Details of the Target
General Information of Target
| Target ID | LDTP04892 | |||||
|---|---|---|---|---|---|---|
| Target Name | Ras-related protein Rab-2A (RAB2A) | |||||
| Gene Name | RAB2A | |||||
| Gene ID | 5862 | |||||
| Synonyms |
RAB2; Ras-related protein Rab-2A; EC 3.6.5.2 |
|||||
| 3D Structure | ||||||
| Sequence |
MAYAYLFKYIIIGDTGVGKSCLLLQFTDKRFQPVHDLTIGVEFGARMITIDGKQIKLQIW
DTAGQESFRSITRSYYRGAAGALLVYDITRRDTFNHLTTWLEDARQHSNSNMVIMLIGNK SDLESRREVKKEEGEAFAREHGLIFMETSAKTASNVEEAFINTAKEIYEKIQEGVFDINN EANGIKIGPQHAATNATHAGNQGGQQAGGGCC |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Small GTPase superfamily, Rab family
|
|||||
| Subcellular location |
Endoplasmic reticulum-Golgi intermediate compartment membrane
|
|||||
| Function |
The small GTPases Rab are key regulators of intracellular membrane trafficking, from the formation of transport vesicles to their fusion with membranes. Rabs cycle between active GTP-bound and inactive GDP-bound states. In their active state, drive transport of vesicular carriers from donor organelles to acceptor organelles to regulate the membrane traffic that maintains organelle identity and morphology. Required for protein transport from the endoplasmic reticulum to the Golgi complex. Regulates the compacted morphology of the Golgi.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
7.14 | LDD0402 | [1] | |
|
STPyne Probe Info |
 |
K165(8.91); K56(6.53) | LDD0277 | [2] | |
|
AZ-9 Probe Info |
 |
E66(0.76); E173(1.12) | LDD2208 | [3] | |
|
IPM Probe Info |
 |
C21(3.40) | LDD2230 | [4] | |
|
OPA-S-S-alkyne Probe Info |
 |
K130(1.45); K131(1.70) | LDD3494 | [5] | |
|
MCL-4 Probe Info |
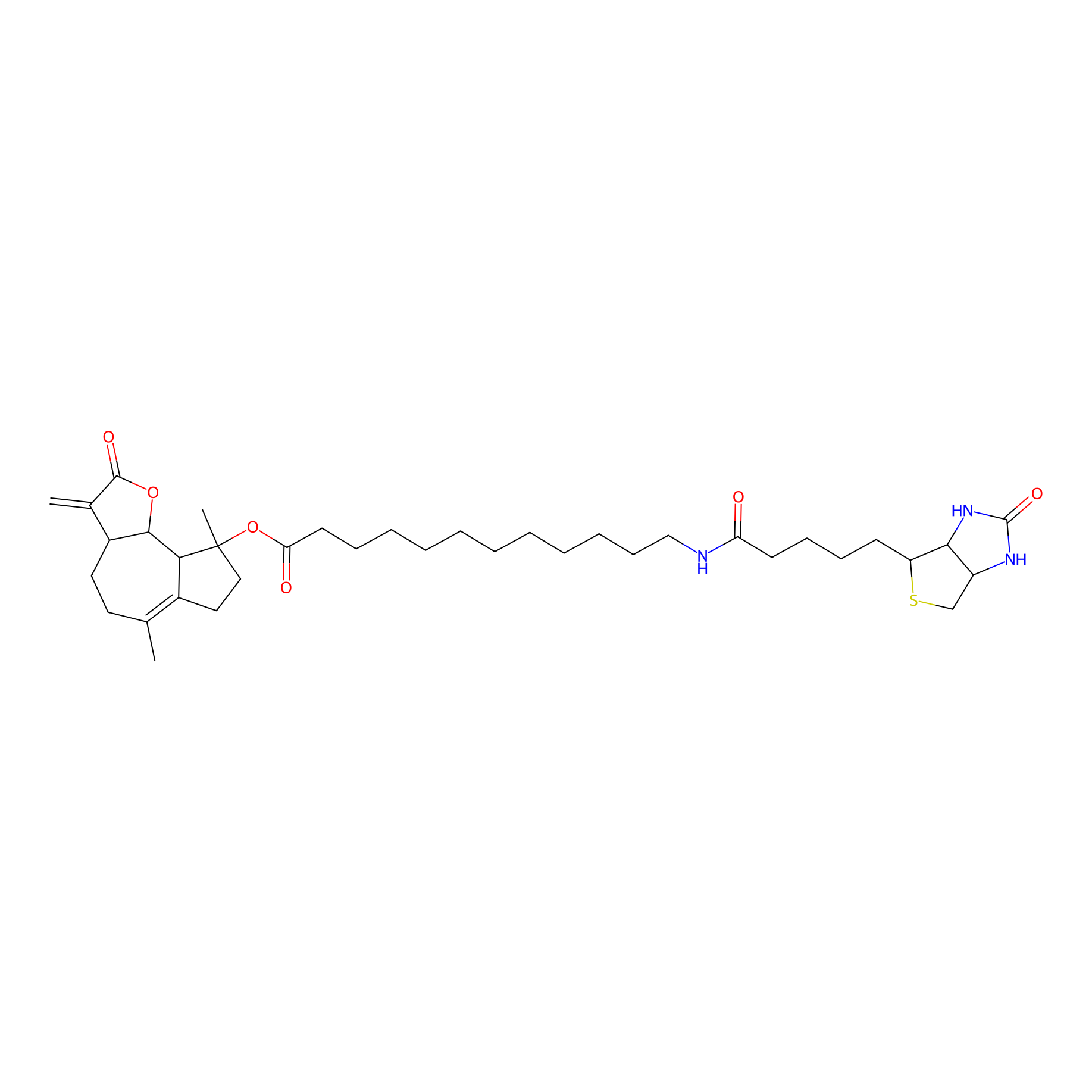 |
7.00 | LDD0049 | [6] | |
|
HPAP Probe Info |
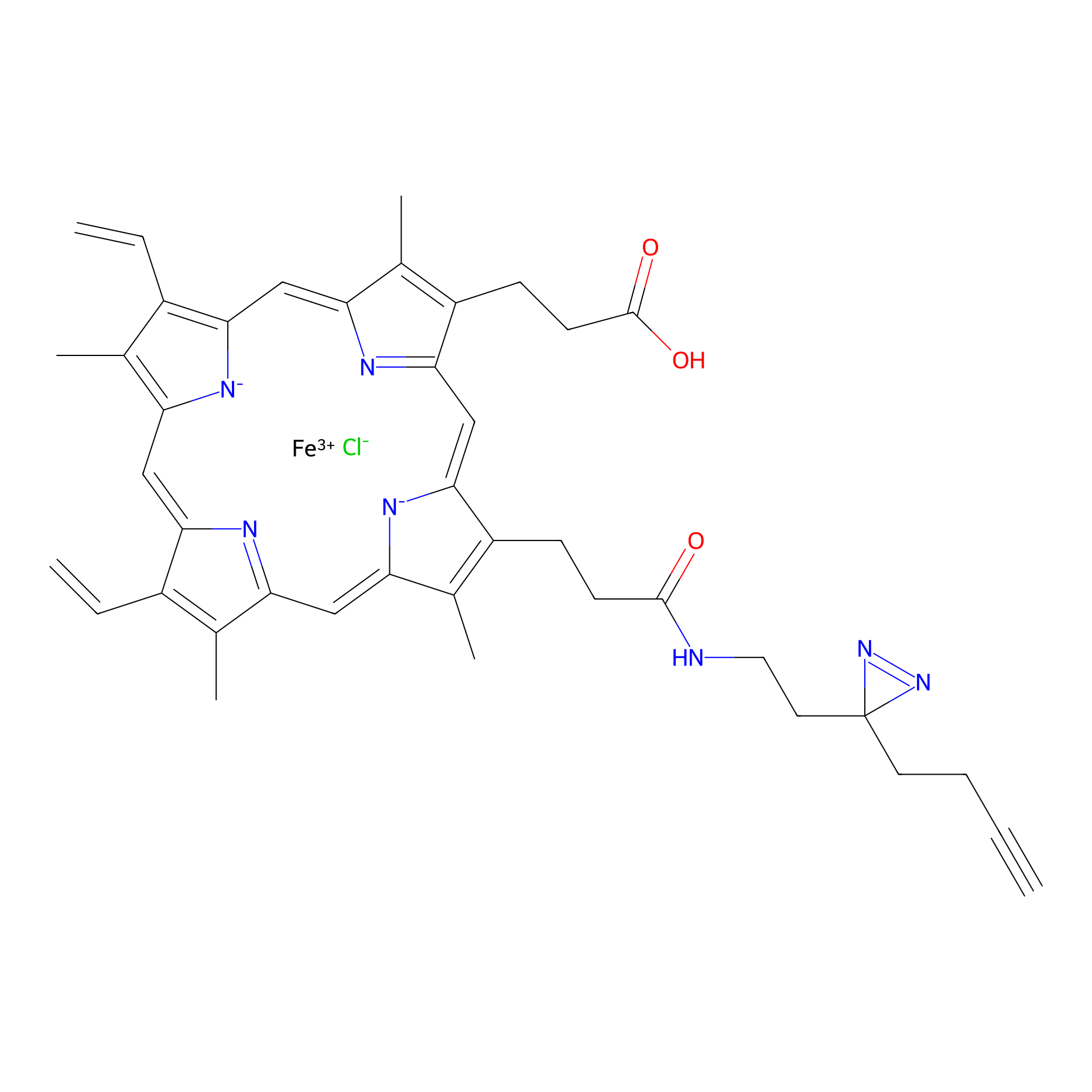 |
3.72 | LDD0062 | [7] | |
|
DBIA Probe Info |
 |
C21(0.83); C212(0.91) | LDD0531 | [8] | |
|
5E-2FA Probe Info |
 |
H141(0.00); H35(0.00) | LDD2235 | [9] | |
|
ATP probe Probe Info |
 |
K29(0.00); K53(0.00); K56(0.00) | LDD0199 | [10] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0032 | [11] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [12] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [13] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [14] | |
|
Acrolein Probe Info |
 |
C21(0.00); H141(0.00) | LDD0217 | [15] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [15] | |
|
MPP-AC Probe Info |
