Details of the Target
General Information of Target
| Target ID | LDTP06331 | |||||
|---|---|---|---|---|---|---|
| Target Name | Pericentriolar material 1 protein (PCM1) | |||||
| Gene Name | PCM1 | |||||
| Gene ID | 5108 | |||||
| Synonyms |
Pericentriolar material 1 protein; PCM-1; hPCM-1 |
|||||
| 3D Structure | ||||||
| Sequence |
MATGGGPFEDGMNDQDLPNWSNENVDDRLNNMDWGAQQKKANRSSEKNKKKFGVESDKRV
TNDISPESSPGVGRRRTKTPHTFPHSRYMSQMSVPEQAELEKLKQRINFSDLDQRSIGSD SQGRATAANNKRQLSENRKPFNFLPMQINTNKSKDASTNPPNRETIGSAQCKELFASALS NDLLQNCQVSEEDGRGEPAMESSQIVSRLVQIRDYITKASSMREDLVEKNERSANVERLT HLIDHLKEQEKSYMKFLKKILARDPQQEPMEEIENLKKQHDLLKRMLQQQEQLRALQGRQ AALLALQHKAEQAIAVMDDSVVAETAGSLSGVSITSELNEELNDLIQRFHNQLRDSQPPA VPDNRRQAESLSLTREVSQSRKPSASERLPDEKVELFSKMRVLQEKKQKMDKLLGELHTL RDQHLNNSSSSPQRSVDQRSTSAPSASVGLAPVVNGESNSLTSSVPYPTASLVSQNESEN EGHLNPSEKLQKLNEVRKRLNELRELVHYYEQTSDMMTDAVNENRKDEETEESEYDSEHE NSEPVTNIRNPQVASTWNEVNSHSNAQCVSNNRDGRTVNSNCEINNRSAANIRALNMPPS LDCRYNREGEQEIHVAQGEDDEEEEEEAEEEGVSGASLSSHRSSLVDEHPEDAEFEQKIN RLMAAKQKLRQLQDLVAMVQDDDAAQGVISASASNLDDFYPAEEDTKQNSNNTRGNANKT QKDTGVNEKAREKFYEAKLQQQQRELKQLQEERKKLIDIQEKIQALQTACPDLQLSAASV GNCPTKKYMPAVTSTPTVNQHETSTSKSVFEPEDSSIVDNELWSEMRRHEMLREELRQRR KQLEALMAEHQRRQGLAETASPVAVSLRSDGSENLCTPQQSRTEKTMATWGGSTQCALDE EGDEDGYLSEGIVRTDEEEEEEQDASSNDNFSVCPSNSVNHNSYNGKETKNRWKNNCPFS ADENYRPLAKTRQQNISMQRQENLRWVSELSYVEEKEQWQEQINQLKKQLDFSVSICQTL MQDQQTLSCLLQTLLTGPYSVMPSNVASPQVHFIMHQLNQCYTQLTWQQNNVQRLKQMLN ELMRQQNQHPEKPGGKERGSSASHPPSPSLFCPFSFPTQPVNLFNIPGFTNFSSFAPGMN FSPLFPSNFGDFSQNISTPSEQQQPLAQNSSGKTEYMAFPKPFESSSSIGAEKPRNKKLP EEEVESSRTPWLYEQEGEVEKPFIKTGFSVSVEKSTSSNRKNQLDTNGRRRQFDEESLES FSSMPDPVDPTTVTKTFKTRKASAQASLASKDKTPKSKSKKRNSTQLKSRVKNIRYESAS MSSTCEPCKSRNRHSAQTEEPVQAKVFSRKNHEQLEKIIKCNRSTEISSETGSDFSMFEA LRDTIYSEVATLISQNESRPHFLIELFHELQLLNTDYLRQRALYALQDIVSRHISESHEK GENVKSVNSGTWIASNSELTPSESLATTDDETFEKNFERETHKISEQNDADNASVLSVSS NFEPFATDDLGNTVIHLDQALARMREYERMKTEAESNSNMRCTCRIIEDGDGAGAGTTVN NLEETPVIENRSSQQPVSEVSTIPCPRIDTQQLDRQIKAIMKEVIPFLKEHMDEVCSSQL LTSVRRMVLTLTQQNDESKEFVKFFHKQLGSILQDSLAKFAGRKLKDCGEDLLVEISEVL FNELAFFKLMQDLDNNSITVKQRCKRKIEATGVIQSCAKEAKRILEDHGSPAGEIDDEDK DKDETETVKQTQTSEVYDGPKNVRSDISDQEEDEESEGCPVSINLSKAETQALTNYGSGE DENEDEEMEEFEEGPVDVQTSLQANTEATEENEHDEQVLQRDFKKTAESKNVPLEREATS KNDQNNCPVKPCYLNILEDEQPLNSAAHKESPPTVDSTQQPNPLPLRLPEMEPLVPRVKE VKSAQETPESSLAGSPDTESPVLVNDYEAESGNISQKSDEEDFVKVEDLPLKLTIYSEAD LRKKMVEEEQKNHLSGEICEMQTEELAGNSETLKEPETVGAQSI |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
PCM1 family
|
|||||
| Subcellular location |
Cytoplasm, cytoskeleton
|
|||||
| Function |
Required for centrosome assembly and function. Essential for the correct localization of several centrosomal proteins including CEP250, CETN3, PCNT and NEK2. Required to anchor microtubules to the centrosome. Also involved in cilium biogenesis by recruiting the BBSome, a ciliary protein complex involved in cilium biogenesis, to the centriolar satellites. Recruits the tubulin polyglutamylase complex (TPGC) to centriolar satellites.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
TG42 Probe Info |
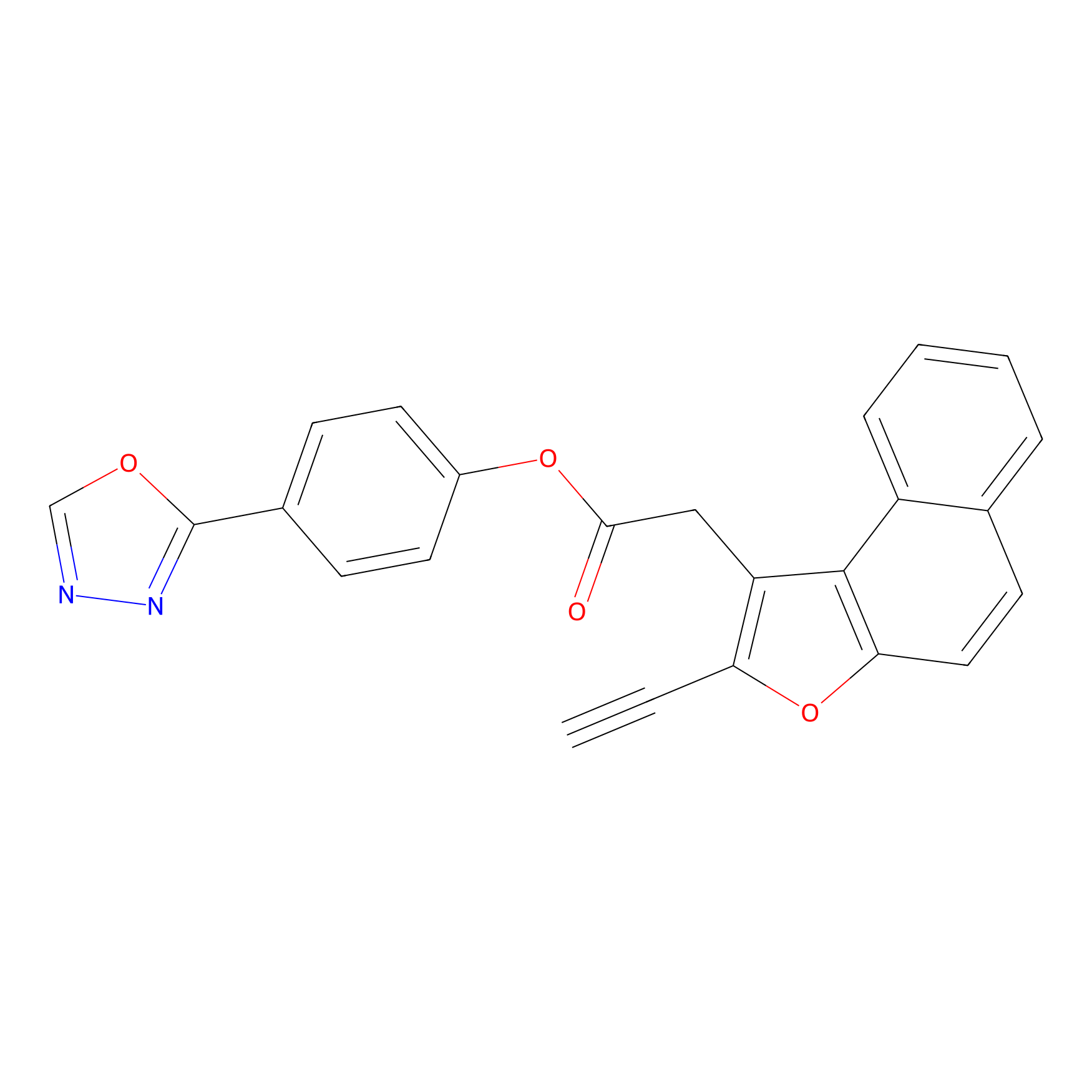 |
5.00 | LDD0043 | [2] | |
|
STPyne Probe Info |
 |
K1076(9.17); K1602(10.00); K1707(10.00); K284(4.52) | LDD0277 | [3] | |
|
ONAyne Probe Info |
 |
K738(10.00) | LDD0275 | [3] | |
|
BTD Probe Info |
 |
C957(0.43) | LDD2096 | [4] | |
|
DA-P3 Probe Info |
 |
4.23 | LDD0181 | [5] | |
|
AHL-Pu-1 Probe Info |
 |
C957(5.58); C876(2.42); C783(4.71); C187(2.78) | LDD0168 | [6] | |
|
HHS-475 Probe Info |
 |
Y1176(0.94) | LDD0265 | [7] | |
|
DBIA Probe Info |
 |
C582(1.17); C568(1.16); C1325(1.15); C1328(1.15) | LDD0078 | [8] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C171(0.00); C1717(0.00); C957(0.00); C568(0.00) | LDD0038 | [9] | |
|
IA-alkyne Probe Info |
 |
C1585(0.00); C582(0.00); C1616(0.00); C171(0.00) | LDD0036 | [9] | |
|
Lodoacetamide azide Probe Info |
 |
C1616(0.00); C171(0.00); C1717(0.00); C957(0.00) | LDD0037 | [9] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [10] | |
|
NAIA_4 Probe Info |
 |
C187(0.00); C1585(0.00); C1717(0.00) | LDD2226 | [11] | |
|
NAIA_5 Probe Info |
 |
C568(0.00); C876(0.00); C171(0.00); C582(0.00) | LDD2224 | [11] | |
|
TFBX Probe Info |
 |
C582(0.00); C1585(0.00) | LDD0027 | [10] | |
|
WYneN Probe Info |
 |
C582(0.00); C876(0.00) | LDD0021 | [12] | |
|
WYneO Probe Info |
 |
C1717(0.00); C582(0.00) | LDD0022 | [12] | |
|
Compound 10 Probe Info |
 |
C1585(0.00); C171(0.00); C1717(0.00); C1872(0.00) | LDD2216 | [13] | |
