | [28] |
| LDCM0226 | AC11 | HEK-293T | C108(0.92); C90(0.94); C31(1.05) | LDD1509 | [28] |
| LDCM0237 | AC12 | HEK-293T | C108(0.97); C90(1.08); C31(1.20) | LDD1510 | [28] |
| LDCM0259 | AC14 | HEK-293T | C108(0.84); C90(0.98) | LDD1512 | [28] |
| LDCM0270 | AC15 | HEK-293T | C108(0.92); C90(0.96) | LDD1513 | [28] |
| LDCM0276 | AC17 | HEK-293T | C108(0.90); C90(1.00) | LDD1515 | [28] |
| LDCM0277 | AC18 | HEK-293T | C108(1.04); C90(1.07); C31(0.99) | LDD1516 | [28] |
| LDCM0278 | AC19 | HEK-293T | C108(0.90); C90(1.12); C31(0.55) | LDD1517 | [28] |
| LDCM0279 | AC2 | HEK-293T | C108(0.97); C90(1.07); C31(0.79) | LDD1518 | [28] |
| LDCM0280 | AC20 | HEK-293T | C108(0.97); C90(1.09); C31(1.13) | LDD1519 | [28] |
| LDCM0281 | AC21 | HEK-293T | C108(0.97); C90(1.00); C31(0.91) | LDD1520 | [28] |
| LDCM0282 | AC22 | HEK-293T | C108(0.97); C90(0.98) | LDD1521 | [28] |
| LDCM0283 | AC23 | HEK-293T | C108(1.04); C90(1.13) | LDD1522 | [28] |
| LDCM0284 | AC24 | HEK-293T | C108(0.97); C90(1.04) | LDD1523 | [28] |
| LDCM0285 | AC25 | HEK-293T | C108(0.98); C90(0.95) | LDD1524 | [28] |
| LDCM0286 | AC26 | HEK-293T | C108(1.03); C90(0.91); C31(0.96) | LDD1525 | [28] |
| LDCM0287 | AC27 | HEK-293T | C108(0.92); C90(1.13); C31(1.07) | LDD1526 | [28] |
| LDCM0288 | AC28 | HEK-293T | C108(1.00); C90(0.98); C31(1.15) | LDD1527 | [28] |
| LDCM0289 | AC29 | HEK-293T | C108(0.96); C90(1.00); C31(1.11) | LDD1528 | [28] |
| LDCM0290 | AC3 | HEK-293T | C108(0.92); C90(0.91); C31(1.06) | LDD1529 | [28] |
| LDCM0291 | AC30 | HEK-293T | C108(1.06); C90(1.04) | LDD1530 | [28] |
| LDCM0292 | AC31 | HEK-293T | C108(1.01); C90(0.93) | LDD1531 | [28] |
| LDCM0293 | AC32 | HEK-293T | C108(1.07); C90(0.96) | LDD1532 | [28] |
| LDCM0294 | AC33 | HEK-293T | C108(0.88); C90(0.83) | LDD1533 | [28] |
| LDCM0295 | AC34 | HEK-293T | C108(0.90); C90(0.93); C31(1.16) | LDD1534 | [28] |
| LDCM0296 | AC35 | HEK-293T | C108(0.89); C90(1.06); C31(1.05) | LDD1535 | [28] |
| LDCM0297 | AC36 | HEK-293T | C108(1.04); C90(0.97); C31(1.15) | LDD1536 | [28] |
| LDCM0298 | AC37 | HEK-293T | C108(0.96); C90(1.01); C31(1.03) | LDD1537 | [28] |
| LDCM0299 | AC38 | HEK-293T | C108(0.86); C90(1.11) | LDD1538 | [28] |
| LDCM0300 | AC39 | HEK-293T | C108(0.96); C90(1.04) | LDD1539 | [28] |
| LDCM0301 | AC4 | HEK-293T | C108(0.98); C90(0.94); C31(1.28) | LDD1540 | [28] |
| LDCM0302 | AC40 | HEK-293T | C108(0.89); C90(1.11) | LDD1541 | [28] |
| LDCM0303 | AC41 | HEK-293T | C108(0.92); C90(0.97) | LDD1542 | [28] |