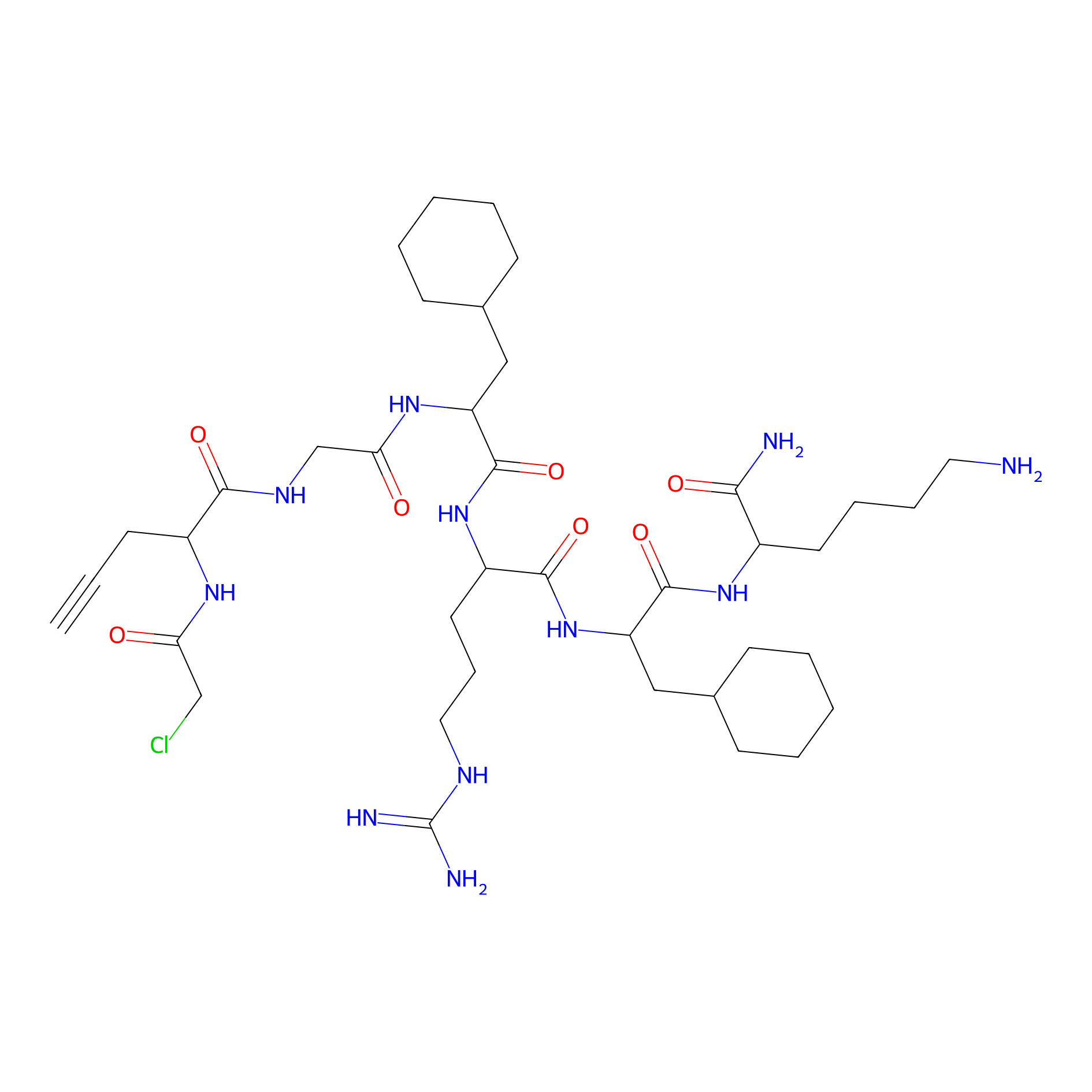 |
N.A. | LDD0428 | [16] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [17] | |
|
TER-AC Probe Info |
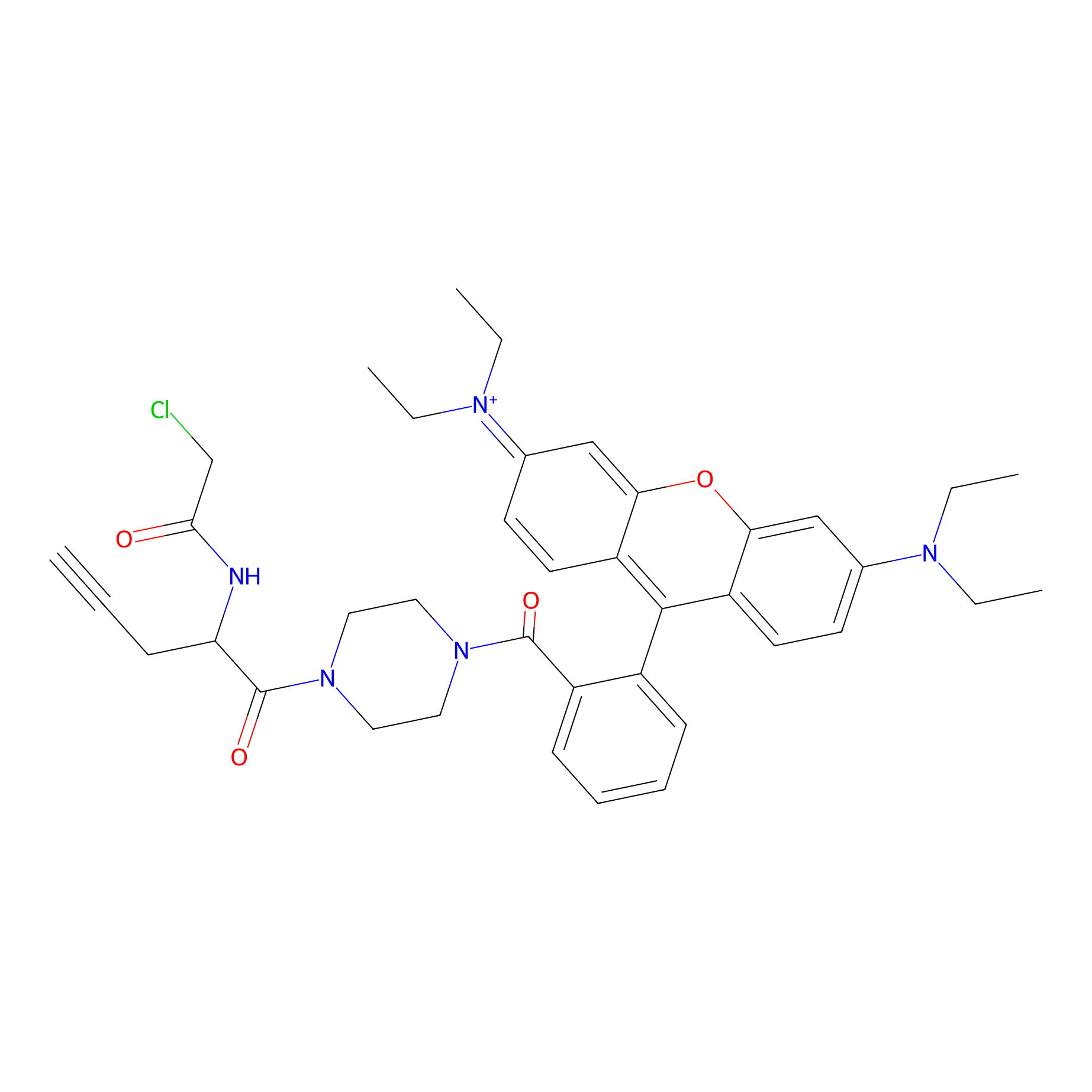 |
N.A. | LDD0426 | [16] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
DR-1 Probe Info |
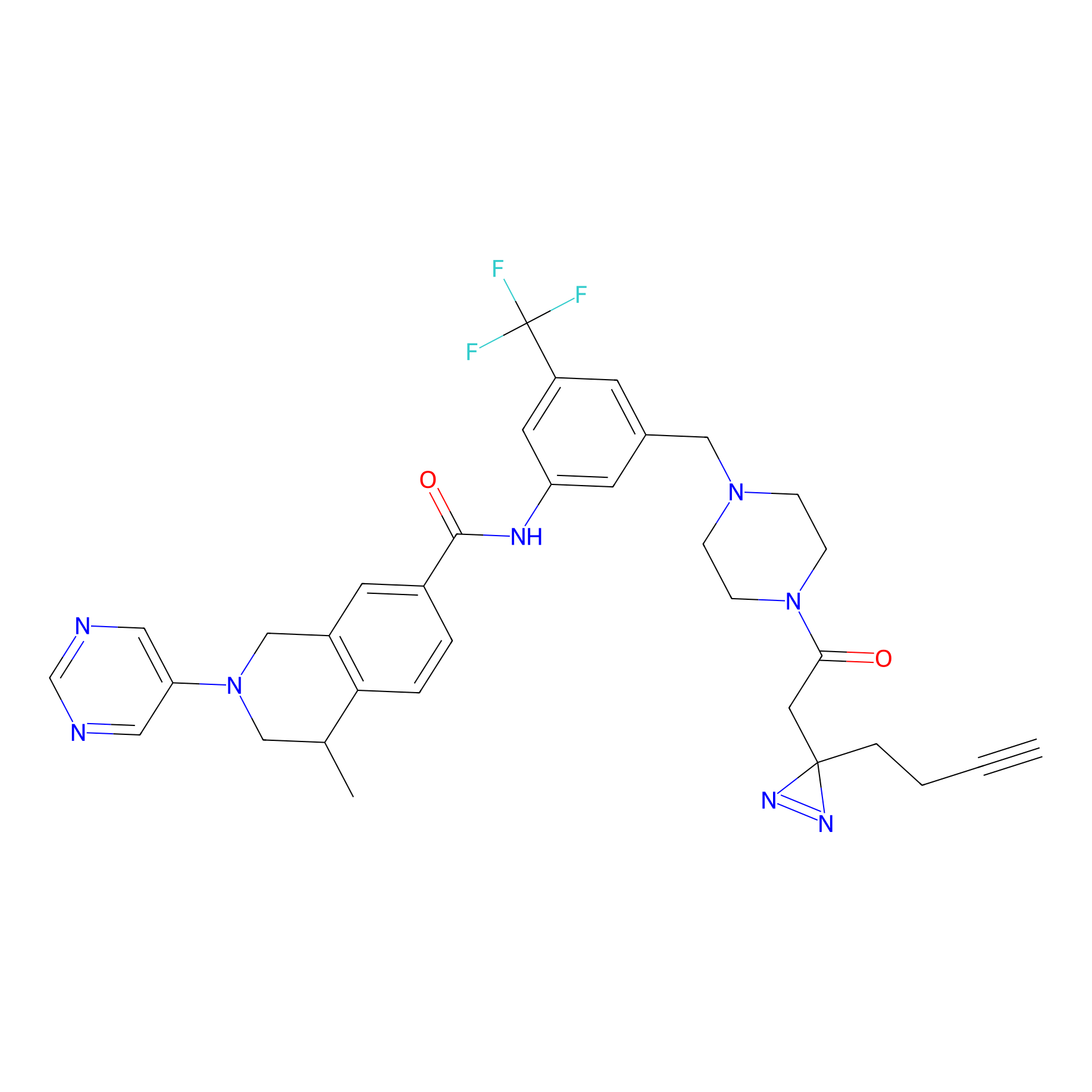 |
2.05 | LDD0398 | [18] | |
|
C040 Probe Info |
 |
7.46 | LDD1740 | [19] | |
|
C228 Probe Info |
 |
16.45 | LDD1901 | [19] | |
|
C232 Probe Info |
 |
38.85 | LDD1905 | [19] | |
|
C235 Probe Info |
 |
28.25 | LDD1908 | [19] | |
|
C349 Probe Info |
 |
8.51 | LDD2010 | [19] | |
|
C362 Probe Info |
 |
27.47 | LDD2023 | [19] | |
|
FFF probe11 Probe Info |
 |
15.44 | LDD0471 | [20] | |
|
FFF probe13 Probe Info |
 |
17.09 | LDD0475 | [20] | |
|
FFF probe14 Probe Info |
 |
14.68 | LDD0477 | [20] | |
|
FFF probe2 Probe Info |
 |
13.30 | LDD0463 | [20] | |
|
FFF probe3 Probe Info |
 |
20.00 | LDD0464 | [20] | |
|
JN0003 Probe Info |
 |
20.00 | LDD0469 | [20] | |
|
OEA-DA Probe Info |
 |
4.75 | LDD0046 | [21] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0214 | AC1 | HCT 116 | C21(0.83); C212(0.91) | LDD0531 | [8] |
| LDCM0215 | AC10 | HCT 116 | C21(1.08) | LDD0532 | [8] |
| LDCM0216 | AC100 | HCT 116 | C21(0.68) | LDD0533 | [8] |
| LDCM0217 | AC101 | HCT 116 | C21(0.66) | LDD0534 | [8] |
| LDCM0218 | AC102 | HCT 116 | C21(0.69) | LDD0535 | [8] |
| LDCM0219 | AC103 | HCT 116 | C21(0.67) | LDD0536 | [8] |
| LDCM0220 | AC104 | HCT 116 | C21(0.72) | LDD0537 | [8] |
| LDCM0221 | AC105 | HCT 116 | C21(0.66) | LDD0538 | [8] |
| LDCM0222 | AC106 | HCT 116 | C21(0.70) | LDD0539 | [8] |
| LDCM0223 | AC107 | HCT 116 | C21(0.73) | LDD0540 | [8] |
| LDCM0224 | AC108 | HCT 116 | C21(0.73) | LDD0541 | [8] |
| LDCM0225 | AC109 | HCT 116 | C21(0.71) | LDD0542 | [8] |
| LDCM0226 | AC11 | HCT 116 | C21(1.05) | LDD0543 | [8] |
| LDCM0227 | AC110 | HCT 116 | C21(0.76) | LDD0544 | [8] |
| LDCM0228 | AC111 | HCT 116 | C21(0.65) | LDD0545 | [8] |
| LDCM0229 | AC112 | HCT 116 | C21(0.71) | LDD0546 | [8] |
| LDCM0230 | AC113 | HCT 116 | C212(1.20) | LDD0547 | [8] |
| LDCM0231 | AC114 | HCT 116 | C212(1.08) | LDD0548 | [8] |
| LDCM0232 | AC115 | HCT 116 | C212(0.95) | LDD0549 | [8] |
| LDCM0233 | AC116 | HCT 116 | C212(1.09) | LDD0550 | [8] |
| LDCM0234 | AC117 | HCT 116 | C212(1.09) | LDD0551 | [8] |
| LDCM0235 | AC118 | HCT 116 | C212(1.35) | LDD0552 | [8] |