|
Compound 11 Probe Info |
 |
C1585(0.00); C171(0.00); C1872(0.00); C568(0.00) | LDD2213 | [13] | |
|
ENE Probe Info |
 |
N.A. | LDD0006 | [12] | |
|
IPM Probe Info |
 |
C1585(0.00); C171(0.00) | LDD0005 | [12] | |
|
VSF Probe Info |
 |
C1717(0.00); C770(0.00); C582(0.00); C783(0.00) | LDD0007 | [12] | |
|
Phosphinate-6 Probe Info |
 |
C582(0.00); C783(0.00); C957(0.00) | LDD0018 | [14] | |
|
Acrolein Probe Info |
 |
C1585(0.00); C582(0.00); C957(0.00); H1334(0.00) | LDD0217 | [15] | |
|
Crotonaldehyde Probe Info |
 |
C582(0.00); C1585(0.00) | LDD0219 | [15] | |
|
Methacrolein Probe Info |
 |
C582(0.00); C876(0.00); C1585(0.00) | LDD0218 | [15] | |
|
W1 Probe Info |
 |
C171(0.00); C582(0.00); C1585(0.00) | LDD0236 | [16] | |
|
AOyne Probe Info |
 |
12.50 | LDD0443 | [17] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C107 Probe Info |
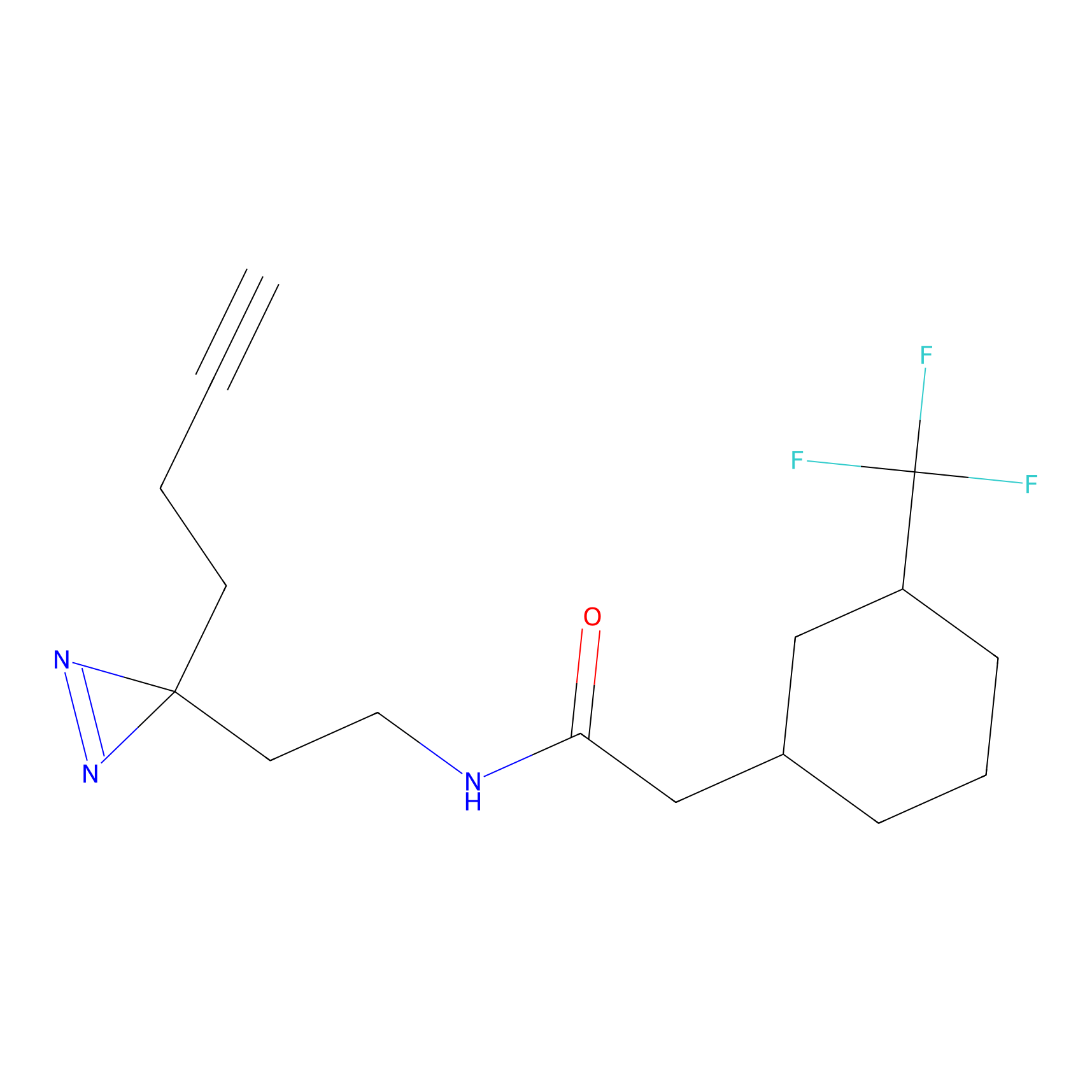 |
5.50 | LDD1794 | [18] | |
|
C219 Probe Info |
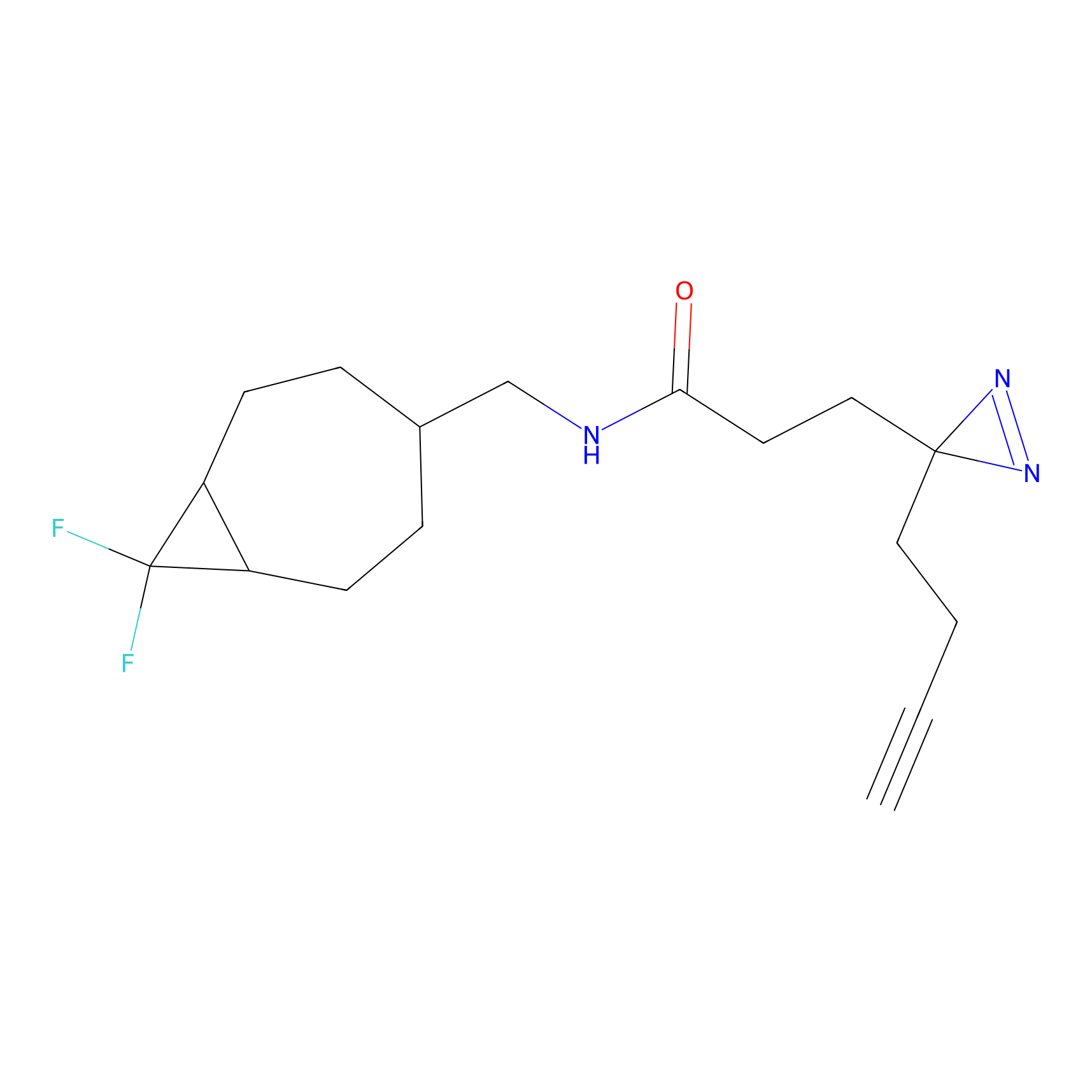 |
5.28 | LDD1893 | [18] | |
|
C383 Probe Info |
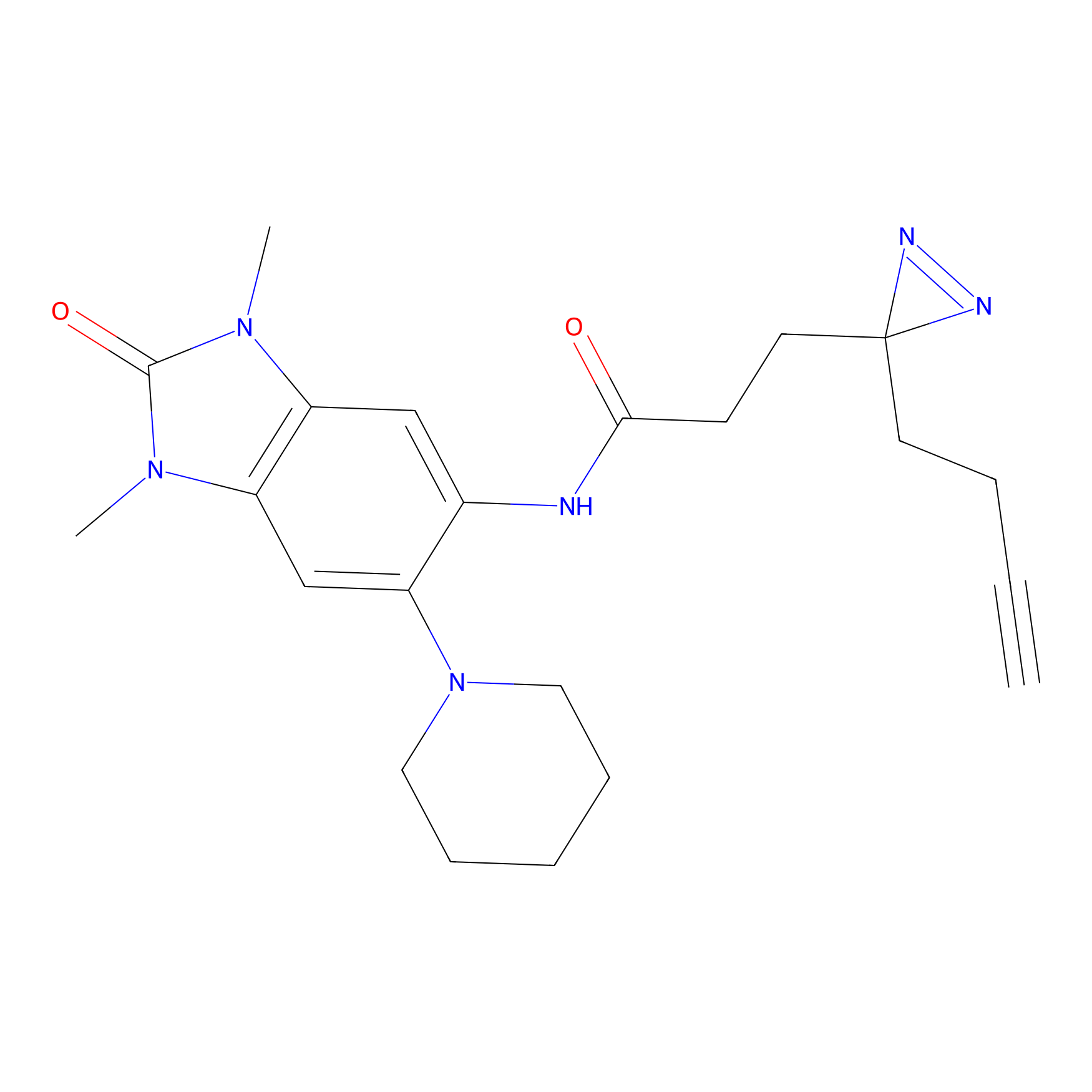 |
11.08 | LDD2042 | [18] | |
|
C388 Probe Info |
 |
35.75 | LDD2047 | [18] | |
|
C407 Probe Info |
 |
11.79 | LDD2064 | [18] | |
|
FFF probe11 Probe Info |
 |
20.00 | LDD0472 | [19] | |
|
FFF probe13 Probe Info |
 |
20.00 | LDD0476 | [19] | |
|
FFF probe14 Probe Info |
 |
20.00 | LDD0477 | [19] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C171(0.99) | LDD2117 | [4] |
| LDCM0025 | 4SU-RNA | HEK-293T | C957(5.58); C876(2.42); C783(4.71); C187(2.78) | LDD0168 | [6] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C957(3.50); C876(2.53); C783(3.25); C770(2.00) | LDD0169 | [6] |
| LDCM0214 | AC1 | HCT 116 | C770(0.84); C783(0.84); C1585(0.96); C1867(0.87) | LDD0531 | [8] |
| LDCM0215 | AC10 | HCT 116 | C770(1.05); C783(1.05); C1585(1.09); C957(1.06) | LDD0532 | [8] |
| LDCM0216 | AC100 | HCT 116 | C770(1.02); C783(1.02); C1585(1.00); C957(1.05) | LDD0533 | [8] |
| LDCM0217 | AC101 | HCT 116 | C770(1.18); C783(1.18); C1585(1.08); C957(1.12) | LDD0534 | [8] |
| LDCM0218 | AC102 | HCT 116 | C770(1.29); C783(1.29); C1585(1.12); C957(0.97) | LDD0535 | [8] |
| LDCM0219 | AC103 | HCT 116 | C770(1.54); C783(1.54); C1585(1.31); C957(1.40) | LDD0536 | [8] |
| LDCM0220 | AC104 | HCT 116 | C770(1.42); C783(1.42); C1585(1.16); C957(1.23) | LDD0537 | [8] |
| LDCM0221 | AC105 | HCT 116 | C770(1.38); C783(1.38); C1585(1.39); C957(1.13) | LDD0538 | [8] |
| LDCM0222 | AC106 | HCT 116 | C770(1.41); C783(1.41); C1585(1.14); C957(1.13) | LDD0539 | [8] |
| LDCM0223 | AC107 | HCT 116 | C770(1.05); C783(1.36); C1585(1.30); C957(1.19) | LDD0540 | [8] |
| LDCM0224 | AC108 | HCT 116 | C770(0.89); C783(0.97); C1585(1.06); C957(0.87) | LDD0541 | [8] |
| LDCM0225 | AC109 | HCT 116 | C770(0.94); C783(1.00); C1585(1.04); C957(0.95) | LDD0542 | [8] |
| LDCM0226 | AC11 | HCT 116 | C770(1.20); C783(1.20); C1585(1.13); C957(1.07) | LDD0543 | [8] |
| LDCM0227 | AC110 | HCT 116 | C770(1.09); C783(1.13); C1585(1.09); C957(1.13) | LDD0544 | [8] |
| LDCM0228 | AC111 | HCT 116 | C770(1.02); C783(1.12); C1585(1.04); C957(1.03) | LDD0545 | [8] |
| LDCM0229 | AC112 | HCT 116 | C770(1.23); C783(1.46); C1585(1.12); C957(1.16) | LDD0546 | [8] |
| LDCM0230 | AC113 | HCT 116 | C770(1.00); C783(1.00); C1585(1.00); C582(0.96) | LDD0547 | [8] |
| LDCM0231 | AC114 | HCT 116 | C770(1.23); C783(1.23); C1585(1.07); C582(1.18) | LDD0548 | [8] |
| LDCM0232 | AC115 | HCT 116 | C770(1.51); C783(1.51); C1585(1.13); C582(1.73) | LDD0549 | [8] |
| LDCM0233 | AC116 | HCT 116 | C770(1.40); C783(1.40); C1585(1.21); C582(1.30) | LDD0550 | [8] |
| LDCM0234 | AC117 | HCT 116 | C770(1.12); C783(1.12); C1585(1.06); C582(1.16) | LDD0551 | [8] |
| LDCM0235 | AC118 | HCT 116 | C770(1.03); C783(1.03); C1585(1.05); C582(1.31) | LDD0552 | [8] |
| LDCM0236 | AC119 | HCT 116 | C770(1.29); C783(1.29); C1585(1.04); C582(1.24) | LDD0553 | [8] |
| LDCM0237 | AC12 | HCT 116 | C770(0.96); C783(0.96); C1585(1.08); C957(0.75) | LDD0554 | [8] |
| LDCM0238 | AC120 | HCT 116 | C770(1.26); C783(1.26); C1585(0.96); C582(1.18) | LDD0555 | [8] |
| LDCM0239 | AC121 | HCT 116 | C770(1.03); C783(1.03); C1585(0.99); C582(1.38) | LDD0556 | [8] |
| LDCM0240 | AC122 | HCT 116 | C770(1.09); C783(1.09); C1585(0.95); C582(1.45) | LDD0557 | [8] |
| LDCM0241 | AC123 | HCT 116 | C770(1.20); C783(1.20); C1585(0.88); C582(1.14) | LDD0558 | [8] |
| LDCM0242 | AC124 | HCT 116 | C770(1.15); C783(1.15); C1585(0.95); C582(1.25) | LDD0559 | [8] |