| LDCM0304 | AC42 | HEK-293T | C108(0.82); C90(0.95); C31(1.16) | LDD1543 | [28] |
| LDCM0305 | AC43 | HEK-293T | C108(0.90); C90(1.06); C31(1.23) | LDD1544 | [28] |
| LDCM0306 | AC44 | HEK-293T | C108(1.03); C90(1.06); C31(1.09) | LDD1545 | [28] |
| LDCM0307 | AC45 | HEK-293T | C108(0.89); C90(0.89); C31(0.98) | LDD1546 | [28] |
| LDCM0308 | AC46 | HEK-293T | C108(0.86); C90(1.03) | LDD1547 | [28] |
| LDCM0309 | AC47 | HEK-293T | C108(0.92); C90(1.00) | LDD1548 | [28] |
| LDCM0310 | AC48 | HEK-293T | C108(0.88); C90(0.94) | LDD1549 | [28] |
| LDCM0311 | AC49 | HEK-293T | C108(0.90); C90(1.11) | LDD1550 | [28] |
| LDCM0312 | AC5 | HEK-293T | C108(0.90); C90(1.05); C31(1.00) | LDD1551 | [28] |
| LDCM0313 | AC50 | HEK-293T | C108(0.90); C90(1.01); C31(1.00) | LDD1552 | [28] |
| LDCM0314 | AC51 | HEK-293T | C108(0.99); C90(1.15); C31(0.95) | LDD1553 | [28] |
| LDCM0315 | AC52 | HEK-293T | C108(0.98); C90(0.92); C31(1.02) | LDD1554 | [28] |
| LDCM0316 | AC53 | HEK-293T | C108(0.97); C90(1.09); C31(0.99) | LDD1555 | [28] |
| LDCM0317 | AC54 | HEK-293T | C108(0.90); C90(1.07) | LDD1556 | [28] |
| LDCM0318 | AC55 | HEK-293T | C108(0.98); C90(0.94) | LDD1557 | [28] |
| LDCM0319 | AC56 | HEK-293T | C108(0.90); C90(0.98) | LDD1558 | [28] |
| LDCM0320 | AC57 | HEK-293T | C108(0.88); C90(0.98) | LDD1559 | [28] |
| LDCM0321 | AC58 | HEK-293T | C108(0.92); C90(0.91); C31(1.00) | LDD1560 | [28] |
| LDCM0322 | AC59 | HEK-293T | C108(0.96); C90(1.00); C31(1.04) | LDD1561 | [28] |
| LDCM0323 | AC6 | HEK-293T | C108(0.88); C90(0.96) | LDD1562 | [28] |
| LDCM0324 | AC60 | HEK-293T | C108(1.02); C90(0.95); C31(1.13) | LDD1563 | [28] |
| LDCM0325 | AC61 | HEK-293T | C108(0.91); C90(1.01); C31(1.08) | LDD1564 | [28] |
| LDCM0326 | AC62 | HEK-293T | C108(0.90); C90(1.03) | LDD1565 | [28] |
| LDCM0327 | AC63 | HEK-293T | C108(0.92); C90(1.02) | LDD1566 | [28] |
| LDCM0328 | AC64 | HEK-293T | C108(0.96); C90(1.02) | LDD1567 | [28] |
| LDCM0334 | AC7 | HEK-293T | C108(1.02); C90(0.94) | LDD1568 | [28] |
| LDCM0345 | AC8 | HEK-293T | C108(0.89); C90(0.87) | LDD1569 | [28] |
| LDCM0248 | AKOS034007472 | HEK-293T | C108(0.97); C90(1.05); C31(1.19) | LDD1511 | [28] |
| LDCM0356 | AKOS034007680 | HEK-293T | C108(0.89); C90(0.92) | LDD1570 | [28] |
| LDCM0275 | AKOS034007705 | HEK-293T | C108(0.89); C90(0.95) | LDD1514 | [28] |
| LDCM0020 | ARS-1620 | HCC44 | C90(0.88) | LDD0078 | [11] |
| LDCM0108 | Chloroacetamide | HeLa | C90(0.00); H58(0.00); C108(0.00); H41(0.00) | LDD0222 | [22] |