| LDCM0236 | AC119 | HCT 116 | C212(1.18) | LDD0553 | [8] |
| LDCM0237 | AC12 | HCT 116 | C21(1.13) | LDD0554 | [8] |
| LDCM0238 | AC120 | HCT 116 | C212(1.02) | LDD0555 | [8] |
| LDCM0239 | AC121 | HCT 116 | C212(1.23) | LDD0556 | [8] |
| LDCM0240 | AC122 | HCT 116 | C212(1.33) | LDD0557 | [8] |
| LDCM0241 | AC123 | HCT 116 | C212(1.04) | LDD0558 | [8] |
| LDCM0242 | AC124 | HCT 116 | C212(0.72) | LDD0559 | [8] |
| LDCM0243 | AC125 | HCT 116 | C212(1.23) | LDD0560 | [8] |
| LDCM0244 | AC126 | HCT 116 | C212(1.23) | LDD0561 | [8] |
| LDCM0245 | AC127 | HCT 116 | C212(0.86) | LDD0562 | [8] |
| LDCM0259 | AC14 | HCT 116 | C21(1.09) | LDD0576 | [8] |
| LDCM0270 | AC15 | HCT 116 | C21(1.07) | LDD0587 | [8] |
| LDCM0276 | AC17 | HCT 116 | C21(1.05); C212(1.21) | LDD0593 | [8] |
| LDCM0277 | AC18 | HCT 116 | C212(1.19); C21(1.33) | LDD0594 | [8] |
| LDCM0278 | AC19 | HCT 116 | C212(1.37); C21(1.49) | LDD0595 | [8] |
| LDCM0279 | AC2 | HCT 116 | C21(1.04); C212(1.14) | LDD0596 | [8] |
| LDCM0280 | AC20 | HCT 116 | C212(1.14); C21(1.24) | LDD0597 | [8] |
| LDCM0281 | AC21 | HCT 116 | C21(1.02); C212(1.29) | LDD0598 | [8] |
| LDCM0282 | AC22 | HCT 116 | C21(1.03); C212(1.22) | LDD0599 | [8] |
| LDCM0283 | AC23 | HCT 116 | C21(1.04); C212(1.45) | LDD0600 | [8] |
| LDCM0284 | AC24 | HCT 116 | C212(0.81); C21(1.22) | LDD0601 | [8] |
| LDCM0285 | AC25 | HCT 116 | C21(1.02) | LDD0602 | [8] |
| LDCM0286 | AC26 | HCT 116 | C21(0.99) | LDD0603 | [8] |
| LDCM0287 | AC27 | HCT 116 | C21(1.08) | LDD0604 | [8] |
| LDCM0288 | AC28 | HCT 116 | C21(0.96) | LDD0605 | [8] |
| LDCM0289 | AC29 | HCT 116 | C21(0.97) | LDD0606 | [8] |
| LDCM0290 | AC3 | HCT 116 | C21(0.94); C212(0.96) | LDD0607 | [8] |
| LDCM0291 | AC30 | HCT 116 | C21(1.03) | LDD0608 | [8] |
| LDCM0292 | AC31 | HCT 116 | C21(0.98) | LDD0609 | [8] |
| LDCM0293 | AC32 | HCT 116 | C21(0.94) | LDD0610 | [8] |
| LDCM0294 | AC33 | HCT 116 | C21(0.92) | LDD0611 | [8] |
| LDCM0295 | AC34 | HCT 116 | C21(0.94) | LDD0612 | [8] |
| LDCM0301 | AC4 | HCT 116 | C212(0.83); C21(0.93) | LDD0618 | [8] |
| LDCM0308 | AC46 | HCT 116 | C21(1.05); C212(2.57) | LDD0625 | [8] |
| LDCM0309 | AC47 | HCT 116 | C21(0.94); C212(1.69) | LDD0626 | [8] |
| LDCM0310 | AC48 | HCT 116 | C21(0.81); C212(1.66) | LDD0627 | [8] |
| LDCM0311 | AC49 | HCT 116 | C21(0.70); C212(1.23) | LDD0628 | [8] |
| LDCM0312 | AC5 | HCT 116 | C21(0.87); C212(0.90) | LDD0629 | [8] |