| LDCM0243 | AC125 | HCT 116 | C770(1.03); C783(1.03); C1585(0.95); C582(1.20) | LDD0560 | [8] |
| LDCM0244 | AC126 | HCT 116 | C770(1.41); C783(1.41); C1585(1.26); C582(1.74) | LDD0561 | [8] |
| LDCM0245 | AC127 | HCT 116 | C770(1.47); C783(1.47); C1585(1.18); C582(1.35) | LDD0562 | [8] |
| LDCM0246 | AC128 | HCT 116 | C770(0.81); C783(0.81); C1585(0.63); C582(0.17) | LDD0563 | [8] |
| LDCM0247 | AC129 | HCT 116 | C770(0.61); C783(0.61); C1585(0.57); C582(0.17) | LDD0564 | [8] |
| LDCM0249 | AC130 | HCT 116 | C770(0.65); C783(0.65); C1585(0.59); C582(0.19) | LDD0566 | [8] |
| LDCM0250 | AC131 | HCT 116 | C770(0.76); C783(0.76); C1585(0.67); C582(0.20) | LDD0567 | [8] |
| LDCM0251 | AC132 | HCT 116 | C770(0.48); C783(0.48); C1585(0.78); C582(0.22) | LDD0568 | [8] |
| LDCM0252 | AC133 | HCT 116 | C770(0.58); C783(0.58); C1585(0.69); C582(0.22) | LDD0569 | [8] |
| LDCM0253 | AC134 | HCT 116 | C770(0.80); C783(0.80); C1585(0.85); C582(0.23) | LDD0570 | [8] |
| LDCM0254 | AC135 | HCT 116 | C770(0.56); C783(0.56); C1585(0.74); C582(0.27) | LDD0571 | [8] |
| LDCM0255 | AC136 | HCT 116 | C770(0.62); C783(0.62); C1585(0.72); C582(0.25) | LDD0572 | [8] |
| LDCM0256 | AC137 | HCT 116 | C770(0.60); C783(0.60); C1585(0.70); C582(0.21) | LDD0573 | [8] |
| LDCM0257 | AC138 | HCT 116 | C770(0.77); C783(0.77); C1585(0.84); C582(0.23) | LDD0574 | [8] |
| LDCM0258 | AC139 | HCT 116 | C770(0.61); C783(0.61); C1585(0.80); C582(0.23) | LDD0575 | [8] |
| LDCM0259 | AC14 | HCT 116 | C770(0.96); C783(0.96); C1585(1.11); C957(0.83) | LDD0576 | [8] |
| LDCM0260 | AC140 | HCT 116 | C770(0.71); C783(0.71); C1585(1.01); C582(0.23) | LDD0577 | [8] |
| LDCM0261 | AC141 | HCT 116 | C770(0.70); C783(0.70); C1585(0.85); C582(0.24) | LDD0578 | [8] |
| LDCM0262 | AC142 | HCT 116 | C770(0.53); C783(0.53); C1585(0.82); C582(0.22) | LDD0579 | [8] |
| LDCM0263 | AC143 | HCT 116 | C770(1.11); C783(1.11); C1585(1.30); C1867(0.87) | LDD0580 | [8] |
| LDCM0264 | AC144 | HCT 116 | C1325(0.83); C1328(0.83); C876(0.97); C1585(0.99) | LDD0581 | [8] |
| LDCM0265 | AC145 | HCT 116 | C1325(0.71); C1328(0.71); C1717(0.93); C582(0.93) | LDD0582 | [8] |
| LDCM0266 | AC146 | HCT 116 | C1325(0.80); C1328(0.80); C876(0.84); C1717(1.03) | LDD0583 | [8] |
| LDCM0267 | AC147 | HCT 116 | C1325(0.72); C1328(0.72); C1717(0.82); C1585(1.05) | LDD0584 | [8] |
| LDCM0268 | AC148 | HCT 116 | C1717(0.80); C1325(0.91); C1328(0.91); C876(0.97) | LDD0585 | [8] |
| LDCM0269 | AC149 | HCT 116 | C876(0.83); C1717(0.94); C1325(1.01); C1328(1.01) | LDD0586 | [8] |
| LDCM0270 | AC15 | HCT 116 | C1717(0.71); C876(0.81); C957(0.81); C770(1.02) | LDD0587 | [8] |
| LDCM0271 | AC150 | HCT 116 | C1325(0.82); C1328(0.82); C876(0.95); C1717(0.98) | LDD0588 | [8] |
| LDCM0272 | AC151 | HCT 116 | C1717(0.81); C1325(0.81); C1328(0.81); C876(0.85) | LDD0589 | [8] |
| LDCM0273 | AC152 | HCT 116 | C1325(0.77); C1328(0.77); C876(0.87); C1717(0.98) | LDD0590 | [8] |
| LDCM0274 | AC153 | HCT 116 | C876(0.66); C1325(0.73); C1328(0.73); C1717(1.29) | LDD0591 | [8] |
| LDCM0621 | AC154 | HCT 116 | C770(1.30); C783(1.30); C1585(1.13); C1867(2.55) | LDD2158 | [8] |
| LDCM0622 | AC155 | HCT 116 | C770(1.65); C783(1.65); C1585(1.16); C1867(1.97) | LDD2159 | [8] |
| LDCM0623 | AC156 | HCT 116 | C770(1.12); C783(1.12); C1585(1.22); C1867(1.44) | LDD2160 | [8] |
| LDCM0624 | AC157 | HCT 116 | C770(1.43); C783(1.43); C1585(1.10); C1867(1.68) | LDD2161 | [8] |
| LDCM0276 | AC17 | HCT 116 | C582(0.52); C957(0.92); C1585(1.02); C876(1.05) | LDD0593 | [8] |
| LDCM0277 | AC18 | HCT 116 | C582(0.56); C876(0.84); C1585(0.90); C568(0.90) | LDD0594 | [8] |
| LDCM0278 | AC19 | HCT 116 | C582(0.54); C876(0.96); C1585(0.98); C957(1.06) | LDD0595 | [8] |
| LDCM0279 | AC2 | HCT 116 | C770(0.80); C783(0.80); C957(0.82); C876(0.83) | LDD0596 | [8] |
| LDCM0280 | AC20 | HCT 116 | C582(0.52); C1585(1.05); C171(1.07); C876(1.16) | LDD0597 | [8] |
| LDCM0281 | AC21 | HCT 116 | C582(0.48); C1585(0.97); C876(1.12); C171(1.25) | LDD0598 | [8] |
| LDCM0282 | AC22 | HCT 116 | C582(0.46); C568(0.95); C171(1.02); C1585(1.03) | LDD0599 | [8] |
| LDCM0283 | AC23 | HCT 116 | C582(0.45); C876(0.83); C1585(0.93); C171(1.00) | LDD0600 | [8] |
| LDCM0284 | AC24 | HCT 116 | C582(0.36); C876(0.96); C957(1.00); C171(1.03) | LDD0601 | [8] |
| LDCM0285 | AC25 | HCT 116 | C568(0.86); C957(0.89); C876(1.07); C1585(1.08) | LDD0602 | [8] |
| LDCM0286 | AC26 | HCT 116 | C568(0.89); C957(1.08); C1325(1.08); C1328(1.08) | LDD0603 | [8] |