| LDCM0367 | CL1 | HEK-293T | C108(0.91); C90(0.93); C31(1.32) | LDD1571 | [28] |
| LDCM0368 | CL10 | HEK-293T | C108(0.76); C90(1.04) | LDD1572 | [28] |
| LDCM0369 | CL100 | HEK-293T | C108(0.91); C90(0.97) | LDD1573 | [28] |
| LDCM0370 | CL101 | HEK-293T | C108(0.88); C90(0.90); C31(0.97) | LDD1574 | [28] |
| LDCM0371 | CL102 | HEK-293T | C108(0.82); C90(0.91); C31(1.08) | LDD1575 | [28] |
| LDCM0372 | CL103 | HEK-293T | C108(0.94); C90(1.01) | LDD1576 | [28] |
| LDCM0373 | CL104 | HEK-293T | C108(0.91); C90(0.96) | LDD1577 | [28] |
| LDCM0374 | CL105 | HEK-293T | C108(0.91); C90(0.98); C31(1.11) | LDD1578 | [28] |
| LDCM0375 | CL106 | HEK-293T | C108(0.94); C90(0.87); C31(0.83) | LDD1579 | [28] |
| LDCM0376 | CL107 | HEK-293T | C108(1.03); C90(0.90) | LDD1580 | [28] |
| LDCM0377 | CL108 | HEK-293T | C108(0.95); C90(1.04) | LDD1581 | [28] |
| LDCM0378 | CL109 | HEK-293T | C108(0.95); C90(0.93); C31(1.12) | LDD1582 | [28] |
| LDCM0379 | CL11 | HEK-293T | C108(0.93); C90(0.96) | LDD1583 | [28] |
| LDCM0380 | CL110 | HEK-293T | C108(0.96); C90(0.83); C31(0.83) | LDD1584 | [28] |
| LDCM0381 | CL111 | HEK-293T | C108(0.97); C90(0.94) | LDD1585 | [28] |
| LDCM0382 | CL112 | HEK-293T | C108(0.94); C90(0.84) | LDD1586 | [28] |
| LDCM0383 | CL113 | HEK-293T | C108(0.90); C90(0.94); C31(0.98) | LDD1587 | [28] |
| LDCM0384 | CL114 | HEK-293T | C108(0.78); C90(0.80); C31(0.89) | LDD1588 | [28] |
| LDCM0385 | CL115 | HEK-293T | C108(0.88); C90(1.07) | LDD1589 | [28] |
| LDCM0386 | CL116 | HEK-293T | C108(0.94); C90(0.91) | LDD1590 | [28] |
| LDCM0387 | CL117 | HEK-293T | C108(0.92); C90(0.93); C31(1.21) | LDD1591 | [28] |
| LDCM0388 | CL118 | HEK-293T | C108(0.90); C90(0.95); C31(1.00) | LDD1592 | [28] |
| LDCM0389 | CL119 | HEK-293T | C108(0.95); C90(1.01) | LDD1593 | [28] |
| LDCM0390 | CL12 | HEK-293T | C108(0.90); C90(0.95) | LDD1594 | [28] |
| LDCM0391 | CL120 | HEK-293T | C108(0.91); C90(1.00) | LDD1595 | [28] |
| LDCM0392 | CL121 | HEK-293T | C108(0.87); C90(0.96); C31(1.29) | LDD1596 | [28] |
| LDCM0393 | CL122 | HEK-293T | C108(0.86); C90(0.88); C31(0.96) | LDD1597 | [28] |
| LDCM0394 | CL123 | HEK-293T | C108(0.87); C90(0.89) | LDD1598 | [28] |
| LDCM0395 | CL124 | HEK-293T | C108(0.90); C90(0.95) | LDD1599 | [28] |
| LDCM0396 | CL125 | HEK-293T | C108(0.89); C90(0.97); C31(1.48) | LDD1600 | [28] |
| LDCM0397 | CL126 | HEK-293T | C108(0.89); C90(0.90); C31(1.01) | LDD1601 | [28] |
| LDCM0398 | CL127 | HEK-293T | C108(0.90); C90(0.97) | LDD1602 | [28] |