| LDCM0313 | AC50 | HCT 116 | C21(0.80); C212(1.93) | LDD0630 | [8] |
| LDCM0314 | AC51 | HCT 116 | C21(0.90); C212(1.56) | LDD0631 | [8] |
| LDCM0315 | AC52 | HCT 116 | C21(0.80); C212(2.05) | LDD0632 | [8] |
| LDCM0316 | AC53 | HCT 116 | C21(0.92); C212(1.73) | LDD0633 | [8] |
| LDCM0317 | AC54 | HCT 116 | C21(0.79); C212(0.94) | LDD0634 | [8] |
| LDCM0318 | AC55 | HCT 116 | C21(0.74); C212(0.99) | LDD0635 | [8] |
| LDCM0319 | AC56 | HCT 116 | C21(0.62); C212(0.96) | LDD0636 | [8] |
| LDCM0320 | AC57 | HCT 116 | C212(0.70) | LDD0637 | [8] |
| LDCM0321 | AC58 | HCT 116 | C212(0.77) | LDD0638 | [8] |
| LDCM0322 | AC59 | HCT 116 | C212(0.92) | LDD0639 | [8] |
| LDCM0323 | AC6 | HCT 116 | C21(1.06) | LDD0640 | [8] |
| LDCM0324 | AC60 | HCT 116 | C212(1.13) | LDD0641 | [8] |
| LDCM0325 | AC61 | HCT 116 | C212(0.66) | LDD0642 | [8] |
| LDCM0326 | AC62 | HCT 116 | C212(0.92) | LDD0643 | [8] |
| LDCM0327 | AC63 | HCT 116 | C212(1.10) | LDD0644 | [8] |
| LDCM0328 | AC64 | HCT 116 | C212(0.88) | LDD0645 | [8] |
| LDCM0329 | AC65 | HCT 116 | C212(1.03) | LDD0646 | [8] |
| LDCM0330 | AC66 | HCT 116 | C212(0.66) | LDD0647 | [8] |
| LDCM0331 | AC67 | HCT 116 | C212(0.93) | LDD0648 | [8] |
| LDCM0332 | AC68 | HCT 116 | C21(1.05) | LDD0649 | [8] |
| LDCM0333 | AC69 | HCT 116 | C21(1.01) | LDD0650 | [8] |
| LDCM0334 | AC7 | HCT 116 | C21(1.07) | LDD0651 | [8] |
| LDCM0335 | AC70 | HCT 116 | C21(0.96) | LDD0652 | [8] |
| LDCM0336 | AC71 | HCT 116 | C21(1.18) | LDD0653 | [8] |
| LDCM0337 | AC72 | HCT 116 | C21(1.06) | LDD0654 | [8] |
| LDCM0338 | AC73 | HCT 116 | C21(0.89) | LDD0655 | [8] |
| LDCM0339 | AC74 | HCT 116 | C21(1.08) | LDD0656 | [8] |
| LDCM0340 | AC75 | HCT 116 | C21(0.88) | LDD0657 | [8] |
| LDCM0341 | AC76 | HCT 116 | C21(1.53) | LDD0658 | [8] |
| LDCM0342 | AC77 | HCT 116 | C21(1.39) | LDD0659 | [8] |
| LDCM0343 | AC78 | HCT 116 | C21(1.00) | LDD0660 | [8] |
| LDCM0344 | AC79 | HCT 116 | C21(1.33) | LDD0661 | [8] |
| LDCM0345 | AC8 | HCT 116 | C21(1.10) | LDD0662 | [8] |
| LDCM0346 | AC80 | HCT 116 | C21(1.11) | LDD0663 | [8] |
| LDCM0347 | AC81 | HCT 116 | C21(1.30) | LDD0664 | [8] |
| LDCM0348 | AC82 | HCT 116 | C21(0.58) | LDD0665 | [8] |
| LDCM0349 | AC83 | HCT 116 | C212(1.05) | LDD0666 | [8] |
| LDCM0350 | AC84 | HCT 116 | C212(1.54) | LDD0667 | [8] |
| LDCM0351 | AC85 | HCT 116 | C212(1.68) | LDD0668 | [8] |
| LDCM0352 | AC86 | HCT 116 | C212(1.26) | LDD0669 | [8] |