| LDCM0287 | AC27 | HCT 116 | C568(0.97); C957(1.10); C1585(1.20); C876(1.23) | LDD0604 | [8] |
| LDCM0288 | AC28 | HCT 116 | C568(0.90); C957(0.97); C1325(1.21); C1328(1.21) | LDD0605 | [8] |
| LDCM0289 | AC29 | HCT 116 | C568(1.05); C957(1.23); C1585(1.26); C876(1.35) | LDD0606 | [8] |
| LDCM0290 | AC3 | HCT 116 | C957(0.74); C770(0.75); C783(0.75); C876(0.78) | LDD0607 | [8] |
| LDCM0291 | AC30 | HCT 116 | C568(1.08); C957(1.09); C1325(1.23); C1328(1.23) | LDD0608 | [8] |
| LDCM0292 | AC31 | HCT 116 | C568(0.94); C957(1.03); C171(1.24); C770(1.27) | LDD0609 | [8] |
| LDCM0293 | AC32 | HCT 116 | C568(1.12); C957(1.19); C1585(1.32); C1325(1.33) | LDD0610 | [8] |
| LDCM0294 | AC33 | HCT 116 | C568(1.04); C957(1.19); C876(1.35); C1585(1.38) | LDD0611 | [8] |
| LDCM0295 | AC34 | HCT 116 | C568(0.92); C876(1.18); C1585(1.21); C957(1.38) | LDD0612 | [8] |
| LDCM0296 | AC35 | HCT 116 | C876(0.71); C770(0.84); C783(0.84); C171(0.85) | LDD0613 | [8] |
| LDCM0297 | AC36 | HCT 116 | C770(0.76); C783(0.76); C876(0.76); C1585(0.83) | LDD0614 | [8] |
| LDCM0298 | AC37 | HCT 116 | C770(0.78); C783(0.78); C171(0.82); C1585(0.82) | LDD0615 | [8] |
| LDCM0299 | AC38 | HCT 116 | C876(0.78); C770(0.86); C783(0.86); C1585(1.01) | LDD0616 | [8] |
| LDCM0300 | AC39 | HCT 116 | C770(0.90); C783(0.90); C1585(1.04); C876(1.06) | LDD0617 | [8] |
| LDCM0301 | AC4 | HCT 116 | C876(0.64); C582(0.82); C770(0.82); C783(0.82) | LDD0618 | [8] |
| LDCM0302 | AC40 | HCT 116 | C1585(0.94); C770(1.01); C783(1.01); C1325(1.14) | LDD0619 | [8] |
| LDCM0303 | AC41 | HCT 116 | C876(0.77); C1585(0.88); C770(0.90); C783(0.90) | LDD0620 | [8] |
| LDCM0304 | AC42 | HCT 116 | C876(0.89); C770(0.92); C783(0.92); C1585(1.10) | LDD0621 | [8] |
| LDCM0305 | AC43 | HCT 116 | C876(0.88); C1585(1.01); C770(1.05); C783(1.05) | LDD0622 | [8] |
| LDCM0306 | AC44 | HCT 116 | C770(0.84); C783(0.84); C1585(0.89); C876(0.93) | LDD0623 | [8] |
| LDCM0307 | AC45 | HCT 116 | C1585(0.95); C770(1.11); C783(1.11); C568(1.12) | LDD0624 | [8] |
| LDCM0308 | AC46 | HCT 116 | C171(1.06); C770(1.11); C783(1.11); C1325(1.24) | LDD0625 | [8] |
| LDCM0309 | AC47 | HCT 116 | C171(1.08); C1585(1.14); C770(1.19); C783(1.19) | LDD0626 | [8] |
| LDCM0310 | AC48 | HCT 116 | C770(1.04); C783(1.04); C171(1.07); C1325(1.13) | LDD0627 | [8] |
| LDCM0311 | AC49 | HCT 116 | C957(1.07); C171(1.33); C1325(1.39); C1328(1.39) | LDD0628 | [8] |
| LDCM0312 | AC5 | HCT 116 | C876(0.49); C770(0.68); C783(0.68); C957(0.76) | LDD0629 | [8] |
| LDCM0313 | AC50 | HCT 116 | C957(1.13); C1325(1.17); C1328(1.17); C171(1.35) | LDD0630 | [8] |
| LDCM0314 | AC51 | HCT 116 | C171(0.82); C770(0.93); C783(0.93); C1325(1.20) | LDD0631 | [8] |
| LDCM0315 | AC52 | HCT 116 | C171(0.88); C770(1.14); C783(1.14); C957(1.15) | LDD0632 | [8] |
| LDCM0316 | AC53 | HCT 116 | C171(1.28); C1585(1.29); C876(1.31); C770(1.42) | LDD0633 | [8] |
| LDCM0317 | AC54 | HCT 116 | C171(1.10); C1585(1.32); C770(1.43); C783(1.43) | LDD0634 | [8] |
| LDCM0318 | AC55 | HCT 116 | C1585(1.20); C171(1.41); C876(1.54); C957(1.72) | LDD0635 | [8] |
| LDCM0319 | AC56 | HCT 116 | C1585(1.30); C957(1.30); C171(1.71); C1325(2.15) | LDD0636 | [8] |
| LDCM0320 | AC57 | HCT 116 | C568(0.55); C1585(0.88); C770(1.11); C783(1.11) | LDD0637 | [8] |
| LDCM0321 | AC58 | HCT 116 | C568(0.81); C1585(0.99); C957(1.12); C770(1.27) | LDD0638 | [8] |
| LDCM0322 | AC59 | HCT 116 | C568(0.83); C1585(1.03); C770(1.28); C783(1.28) | LDD0639 | [8] |
| LDCM0323 | AC6 | HCT 116 | C1717(0.88); C876(0.97); C957(1.02); C1585(1.05) | LDD0640 | [8] |
| LDCM0324 | AC60 | HCT 116 | C568(0.66); C1585(0.98); C770(1.15); C783(1.15) | LDD0641 | [8] |
| LDCM0325 | AC61 | HCT 116 | C568(0.68); C770(1.03); C783(1.03); C957(1.06) | LDD0642 | [8] |
| LDCM0326 | AC62 | HCT 116 | C568(0.53); C1585(0.83); C957(1.14); C171(1.24) | LDD0643 | [8] |
| LDCM0327 | AC63 | HCT 116 | C568(0.50); C1585(0.84); C876(1.21); C770(1.32) | LDD0644 | [8] |
| LDCM0328 | AC64 | HCT 116 | C568(0.54); C1585(0.76); C770(1.16); C783(1.16) | LDD0645 | [8] |
| LDCM0329 | AC65 | HCT 116 | C568(0.65); C770(0.79); C783(0.79); C876(0.80) | LDD0646 | [8] |
| LDCM0330 | AC66 | HCT 116 | C568(0.69); C876(0.77); C770(0.84); C783(0.84) | LDD0647 | [8] |
| LDCM0331 | AC67 | HCT 116 | C568(0.63); C1585(0.86); C770(0.91); C783(0.91) | LDD0648 | [8] |
| LDCM0332 | AC68 | HCT 116 | C957(0.79); C1328(1.03); C582(1.05); C568(1.09) | LDD0649 | [8] |