| LDCM0399 | CL128 | HEK-293T | C108(0.90); C90(1.00) | LDD1603 | [28] |
| LDCM0400 | CL13 | HEK-293T | C108(0.91); C90(0.98); C31(1.12) | LDD1604 | [28] |
| LDCM0401 | CL14 | HEK-293T | C108(0.96); C90(0.92); C31(1.12) | LDD1605 | [28] |
| LDCM0402 | CL15 | HEK-293T | C108(0.82); C90(0.82) | LDD1606 | [28] |
| LDCM0403 | CL16 | HEK-293T | C108(0.99); C90(0.97) | LDD1607 | [28] |
| LDCM0404 | CL17 | HEK-293T | C108(0.85); C90(0.72) | LDD1608 | [28] |
| LDCM0405 | CL18 | HEK-293T | C108(0.93); C90(0.85); C31(0.97) | LDD1609 | [28] |
| LDCM0406 | CL19 | HEK-293T | C108(0.90); C90(0.98); C31(1.06) | LDD1610 | [28] |
| LDCM0407 | CL2 | HEK-293T | C108(0.87); C90(0.91); C31(0.99) | LDD1611 | [28] |
| LDCM0408 | CL20 | HEK-293T | C108(1.03); C90(0.89); C31(1.06) | LDD1612 | [28] |
| LDCM0409 | CL21 | HEK-293T | C108(0.83); C90(0.94); C31(0.81) | LDD1613 | [28] |
| LDCM0410 | CL22 | HEK-293T | C108(0.84); C90(1.16) | LDD1614 | [28] |
| LDCM0411 | CL23 | HEK-293T | C108(0.95); C90(0.96) | LDD1615 | [28] |
| LDCM0412 | CL24 | HEK-293T | C108(0.90); C90(1.01) | LDD1616 | [28] |
| LDCM0413 | CL25 | HEK-293T | C108(0.97); C90(1.02); C31(1.34) | LDD1617 | [28] |
| LDCM0414 | CL26 | HEK-293T | C108(1.05); C90(0.93); C31(0.89) | LDD1618 | [28] |
| LDCM0415 | CL27 | HEK-293T | C108(1.00); C90(1.00) | LDD1619 | [28] |
| LDCM0416 | CL28 | HEK-293T | C108(0.99); C90(1.02) | LDD1620 | [28] |
| LDCM0417 | CL29 | HEK-293T | C108(1.00); C90(0.88) | LDD1621 | [28] |
| LDCM0418 | CL3 | HEK-293T | C108(0.84); C90(1.03) | LDD1622 | [28] |
| LDCM0419 | CL30 | HEK-293T | C108(1.01); C90(1.00); C31(0.91) | LDD1623 | [28] |
| LDCM0420 | CL31 | HEK-293T | C108(0.93); C90(1.04); C31(0.90) | LDD1624 | [28] |
| LDCM0421 | CL32 | HEK-293T | C108(1.02); C90(1.07); C31(1.03) | LDD1625 | [28] |
| LDCM0422 | CL33 | HEK-293T | C108(0.87); C90(1.03); C31(0.79) | LDD1626 | [28] |
| LDCM0423 | CL34 | HEK-293T | C108(1.06); C90(0.98) | LDD1627 | [28] |
| LDCM0424 | CL35 | HEK-293T | C108(1.04); C90(1.05) | LDD1628 | [28] |
| LDCM0425 | CL36 | HEK-293T | C108(0.97); C90(0.98) | LDD1629 | [28] |
| LDCM0426 | CL37 | HEK-293T | C108(1.01); C90(1.02); C31(1.16) | LDD1630 | [28] |
| LDCM0428 | CL39 | HEK-293T | C108(1.02); C90(1.02) | LDD1632 | [28] |
| LDCM0429 | CL4 | HEK-293T | C108(0.93); C90(0.89) | LDD1633 | [28] |
| LDCM0430 | CL40 | HEK-293T | C108(1.02); C90(1.03) | LDD1634 | [28] |
| LDCM0431 | CL41 | HEK-293T | C108(0.99); C90(0.92) | LDD1635 | [28] |
| LDCM0432 | CL42 | HEK-293T | C108(1.05); C90(0.96); C31(0.85) | LDD1636 | [28] |