| LDCM0353 | AC87 | HCT 116 | C212(1.23) | LDD0670 | [8] |
| LDCM0354 | AC88 | HCT 116 | C212(1.03) | LDD0671 | [8] |
| LDCM0355 | AC89 | HCT 116 | C212(1.12) | LDD0672 | [8] |
| LDCM0357 | AC90 | HCT 116 | C212(1.53) | LDD0674 | [8] |
| LDCM0358 | AC91 | HCT 116 | C212(1.34) | LDD0675 | [8] |
| LDCM0359 | AC92 | HCT 116 | C212(1.43) | LDD0676 | [8] |
| LDCM0360 | AC93 | HCT 116 | C212(1.57) | LDD0677 | [8] |
| LDCM0361 | AC94 | HCT 116 | C212(1.81) | LDD0678 | [8] |
| LDCM0362 | AC95 | HCT 116 | C212(1.36) | LDD0679 | [8] |
| LDCM0363 | AC96 | HCT 116 | C212(2.33) | LDD0680 | [8] |
| LDCM0364 | AC97 | HCT 116 | C212(1.22) | LDD0681 | [8] |
| LDCM0365 | AC98 | HCT 116 | C21(0.28) | LDD0682 | [8] |
| LDCM0366 | AC99 | HCT 116 | C21(0.76) | LDD0683 | [8] |
| LDCM0248 | AKOS034007472 | HCT 116 | C21(1.19) | LDD0565 | [8] |
| LDCM0356 | AKOS034007680 | HCT 116 | C21(1.13) | LDD0673 | [8] |
| LDCM0275 | AKOS034007705 | HCT 116 | C21(1.02) | LDD0592 | [8] |
| LDCM0156 | Aniline | NCI-H1299 | 11.36 | LDD0403 | [1] |
| LDCM0108 | Chloroacetamide | HeLa | H141(0.00); C21(0.00); H96(0.00) | LDD0222 | [15] |
| LDCM0367 | CL1 | HCT 116 | C212(0.92); C21(0.93) | LDD0684 | [8] |
| LDCM0368 | CL10 | HCT 116 | C212(0.82); C21(0.86) | LDD0685 | [8] |
| LDCM0369 | CL100 | HCT 116 | C21(0.90); C212(1.15) | LDD0686 | [8] |
| LDCM0370 | CL101 | HCT 116 | C21(1.12) | LDD0687 | [8] |
| LDCM0371 | CL102 | HCT 116 | C21(1.14) | LDD0688 | [8] |
| LDCM0372 | CL103 | HCT 116 | C21(0.99) | LDD0689 | [8] |
| LDCM0373 | CL104 | HCT 116 | C21(1.02) | LDD0690 | [8] |
| LDCM0374 | CL105 | HCT 116 | C212(0.91); C21(1.20) | LDD0691 | [8] |
| LDCM0375 | CL106 | HCT 116 | C21(0.94); C212(1.13) | LDD0692 | [8] |
| LDCM0376 | CL107 | HCT 116 | C21(1.05); C212(1.15) | LDD0693 | [8] |
| LDCM0377 | CL108 | HCT 116 | C212(0.79); C21(1.15) | LDD0694 | [8] |
| LDCM0378 | CL109 | HCT 116 | C212(0.84); C21(1.05) | LDD0695 | [8] |
| LDCM0379 | CL11 | HCT 116 | C212(0.69); C21(0.87) | LDD0696 | [8] |
| LDCM0380 | CL110 | HCT 116 | C212(0.70); C21(1.14) | LDD0697 | [8] |
| LDCM0381 | CL111 | HCT 116 | C21(1.06); C212(1.17) | LDD0698 | [8] |
| LDCM0382 | CL112 | HCT 116 | C21(1.06) | LDD0699 | [8] |
| LDCM0383 | CL113 | HCT 116 | C21(0.94) | LDD0700 | [8] |
| LDCM0384 | CL114 | HCT 116 | C21(0.85) | LDD0701 | [8] |
| LDCM0385 | CL115 | HCT 116 | C21(0.97) | LDD0702 | [8] |