| LDCM0333 | AC69 | HCT 116 | C957(1.00); C568(1.01); C171(1.08); C876(1.23) | LDD0650 | [8] |
| LDCM0334 | AC7 | HCT 116 | C876(0.85); C1717(0.88); C568(0.94); C770(0.94) | LDD0651 | [8] |
| LDCM0335 | AC70 | HCT 116 | C171(1.12); C957(1.13); C1328(1.15); C568(1.15) | LDD0652 | [8] |
| LDCM0336 | AC71 | HCT 116 | C568(0.78); C957(0.94); C582(1.01); C1328(1.09) | LDD0653 | [8] |
| LDCM0337 | AC72 | HCT 116 | C568(1.06); C957(1.17); C171(1.22); C1328(1.26) | LDD0654 | [8] |
| LDCM0338 | AC73 | HCT 116 | C568(0.93); C957(1.19); C1585(1.74); C171(1.78) | LDD0655 | [8] |
| LDCM0339 | AC74 | HCT 116 | C568(0.83); C1585(1.27); C957(1.35); C876(1.49) | LDD0656 | [8] |
| LDCM0340 | AC75 | HCT 116 | C568(0.96); C957(1.34); C1585(1.82); C171(1.88) | LDD0657 | [8] |
| LDCM0341 | AC76 | HCT 116 | C568(0.71); C957(0.90); C1328(1.28); C171(1.32) | LDD0658 | [8] |
| LDCM0342 | AC77 | HCT 116 | C957(1.09); C568(1.09); C171(1.33); C1328(1.49) | LDD0659 | [8] |
| LDCM0343 | AC78 | HCT 116 | C568(0.79); C171(1.06); C957(1.14); C1328(1.19) | LDD0660 | [8] |
| LDCM0344 | AC79 | HCT 116 | C957(0.97); C568(1.05); C876(1.15); C582(1.18) | LDD0661 | [8] |
| LDCM0345 | AC8 | HCT 116 | C1717(0.76); C876(0.81); C957(0.93); C568(0.96) | LDD0662 | [8] |
| LDCM0346 | AC80 | HCT 116 | C568(0.83); C957(0.83); C1585(1.17); C171(1.21) | LDD0663 | [8] |
| LDCM0347 | AC81 | HCT 116 | C568(0.89); C957(0.99); C171(1.07); C876(1.16) | LDD0664 | [8] |
| LDCM0348 | AC82 | HCT 116 | C568(1.07); C957(1.11); C1585(1.24); C1328(1.94) | LDD0665 | [8] |
| LDCM0349 | AC83 | HCT 116 | C582(1.41); C1585(1.59); C568(1.64); C1325(1.65) | LDD0666 | [8] |
| LDCM0350 | AC84 | HCT 116 | C1325(1.29); C1328(1.29); C582(1.36); C957(1.42) | LDD0667 | [8] |
| LDCM0351 | AC85 | HCT 116 | C1325(1.03); C1328(1.03); C568(1.17); C957(1.28) | LDD0668 | [8] |
| LDCM0352 | AC86 | HCT 116 | C568(1.13); C1585(1.15); C1325(1.33); C1328(1.33) | LDD0669 | [8] |
| LDCM0353 | AC87 | HCT 116 | C582(0.98); C1325(1.00); C1328(1.00); C568(1.01) | LDD0670 | [8] |
| LDCM0354 | AC88 | HCT 116 | C568(1.20); C1325(1.27); C1328(1.27); C171(1.27) | LDD0671 | [8] |
| LDCM0355 | AC89 | HCT 116 | C1585(1.23); C171(1.35); C957(1.39); C568(1.56) | LDD0672 | [8] |
| LDCM0357 | AC90 | HCT 116 | C957(0.81); C568(0.97); C171(1.04); C582(1.09) | LDD0674 | [8] |
| LDCM0358 | AC91 | HCT 116 | C1325(1.64); C1328(1.64); C568(1.67); C876(1.68) | LDD0675 | [8] |
| LDCM0359 | AC92 | HCT 116 | C1585(1.55); C957(1.57); C1325(1.60); C1328(1.60) | LDD0676 | [8] |
| LDCM0360 | AC93 | HCT 116 | C568(1.18); C957(1.22); C876(1.26); C1585(1.30) | LDD0677 | [8] |
| LDCM0361 | AC94 | HCT 116 | C1325(1.08); C1328(1.08); C1585(1.14); C568(1.35) | LDD0678 | [8] |
| LDCM0362 | AC95 | HCT 116 | C1585(1.16); C568(1.19); C876(1.24); C1325(1.26) | LDD0679 | [8] |
| LDCM0363 | AC96 | HCT 116 | C1325(1.33); C1328(1.33); C1585(1.36); C957(1.40) | LDD0680 | [8] |
| LDCM0364 | AC97 | HCT 116 | C1585(1.44); C568(1.55); C957(1.63); C876(1.72) | LDD0681 | [8] |
| LDCM0365 | AC98 | HCT 116 | C568(1.11); C1325(1.22); C957(1.52); C1585(1.60) | LDD0682 | [8] |
| LDCM0366 | AC99 | HCT 116 | C1585(0.87); C957(0.93); C568(0.98); C1717(1.03) | LDD0683 | [8] |
| LDCM0248 | AKOS034007472 | HCT 116 | C770(0.84); C783(0.84); C1585(1.10); C957(0.85) | LDD0565 | [8] |
| LDCM0356 | AKOS034007680 | HCT 116 | C957(0.98); C770(1.00); C783(1.00); C568(1.03) | LDD0673 | [8] |
| LDCM0275 | AKOS034007705 | HCT 116 | C1717(0.79); C876(0.85); C957(1.25); C1585(1.31) | LDD0592 | [8] |
| LDCM0156 | Aniline | NCI-H1299 | 11.60 | LDD0403 | [1] |
| LDCM0020 | ARS-1620 | HCC44 | C582(1.17); C568(1.16); C1325(1.15); C1328(1.15) | LDD0078 | [8] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [15] |
| LDCM0632 | CL-Sc | Hep-G2 | C1717(3.33); C171(1.46); C1717(1.41); C957(0.78) | LDD2227 | [11] |
| LDCM0367 | CL1 | HCT 116 | C876(0.86); C1867(0.94); C582(1.00); C171(1.08) | LDD0684 | [8] |
| LDCM0368 | CL10 | HCT 116 | C957(0.89); C1867(1.02); C1585(1.06); C582(1.15) | LDD0685 | [8] |
| LDCM0369 | CL100 | HCT 116 | C876(0.72); C582(0.85); C957(0.88); C171(0.91) | LDD0686 | [8] |
| LDCM0370 | CL101 | HCT 116 | C1717(0.73); C876(0.78); C957(0.89); C770(1.15) | LDD0687 | [8] |
| LDCM0371 | CL102 | HCT 116 | C876(0.68); C1717(0.86); C568(0.94); C171(1.07) | LDD0688 | [8] |
| LDCM0372 | CL103 | HCT 116 | C876(0.74); C1717(0.76); C957(0.95); C1585(0.99) | LDD0689 | [8] |
| LDCM0373 | CL104 | HCT 116 | C1717(0.79); C876(0.89); C957(0.97); C171(1.03) | LDD0690 | [8] |