| LDCM0433 | CL43 | HEK-293T | C108(1.07); C90(1.04); C31(0.86) | LDD1637 | [28] |
| LDCM0434 | CL44 | HEK-293T | C108(0.99); C90(1.05); C31(0.98) | LDD1638 | [28] |
| LDCM0435 | CL45 | HEK-293T | C108(0.94); C90(0.94); C31(0.84) | LDD1639 | [28] |
| LDCM0436 | CL46 | HEK-293T | C108(0.90); C90(1.08) | LDD1640 | [28] |
| LDCM0437 | CL47 | HEK-293T | C108(0.98); C90(0.96) | LDD1641 | [28] |
| LDCM0438 | CL48 | HEK-293T | C108(1.02); C90(0.94) | LDD1642 | [28] |
| LDCM0439 | CL49 | HEK-293T | C108(0.93); C90(1.01); C31(1.26) | LDD1643 | [28] |
| LDCM0440 | CL5 | HEK-293T | C108(0.94); C90(0.88) | LDD1644 | [28] |
| LDCM0441 | CL50 | HEK-293T | C108(0.90); C90(0.89); C31(1.11) | LDD1645 | [28] |
| LDCM0443 | CL52 | HEK-293T | C108(0.93); C90(0.93) | LDD1646 | [28] |
| LDCM0444 | CL53 | HEK-293T | C108(0.86); C90(0.82) | LDD1647 | [28] |
| LDCM0445 | CL54 | HEK-293T | C108(0.85); C90(0.85); C31(0.87) | LDD1648 | [28] |
| LDCM0446 | CL55 | HEK-293T | C108(0.99); C90(1.12); C31(1.11) | LDD1649 | [28] |
| LDCM0447 | CL56 | HEK-293T | C108(1.03); C90(0.97); C31(1.18) | LDD1650 | [28] |
| LDCM0448 | CL57 | HEK-293T | C108(0.77); C90(0.83); C31(0.81) | LDD1651 | [28] |
| LDCM0449 | CL58 | HEK-293T | C108(0.84); C90(1.08) | LDD1652 | [28] |
| LDCM0450 | CL59 | HEK-293T | C108(0.91); C90(0.96) | LDD1653 | [28] |
| LDCM0451 | CL6 | HEK-293T | C108(0.88); C90(0.97); C31(0.82) | LDD1654 | [28] |
| LDCM0452 | CL60 | HEK-293T | C108(0.87); C90(1.01) | LDD1655 | [28] |
| LDCM0453 | CL61 | HEK-293T | C108(1.01); C90(1.00); C31(1.31) | LDD1656 | [28] |
| LDCM0454 | CL62 | HEK-293T | C108(0.98); C90(0.96); C31(0.91) | LDD1657 | [28] |
| LDCM0455 | CL63 | HEK-293T | C108(0.96); C90(1.04) | LDD1658 | [28] |
| LDCM0456 | CL64 | HEK-293T | C108(0.89); C90(0.92) | LDD1659 | [28] |
| LDCM0457 | CL65 | HEK-293T | C108(0.96); C90(1.10) | LDD1660 | [28] |
| LDCM0458 | CL66 | HEK-293T | C108(0.94); C90(1.03); C31(1.01) | LDD1661 | [28] |
| LDCM0459 | CL67 | HEK-293T | C108(0.95); C90(1.08); C31(1.06) | LDD1662 | [28] |
| LDCM0460 | CL68 | HEK-293T | C108(1.11); C90(1.02); C31(1.36) | LDD1663 | [28] |
| LDCM0461 | CL69 | HEK-293T | C108(0.92); C90(0.97); C31(0.92) | LDD1664 | [28] |
| LDCM0462 | CL7 | HEK-293T | C108(0.84); C90(0.99); C31(0.85) | LDD1665 | [28] |
| LDCM0463 | CL70 | HEK-293T | C108(0.84); C90(1.05) | LDD1666 | [28] |
| LDCM0464 | CL71 | HEK-293T | C108(0.92); C90(0.99) | LDD1667 | [28] |
| LDCM0465 | CL72 | HEK-293T | C108(0.92); C90(1.06) | LDD1668 | [28] |