| LDCM0386 | CL116 | HCT 116 | C21(1.05) | LDD0703 | [8] |
| LDCM0390 | CL12 | HCT 116 | C21(0.85); C212(0.86) | LDD0707 | [8] |
| LDCM0392 | CL121 | HCT 116 | C21(0.75); C212(1.54) | LDD0709 | [8] |
| LDCM0393 | CL122 | HCT 116 | C21(0.90); C212(2.18) | LDD0710 | [8] |
| LDCM0394 | CL123 | HCT 116 | C21(0.95); C212(2.12) | LDD0711 | [8] |
| LDCM0395 | CL124 | HCT 116 | C21(0.94); C212(1.21) | LDD0712 | [8] |
| LDCM0396 | CL125 | HCT 116 | C212(0.85) | LDD0713 | [8] |
| LDCM0397 | CL126 | HCT 116 | C212(0.97) | LDD0714 | [8] |
| LDCM0398 | CL127 | HCT 116 | C212(0.78) | LDD0715 | [8] |
| LDCM0399 | CL128 | HCT 116 | C212(0.78) | LDD0716 | [8] |
| LDCM0400 | CL13 | HCT 116 | C21(0.90); C212(0.93) | LDD0717 | [8] |
| LDCM0401 | CL14 | HCT 116 | C212(0.78); C21(0.91) | LDD0718 | [8] |
| LDCM0402 | CL15 | HCT 116 | C21(0.56); C212(0.95) | LDD0719 | [8] |
| LDCM0403 | CL16 | HCT 116 | C21(0.93) | LDD0720 | [8] |
| LDCM0404 | CL17 | HCT 116 | C21(0.89) | LDD0721 | [8] |
| LDCM0405 | CL18 | HCT 116 | C21(0.87) | LDD0722 | [8] |
| LDCM0406 | CL19 | HCT 116 | C21(1.02) | LDD0723 | [8] |
| LDCM0407 | CL2 | HCT 116 | C212(0.91); C21(1.13) | LDD0724 | [8] |
| LDCM0408 | CL20 | HCT 116 | C21(0.99) | LDD0725 | [8] |
| LDCM0409 | CL21 | HCT 116 | C21(0.86) | LDD0726 | [8] |
| LDCM0410 | CL22 | HCT 116 | C21(0.70) | LDD0727 | [8] |
| LDCM0411 | CL23 | HCT 116 | C21(1.09) | LDD0728 | [8] |
| LDCM0412 | CL24 | HCT 116 | C21(0.95) | LDD0729 | [8] |
| LDCM0413 | CL25 | HCT 116 | C21(1.09) | LDD0730 | [8] |
| LDCM0414 | CL26 | HCT 116 | C21(1.05) | LDD0731 | [8] |
| LDCM0415 | CL27 | HCT 116 | C21(1.09) | LDD0732 | [8] |
| LDCM0416 | CL28 | HCT 116 | C21(0.98) | LDD0733 | [8] |
| LDCM0417 | CL29 | HCT 116 | C21(1.08) | LDD0734 | [8] |
| LDCM0418 | CL3 | HCT 116 | C21(1.06); C212(0.69) | LDD0735 | [8] |
| LDCM0419 | CL30 | HCT 116 | C21(1.13) | LDD0736 | [8] |
| LDCM0420 | CL31 | HCT 116 | C21(0.81) | LDD0737 | [8] |
| LDCM0421 | CL32 | HCT 116 | C21(0.72) | LDD0738 | [8] |
| LDCM0422 | CL33 | HCT 116 | C21(0.73) | LDD0739 | [8] |
| LDCM0423 | CL34 | HCT 116 | C21(0.65) | LDD0740 | [8] |
| LDCM0424 | CL35 | HCT 116 | C21(0.67) | LDD0741 | [8] |
| LDCM0425 | CL36 | HCT 116 | C21(0.80) | LDD0742 | [8] |
| LDCM0426 | CL37 | HCT 116 | C21(0.70) | LDD0743 | [8] |
| LDCM0428 | CL39 | HCT 116 | C21(0.79) | LDD0745 | [8] |
| LDCM0429 | CL4 | HCT 116 | C21(1.00); C212(0.93) | LDD0746 | [8] |