| LDCM0374 | CL105 | HCT 116 | C582(0.59); C1585(0.89); C876(1.00); C568(1.05) | LDD0691 | [8] |
| LDCM0375 | CL106 | HCT 116 | C582(0.50); C1585(0.85); C568(1.22); C876(1.25) | LDD0692 | [8] |
| LDCM0376 | CL107 | HCT 116 | C582(0.44); C1585(0.88); C957(0.90); C876(1.07) | LDD0693 | [8] |
| LDCM0377 | CL108 | HCT 116 | C582(0.40); C1585(0.95); C957(1.03); C876(1.21) | LDD0694 | [8] |
| LDCM0378 | CL109 | HCT 116 | C582(0.92); C1585(1.05); C876(1.10); C957(1.29) | LDD0695 | [8] |
| LDCM0379 | CL11 | HCT 116 | C1867(0.81); C582(0.91); C568(0.95); C876(1.24) | LDD0696 | [8] |
| LDCM0380 | CL110 | HCT 116 | C582(0.47); C1585(0.97); C876(1.15); C957(1.27) | LDD0697 | [8] |
| LDCM0381 | CL111 | HCT 116 | C582(0.45); C1585(0.85); C876(0.95); C568(1.15) | LDD0698 | [8] |
| LDCM0382 | CL112 | HCT 116 | C568(0.77); C876(0.94); C957(0.95); C171(1.07) | LDD0699 | [8] |
| LDCM0383 | CL113 | HCT 116 | C568(1.01); C876(1.23); C1585(1.23); C957(1.24) | LDD0700 | [8] |
| LDCM0384 | CL114 | HCT 116 | C568(1.03); C876(1.13); C1325(1.19); C1328(1.19) | LDD0701 | [8] |
| LDCM0385 | CL115 | HCT 116 | C568(1.11); C1325(1.19); C1328(1.19); C876(1.22) | LDD0702 | [8] |
| LDCM0386 | CL116 | HCT 116 | C568(0.99); C876(1.06); C1325(1.11); C1328(1.11) | LDD0703 | [8] |
| LDCM0387 | CL117 | HCT 116 | C568(1.12); C1325(1.35); C1328(1.35); C1585(1.48) | LDD0704 | [8] |
| LDCM0388 | CL118 | HCT 116 | C770(0.93); C783(0.93); C876(0.95); C1585(1.00) | LDD0705 | [8] |
| LDCM0389 | CL119 | HCT 116 | C1585(1.09); C876(1.10); C770(1.10); C783(1.10) | LDD0706 | [8] |
| LDCM0390 | CL12 | HCT 116 | C957(0.82); C582(0.84); C568(0.93); C1585(0.97) | LDD0707 | [8] |
| LDCM0391 | CL120 | HCT 116 | C770(0.96); C783(0.96); C171(0.96); C1585(1.04) | LDD0708 | [8] |
| LDCM0392 | CL121 | HCT 116 | C171(0.88); C1585(1.08); C876(1.10); C1325(1.66) | LDD0709 | [8] |
| LDCM0393 | CL122 | HCT 116 | C171(1.08); C770(1.16); C783(1.16); C1585(1.30) | LDD0710 | [8] |
| LDCM0394 | CL123 | HCT 116 | C876(1.19); C1585(1.43); C171(1.49); C957(1.74) | LDD0711 | [8] |
| LDCM0395 | CL124 | HCT 116 | C171(1.28); C957(1.30); C1585(1.37); C770(1.62) | LDD0712 | [8] |
| LDCM0396 | CL125 | HCT 116 | C568(0.55); C171(0.81); C876(0.86); C770(0.87) | LDD0713 | [8] |
| LDCM0397 | CL126 | HCT 116 | C568(0.70); C876(0.89); C770(0.95); C783(0.95) | LDD0714 | [8] |
| LDCM0398 | CL127 | HCT 116 | C568(0.70); C770(0.86); C783(0.86); C171(0.96) | LDD0715 | [8] |
| LDCM0399 | CL128 | HCT 116 | C568(0.59); C957(1.00); C1585(1.08); C770(1.23) | LDD0716 | [8] |
| LDCM0400 | CL13 | HCT 116 | C1585(0.85); C568(0.87); C1867(1.01); C582(1.03) | LDD0717 | [8] |
| LDCM0401 | CL14 | HCT 116 | C1585(0.93); C957(1.09); C568(1.10); C876(1.13) | LDD0718 | [8] |
| LDCM0402 | CL15 | HCT 116 | C1867(0.90); C568(0.96); C582(1.02); C1585(1.25) | LDD0719 | [8] |
| LDCM0403 | CL16 | HCT 116 | C582(0.86); C171(0.95); C1717(1.02); C957(1.03) | LDD0720 | [8] |
| LDCM0404 | CL17 | HCT 116 | C1717(0.82); C568(0.82); C1325(1.03); C1328(1.03) | LDD0721 | [8] |
| LDCM0405 | CL18 | HCT 116 | C957(0.95); C1325(0.98); C1328(0.98); C568(1.04) | LDD0722 | [8] |
| LDCM0406 | CL19 | HCT 116 | C1325(0.83); C1328(0.83); C1717(0.90); C957(1.00) | LDD0723 | [8] |
| LDCM0407 | CL2 | HCT 116 | C568(0.79); C876(0.89); C1867(0.92); C770(1.00) | LDD0724 | [8] |
| LDCM0408 | CL20 | HCT 116 | C1325(0.85); C1328(0.85); C957(1.04); C1717(1.05) | LDD0725 | [8] |
| LDCM0409 | CL21 | HCT 116 | C1325(0.86); C1328(0.86); C1717(0.89); C568(1.05) | LDD0726 | [8] |
| LDCM0410 | CL22 | HCT 116 | C1325(0.69); C1328(0.69); C1717(0.90); C568(1.13) | LDD0727 | [8] |
| LDCM0411 | CL23 | HCT 116 | C568(0.67); C957(0.86); C1325(0.96); C1328(0.96) | LDD0728 | [8] |
| LDCM0412 | CL24 | HCT 116 | C957(0.86); C1325(0.89); C1328(0.89); C171(1.02) | LDD0729 | [8] |
| LDCM0413 | CL25 | HCT 116 | C770(3.14); C783(3.14); C1585(1.73); C957(0.93) | LDD0730 | [8] |
| LDCM0414 | CL26 | HCT 116 | C770(1.66); C783(1.66); C1585(1.14); C957(1.05) | LDD0731 | [8] |
| LDCM0415 | CL27 | HCT 116 | C770(1.51); C783(1.51); C1585(1.20); C957(0.90) | LDD0732 | [8] |
| LDCM0416 | CL28 | HCT 116 | C770(2.04); C783(2.04); C1585(1.24); C957(1.23) | LDD0733 | [8] |
| LDCM0417 | CL29 | HCT 116 | C770(1.73); C783(1.73); C1585(1.20); C957(1.07) | LDD0734 | [8] |
| LDCM0418 | CL3 | HCT 116 | C770(1.11); C783(1.11); C1585(1.23); C1867(1.02) | LDD0735 | [8] |
| LDCM0419 | CL30 | HCT 116 | C770(1.45); C783(1.45); C1585(1.15); C957(0.86) | LDD0736 | [8] |