| LDCM0466 | CL73 | HEK-293T | C108(0.89); C90(0.96); C31(1.22) | LDD1669 | [28] |
| LDCM0467 | CL74 | HEK-293T | C108(0.97); C90(0.95); C31(1.03) | LDD1670 | [28] |
| LDCM0469 | CL76 | HEK-293T | C108(0.92); C90(0.93) | LDD1672 | [28] |
| LDCM0470 | CL77 | HEK-293T | C108(0.90); C90(0.94) | LDD1673 | [28] |
| LDCM0471 | CL78 | HEK-293T | C108(0.95); C90(1.08); C31(0.97) | LDD1674 | [28] |
| LDCM0472 | CL79 | HEK-293T | C108(0.99); C90(1.10); C31(0.87) | LDD1675 | [28] |
| LDCM0473 | CL8 | HEK-293T | C108(0.85); C90(0.83); C31(1.04) | LDD1676 | [28] |
| LDCM0474 | CL80 | HEK-293T | C108(1.06); C90(1.09); C31(1.16) | LDD1677 | [28] |
| LDCM0475 | CL81 | HEK-293T | C108(0.95); C90(0.98); C31(0.87) | LDD1678 | [28] |
| LDCM0476 | CL82 | HEK-293T | C108(0.94); C90(1.08) | LDD1679 | [28] |
| LDCM0477 | CL83 | HEK-293T | C108(0.98); C90(1.00) | LDD1680 | [28] |
| LDCM0478 | CL84 | HEK-293T | C108(0.86); C90(0.94) | LDD1681 | [28] |
| LDCM0479 | CL85 | HEK-293T | C108(0.96); C90(1.10); C31(1.38) | LDD1682 | [28] |
| LDCM0480 | CL86 | HEK-293T | C108(0.94); C90(0.90); C31(1.02) | LDD1683 | [28] |
| LDCM0481 | CL87 | HEK-293T | C108(0.91); C90(0.95) | LDD1684 | [28] |
| LDCM0482 | CL88 | HEK-293T | C108(1.02); C90(0.92) | LDD1685 | [28] |
| LDCM0483 | CL89 | HEK-293T | C108(0.97); C90(0.88) | LDD1686 | [28] |
| LDCM0484 | CL9 | HEK-293T | C108(0.93); C90(0.95); C31(0.94) | LDD1687 | [28] |
| LDCM0485 | CL90 | HEK-293T | C108(0.78); C90(0.83); C31(0.77) | LDD1688 | [28] |
| LDCM0486 | CL91 | HEK-293T | C108(0.98); C90(0.97); C31(1.07) | LDD1689 | [28] |
| LDCM0487 | CL92 | HEK-293T | C108(0.98); C90(0.95); C31(1.11) | LDD1690 | [28] |
| LDCM0488 | CL93 | HEK-293T | C108(0.86); C90(0.86); C31(1.06) | LDD1691 | [28] |
| LDCM0489 | CL94 | HEK-293T | C108(0.83); C90(1.11) | LDD1692 | [28] |
| LDCM0490 | CL95 | HEK-293T | C108(0.79); C90(0.76) | LDD1693 | [28] |
| LDCM0491 | CL96 | HEK-293T | C108(0.77); C90(0.97) | LDD1694 | [28] |
| LDCM0492 | CL97 | HEK-293T | C108(0.85); C90(0.83); C31(0.88) | LDD1695 | [28] |
| LDCM0493 | CL98 | HEK-293T | C108(0.81); C90(0.91); C31(0.91) | LDD1696 | [28] |
| LDCM0494 | CL99 | HEK-293T | C108(0.85); C90(0.92) | LDD1697 | [28] |
| LDCM0495 | E2913 | HEK-293T | C108(0.96); C90(0.99) | LDD1698 | [28] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C31(1.90); C108(1.40) | LDD1702 | [7] |
| LDCM0468 | Fragment33 | HEK-293T | C108(0.96); C90(1.04) | LDD1671 | [28] |
| LDCM0427 | Fragment51 | HEK-293T | C108(1.07); C90(0.93); C31(1.06) | LDD1631 | [28] |