| LDCM0430 | CL40 | HCT 116 | C21(0.81) | LDD0747 | [8] |
| LDCM0431 | CL41 | HCT 116 | C21(0.86) | LDD0748 | [8] |
| LDCM0432 | CL42 | HCT 116 | C21(0.49) | LDD0749 | [8] |
| LDCM0433 | CL43 | HCT 116 | C21(0.59) | LDD0750 | [8] |
| LDCM0434 | CL44 | HCT 116 | C21(0.64) | LDD0751 | [8] |
| LDCM0435 | CL45 | HCT 116 | C21(0.62) | LDD0752 | [8] |
| LDCM0436 | CL46 | HCT 116 | C21(1.14) | LDD0753 | [8] |
| LDCM0437 | CL47 | HCT 116 | C21(1.13) | LDD0754 | [8] |
| LDCM0438 | CL48 | HCT 116 | C21(1.03) | LDD0755 | [8] |
| LDCM0439 | CL49 | HCT 116 | C21(1.05) | LDD0756 | [8] |
| LDCM0440 | CL5 | HCT 116 | C21(1.00); C212(1.03) | LDD0757 | [8] |
| LDCM0441 | CL50 | HCT 116 | C21(1.05) | LDD0758 | [8] |
| LDCM0442 | CL51 | HCT 116 | C21(1.08) | LDD0759 | [8] |
| LDCM0443 | CL52 | HCT 116 | C21(1.14) | LDD0760 | [8] |
| LDCM0444 | CL53 | HCT 116 | C21(0.92) | LDD0761 | [8] |
| LDCM0445 | CL54 | HCT 116 | C21(0.99) | LDD0762 | [8] |
| LDCM0446 | CL55 | HCT 116 | C21(1.05) | LDD0763 | [8] |
| LDCM0447 | CL56 | HCT 116 | C21(1.06) | LDD0764 | [8] |
| LDCM0448 | CL57 | HCT 116 | C21(1.05) | LDD0765 | [8] |
| LDCM0449 | CL58 | HCT 116 | C21(1.11) | LDD0766 | [8] |
| LDCM0450 | CL59 | HCT 116 | C21(1.02) | LDD0767 | [8] |
| LDCM0451 | CL6 | HCT 116 | C21(0.86); C212(1.52) | LDD0768 | [8] |
| LDCM0452 | CL60 | HCT 116 | C21(1.01) | LDD0769 | [8] |
| LDCM0453 | CL61 | HCT 116 | C21(1.00) | LDD0770 | [8] |
| LDCM0454 | CL62 | HCT 116 | C21(1.01) | LDD0771 | [8] |
| LDCM0455 | CL63 | HCT 116 | C21(1.03) | LDD0772 | [8] |
| LDCM0456 | CL64 | HCT 116 | C21(0.96) | LDD0773 | [8] |
| LDCM0457 | CL65 | HCT 116 | C21(1.06) | LDD0774 | [8] |
| LDCM0458 | CL66 | HCT 116 | C21(1.02) | LDD0775 | [8] |
| LDCM0459 | CL67 | HCT 116 | C21(0.96) | LDD0776 | [8] |
| LDCM0460 | CL68 | HCT 116 | C21(0.88) | LDD0777 | [8] |
| LDCM0461 | CL69 | HCT 116 | C21(0.82) | LDD0778 | [8] |
| LDCM0462 | CL7 | HCT 116 | C21(0.99); C212(1.48) | LDD0779 | [8] |
| LDCM0463 | CL70 | HCT 116 | C21(0.97) | LDD0780 | [8] |
| LDCM0464 | CL71 | HCT 116 | C21(0.96) | LDD0781 | [8] |
| LDCM0465 | CL72 | HCT 116 | C21(1.03) | LDD0782 | [8] |
| LDCM0466 | CL73 | HCT 116 | C21(0.95) | LDD0783 | [8] |
| LDCM0467 | CL74 | HCT 116 | C21(0.91) | LDD0784 | [8] |
| LDCM0469 | CL76 | HCT 116 | C21(0.83) | LDD0786 | [8] |
| LDCM0470 | CL77 | HCT 116 | C21(0.75) | LDD0787 | [8] |
| LDCM0471 | CL78 | HCT 116 | C21(0.89) | LDD0788 | [8] |