| LDCM0420 | CL31 | HCT 116 | C770(1.31); C783(1.31); C1585(1.10); C582(0.83) | LDD0737 | [8] |
| LDCM0421 | CL32 | HCT 116 | C770(1.32); C783(1.32); C1585(0.99); C582(0.86) | LDD0738 | [8] |
| LDCM0422 | CL33 | HCT 116 | C770(4.64); C783(4.64); C1585(2.78); C582(0.88) | LDD0739 | [8] |
| LDCM0423 | CL34 | HCT 116 | C770(1.78); C783(1.78); C1585(1.45); C582(0.93) | LDD0740 | [8] |
| LDCM0424 | CL35 | HCT 116 | C770(1.88); C783(1.88); C1585(1.08); C582(1.07) | LDD0741 | [8] |
| LDCM0425 | CL36 | HCT 116 | C770(1.73); C783(1.73); C1585(1.05); C582(0.99) | LDD0742 | [8] |
| LDCM0426 | CL37 | HCT 116 | C770(2.04); C783(2.04); C1585(1.53); C582(1.16) | LDD0743 | [8] |
| LDCM0428 | CL39 | HCT 116 | C770(2.06); C783(2.06); C1585(1.26); C582(0.97) | LDD0745 | [8] |
| LDCM0429 | CL4 | HCT 116 | C770(1.89); C783(1.89); C1585(1.06); C1867(0.92) | LDD0746 | [8] |
| LDCM0430 | CL40 | HCT 116 | C770(2.32); C783(2.32); C1585(1.44); C582(0.87) | LDD0747 | [8] |
| LDCM0431 | CL41 | HCT 116 | C770(1.61); C783(1.61); C1585(1.18); C582(0.90) | LDD0748 | [8] |
| LDCM0432 | CL42 | HCT 116 | C770(3.50); C783(3.50); C1585(1.86); C582(0.80) | LDD0749 | [8] |
| LDCM0433 | CL43 | HCT 116 | C770(2.41); C783(2.41); C1585(1.22); C582(0.89) | LDD0750 | [8] |
| LDCM0434 | CL44 | HCT 116 | C770(2.14); C783(2.14); C1585(1.49); C582(0.98) | LDD0751 | [8] |
| LDCM0435 | CL45 | HCT 116 | C770(2.79); C783(2.79); C1585(1.62); C582(0.87) | LDD0752 | [8] |
| LDCM0436 | CL46 | HCT 116 | C770(1.12); C783(1.12); C1585(1.11); C582(0.95) | LDD0753 | [8] |
| LDCM0437 | CL47 | HCT 116 | C770(0.94); C783(0.94); C1585(1.00); C582(0.95) | LDD0754 | [8] |
| LDCM0438 | CL48 | HCT 116 | C770(1.03); C783(1.03); C1585(1.09); C582(1.05) | LDD0755 | [8] |
| LDCM0439 | CL49 | HCT 116 | C770(1.47); C783(1.47); C1585(0.97); C582(0.95) | LDD0756 | [8] |
| LDCM0440 | CL5 | HCT 116 | C770(1.12); C783(1.12); C1585(0.95); C1867(0.98) | LDD0757 | [8] |
| LDCM0441 | CL50 | HCT 116 | C770(1.17); C783(1.17); C1585(1.10); C582(1.01) | LDD0758 | [8] |
| LDCM0442 | CL51 | HCT 116 | C770(1.02); C783(1.02); C1585(1.12); C582(1.09) | LDD0759 | [8] |
| LDCM0443 | CL52 | HCT 116 | C770(1.09); C783(1.09); C1585(1.23); C582(1.07) | LDD0760 | [8] |
| LDCM0444 | CL53 | HCT 116 | C770(1.26); C783(1.26); C1585(1.19); C582(1.19) | LDD0761 | [8] |
| LDCM0445 | CL54 | HCT 116 | C770(1.40); C783(1.40); C1585(1.24); C582(0.88) | LDD0762 | [8] |
| LDCM0446 | CL55 | HCT 116 | C770(1.19); C783(1.19); C1585(0.98); C582(0.67) | LDD0763 | [8] |
| LDCM0447 | CL56 | HCT 116 | C770(1.05); C783(1.05); C1585(0.99); C582(1.10) | LDD0764 | [8] |
| LDCM0448 | CL57 | HCT 116 | C770(1.21); C783(1.21); C1585(1.18); C582(0.95) | LDD0765 | [8] |
| LDCM0449 | CL58 | HCT 116 | C770(1.09); C783(1.09); C1585(1.01); C582(0.99) | LDD0766 | [8] |
| LDCM0450 | CL59 | HCT 116 | C770(1.06); C783(1.06); C1585(1.01); C582(0.85) | LDD0767 | [8] |
| LDCM0451 | CL6 | HCT 116 | C770(1.61); C783(1.61); C1585(1.07); C1867(1.18) | LDD0768 | [8] |
| LDCM0452 | CL60 | HCT 116 | C770(1.12); C783(1.12); C1585(1.07); C582(1.09) | LDD0769 | [8] |
| LDCM0453 | CL61 | HCT 116 | C770(0.96); C783(0.96); C1585(0.90); C1867(1.36) | LDD0770 | [8] |
| LDCM0454 | CL62 | HCT 116 | C770(1.09); C783(1.09); C1585(0.79); C1867(1.44) | LDD0771 | [8] |
| LDCM0455 | CL63 | HCT 116 | C770(1.18); C783(1.18); C1585(0.93); C1867(1.36) | LDD0772 | [8] |
| LDCM0456 | CL64 | HCT 116 | C770(1.63); C783(1.63); C1585(0.87); C1867(0.92) | LDD0773 | [8] |
| LDCM0457 | CL65 | HCT 116 | C770(0.93); C783(0.93); C1585(0.85); C1867(1.29) | LDD0774 | [8] |
| LDCM0458 | CL66 | HCT 116 | C770(1.52); C783(1.52); C1585(0.87); C1867(1.22) | LDD0775 | [8] |
| LDCM0459 | CL67 | HCT 116 | C770(1.34); C783(1.34); C1585(0.84); C1867(1.44) | LDD0776 | [8] |
| LDCM0460 | CL68 | HCT 116 | C770(1.35); C783(1.35); C1585(0.86); C1867(1.39) | LDD0777 | [8] |
| LDCM0461 | CL69 | HCT 116 | C770(1.52); C783(1.52); C1585(0.82); C1867(1.43) | LDD0778 | [8] |
| LDCM0462 | CL7 | HCT 116 | C770(1.32); C783(1.32); C1585(0.83); C1867(1.09) | LDD0779 | [8] |
| LDCM0463 | CL70 | HCT 116 | C770(1.20); C783(1.20); C1585(0.86); C1867(1.37) | LDD0780 | [8] |
| LDCM0464 | CL71 | HCT 116 | C770(1.47); C783(1.47); C1585(0.84); C1867(1.23) | LDD0781 | [8] |
| LDCM0465 | CL72 | HCT 116 | C770(1.17); C783(1.17); C1585(0.84); C1867(1.21) | LDD0782 | [8] |
| LDCM0466 | CL73 | HCT 116 | C770(1.32); C783(1.32); C1585(0.87); C1867(1.36) | LDD0783 | [8] |
| LDCM0467 | CL74 | HCT 116 | C770(1.41); C783(1.41); C1585(0.88); C1867(1.67) | LDD0784 | [8] |