| LDCM0116 | HHS-0101 | DM93 | Y540(0.13); Y665(0.53); Y249(1.15); Y226(1.90) | LDD0264 | [9] |
| LDCM0117 | HHS-0201 | DM93 | Y665(0.47); Y249(0.92); Y237(1.82); Y226(2.16) | LDD0265 | [9] |
| LDCM0118 | HHS-0301 | DM93 | Y540(0.13); Y665(0.48); Y237(1.32); Y249(1.78) | LDD0266 | [9] |
| LDCM0119 | HHS-0401 | DM93 | Y249(0.72); Y665(0.86); Y528(1.71); Y237(2.16) | LDD0267 | [9] |
| LDCM0120 | HHS-0701 | DM93 | Y540(0.10); Y665(0.28); Y528(1.03); Y249(1.62) | LDD0268 | [9] |
| LDCM0107 | IAA | HeLa | C90(0.00); H58(0.00); C108(0.00); H56(0.00) | LDD0221 | [22] |
| LDCM0022 | KB02 | 22RV1 | C108(1.31); C31(0.92); C90(1.36) | LDD2243 | [29] |
| LDCM0023 | KB03 | MDA-MB-231 | C108(4.23); C90(1.49) | LDD1701 | [7] |
| LDCM0024 | KB05 | COLO792 | C108(1.24); C31(0.84); C90(1.29) | LDD3310 | [29] |
| LDCM0509 | N-(4-bromo-3,5-dimethylphenyl)-2-nitroacetamide | MDA-MB-231 | C108(0.79) | LDD2102 | [7] |
| LDCM0109 | NEM | HeLa | H533(0.00); H58(0.00); H56(0.00); H642(0.00) | LDD0223 | [22] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C108(0.90) | LDD2090 | [7] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C108(0.85) | LDD2101 | [7] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C108(1.17) | LDD2105 | [7] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C108(1.23) | LDD2107 | [7] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C108(0.54) | LDD2115 | [7] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C108(0.77) | LDD2120 | [7] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C108(1.05) | LDD2123 | [7] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C108(1.27); C90(0.77) | LDD2125 | [7] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C108(0.70) | LDD2134 | [7] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C108(1.31) | LDD2136 | [7] |
| LDCM0553 | Nucleophilic fragment 6b | MDA-MB-231 | C108(2.87) | LDD2147 | [7] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C108(0.75) | LDD2148 | [7] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C90(1.10) | LDD2206 | [30] |
| LDCM0014 | Panhematin | K562 | 3.37 | LDD0063 | [8] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Wolframin (WFS1) | . | O76024 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| NF-kappa-B inhibitor delta (NFKBID) | NF-kappa-B inhibitor family | Q8NI38 | |||
Cytokine and receptor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Cytokine receptor-like factor 3 (CRLF3) | Cytokine receptor-like factor 3 family | Q8IUI8 | |||
Other
References