| LDCM0472 | CL79 | HCT 116 | C21(0.89) | LDD0789 | [8] |
| LDCM0473 | CL8 | HCT 116 | C21(0.89); C212(0.81) | LDD0790 | [8] |
| LDCM0474 | CL80 | HCT 116 | C21(0.82) | LDD0791 | [8] |
| LDCM0475 | CL81 | HCT 116 | C21(0.88) | LDD0792 | [8] |
| LDCM0476 | CL82 | HCT 116 | C21(0.78) | LDD0793 | [8] |
| LDCM0477 | CL83 | HCT 116 | C21(0.89) | LDD0794 | [8] |
| LDCM0478 | CL84 | HCT 116 | C21(0.70) | LDD0795 | [8] |
| LDCM0479 | CL85 | HCT 116 | C21(0.94) | LDD0796 | [8] |
| LDCM0480 | CL86 | HCT 116 | C21(1.00) | LDD0797 | [8] |
| LDCM0481 | CL87 | HCT 116 | C21(0.93) | LDD0798 | [8] |
| LDCM0482 | CL88 | HCT 116 | C21(0.84) | LDD0799 | [8] |
| LDCM0483 | CL89 | HCT 116 | C21(0.58) | LDD0800 | [8] |
| LDCM0484 | CL9 | HCT 116 | C21(0.96); C212(1.69) | LDD0801 | [8] |
| LDCM0485 | CL90 | HCT 116 | C21(0.83) | LDD0802 | [8] |
| LDCM0486 | CL91 | HCT 116 | C21(0.91); C212(1.49) | LDD0803 | [8] |
| LDCM0487 | CL92 | HCT 116 | C21(0.86); C212(1.20) | LDD0804 | [8] |
| LDCM0488 | CL93 | HCT 116 | C21(0.83); C212(1.02) | LDD0805 | [8] |
| LDCM0489 | CL94 | HCT 116 | C21(0.87); C212(0.89) | LDD0806 | [8] |
| LDCM0490 | CL95 | HCT 116 | C21(0.86); C212(1.57) | LDD0807 | [8] |
| LDCM0491 | CL96 | HCT 116 | C21(0.89); C212(1.42) | LDD0808 | [8] |
| LDCM0492 | CL97 | HCT 116 | C21(0.95); C212(1.23) | LDD0809 | [8] |
| LDCM0493 | CL98 | HCT 116 | C21(1.00); C212(1.14) | LDD0810 | [8] |
| LDCM0494 | CL99 | HCT 116 | C21(0.97); C212(1.05) | LDD0811 | [8] |
| LDCM0468 | Fragment33 | HCT 116 | C21(1.00) | LDD0785 | [8] |
| LDCM0427 | Fragment51 | HCT 116 | C21(0.57) | LDD0744 | [8] |
| LDCM0107 | IAA | HeLa | H96(0.00); H141(0.00) | LDD0221 | [15] |
| LDCM0006 | Micheliolide | M9-ENL1 | 7.00 | LDD0049 | [6] |
| LDCM0109 | NEM | HeLa | H141(0.00); H96(0.00) | LDD0223 | [15] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C21(0.17) | LDD2207 | [22] |
| LDCM0014 | Panhematin | HEK-293T | 3.72 | LDD0062 | [7] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| AMSH-like protease (STAMBPL1) | Peptidase M67C family | Q96FJ0 | |||
| E3 ubiquitin-protein ligase TRIM7 (TRIM7) | TRIM/RBCC family | Q9C029 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Transmembrane protein 70, mitochondrial (TMEM70) | TMEM70 family | Q9BUB7 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Golgin-45 (BLZF1) | . | Q9H2G9 | |||
| Transcription cofactor HES-6 (HES6) | . | Q96HZ4 | |||
Other
References