| LDCM0469 | CL76 | HCT 116 | C770(0.90); C783(0.90); C1585(1.26); C582(1.44) | LDD0786 | [8] |
| LDCM0470 | CL77 | HCT 116 | C770(0.99); C783(0.99); C1585(1.04); C582(0.70) | LDD0787 | [8] |
| LDCM0471 | CL78 | HCT 116 | C770(1.15); C783(1.15); C1585(1.08); C582(1.62) | LDD0788 | [8] |
| LDCM0472 | CL79 | HCT 116 | C770(0.92); C783(0.92); C1585(1.19); C582(1.44) | LDD0789 | [8] |
| LDCM0473 | CL8 | HCT 116 | C770(8.44); C783(8.44); C1585(4.68); C1867(0.99) | LDD0790 | [8] |
| LDCM0474 | CL80 | HCT 116 | C770(0.94); C783(0.94); C1585(0.88); C582(1.09) | LDD0791 | [8] |
| LDCM0475 | CL81 | HCT 116 | C770(0.88); C783(0.88); C1585(1.36); C582(1.78) | LDD0792 | [8] |
| LDCM0476 | CL82 | HCT 116 | C770(0.74); C783(0.74); C1585(1.74); C582(3.07) | LDD0793 | [8] |
| LDCM0477 | CL83 | HCT 116 | C770(1.49); C783(1.49); C1585(1.81); C582(1.68) | LDD0794 | [8] |
| LDCM0478 | CL84 | HCT 116 | C770(1.37); C783(1.37); C1585(1.68); C582(2.13) | LDD0795 | [8] |
| LDCM0479 | CL85 | HCT 116 | C770(1.26); C783(1.26); C1585(0.90); C582(0.91) | LDD0796 | [8] |
| LDCM0480 | CL86 | HCT 116 | C770(1.09); C783(1.09); C1585(0.73); C582(0.50) | LDD0797 | [8] |
| LDCM0481 | CL87 | HCT 116 | C770(1.46); C783(1.46); C1585(1.10); C582(0.91) | LDD0798 | [8] |
| LDCM0482 | CL88 | HCT 116 | C770(1.04); C783(1.04); C1585(1.26); C582(1.57) | LDD0799 | [8] |
| LDCM0483 | CL89 | HCT 116 | C770(1.08); C783(1.08); C1585(1.94); C582(2.91) | LDD0800 | [8] |
| LDCM0484 | CL9 | HCT 116 | C770(1.42); C783(1.42); C1585(0.95); C1867(0.83) | LDD0801 | [8] |
| LDCM0485 | CL90 | HCT 116 | C770(3.10); C783(3.10); C1585(1.58); C582(1.08) | LDD0802 | [8] |
| LDCM0486 | CL91 | HCT 116 | C770(0.98); C783(0.98); C1585(0.95); C1867(0.81) | LDD0803 | [8] |
| LDCM0487 | CL92 | HCT 116 | C770(1.33); C783(1.33); C1585(1.11); C1867(0.79) | LDD0804 | [8] |
| LDCM0488 | CL93 | HCT 116 | C770(0.97); C783(0.97); C1585(0.93); C1867(0.82) | LDD0805 | [8] |
| LDCM0489 | CL94 | HCT 116 | C770(1.50); C783(1.50); C1585(1.03); C1867(1.11) | LDD0806 | [8] |
| LDCM0490 | CL95 | HCT 116 | C770(1.58); C783(1.58); C1585(1.26); C1867(0.91) | LDD0807 | [8] |
| LDCM0491 | CL96 | HCT 116 | C770(1.01); C783(1.01); C1585(0.95); C1867(0.98) | LDD0808 | [8] |
| LDCM0492 | CL97 | HCT 116 | C770(0.95); C783(0.95); C1585(1.08); C1867(1.11) | LDD0809 | [8] |
| LDCM0493 | CL98 | HCT 116 | C770(0.97); C783(0.97); C1585(1.13); C1867(0.97) | LDD0810 | [8] |
| LDCM0494 | CL99 | HCT 116 | C770(0.93); C783(0.93); C1585(0.93); C1867(0.83) | LDD0811 | [8] |
| LDCM0495 | E2913 | HEK-293T | C582(0.93); C1867(0.71); C1585(0.95); C957(1.53) | LDD1698 | [20] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C1717(2.09); C957(1.64) | LDD1702 | [4] |
| LDCM0031 | Epigallocatechin gallate | HEK-293T | 14.29 | LDD0183 | [5] |
| LDCM0468 | Fragment33 | HCT 116 | C770(1.24); C783(1.24); C1585(0.80); C1867(1.26) | LDD0785 | [8] |
| LDCM0427 | Fragment51 | HCT 116 | C770(4.17); C783(4.17); C1585(2.58); C582(0.87) | LDD0744 | [8] |
| LDCM0117 | HHS-0201 | DM93 | Y1176(0.94) | LDD0265 | [7] |
| LDCM0119 | HHS-0401 | DM93 | Y1424(14.06) | LDD0267 | [7] |
| LDCM0120 | HHS-0701 | DM93 | Y1176(1.82); Y1424(11.15) | LDD0268 | [7] |
| LDCM0022 | KB02 | HCT 116 | C171(5.98); C770(9.32); C783(9.32); C1325(5.51) | LDD0080 | [8] |
| LDCM0023 | KB03 | HCT 116 | C171(3.90); C770(11.50); C783(11.50); C1325(1.91) | LDD0081 | [8] |
| LDCM0024 | KB05 | HCT 116 | C171(20.00); C770(13.61); C783(13.61); C1325(3.66) | LDD0082 | [8] |
| LDCM0503 | Nucleophilic fragment 14b | MDA-MB-231 | C957(0.43) | LDD2096 | [4] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C171(0.72) | LDD2099 | [4] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C171(0.53) | LDD2123 | [4] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C171(0.73) | LDD2125 | [4] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C957(1.37) | LDD2128 | [4] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C171(1.01) | LDD2136 | [4] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C171(0.53) | LDD2140 | [4] |
| LDCM0029 | Quercetin | HEK-293T | 4.23 | LDD0181 | [5] |
| LDCM0021 | THZ1 | HCT 116 | C1867(1.45); C1872(1.45); C770(1.13); C783(1.13) | LDD2173 | [8] |
| LDCM0111 | W14 | Hep-G2 | C171(10.48) | LDD0238 | [16] |
| LDCM0112 | W16 | Hep-G2 | C171(0.81) | LDD0239 | [16] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| RalA-binding protein 1 (RALBP1) | . | Q15311 | |||
Transporter and channel
Other
References
