Details of the Target
General Information of Target
| Target ID | LDTP02768 | |||||
|---|---|---|---|---|---|---|
| Target Name | Ras-related C3 botulinum toxin substrate 2 (RAC2) | |||||
| Gene Name | RAC2 | |||||
| Gene ID | 5880 | |||||
| Synonyms |
Ras-related C3 botulinum toxin substrate 2; GX; Small G protein; p21-Rac2 |
|||||
| 3D Structure | ||||||
| Sequence |
MQAIKCVVVGDGAVGKTCLLISYTTNAFPGEYIPTVFDNYSANVMVDSKPVNLGLWDTAG
QEDYDRLRPLSYPQTDVFLICFSLVSPASYENVRAKWFPEVRHHCPSTPIILVGTKLDLR DDKDTIEKLKEKKLAPITYPQGLALAKEIDSVKYLECSALTQRGLKTVFDEAIRAVLCPQ PTRQQKRACSLL |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Small GTPase superfamily, Rho family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Plasma membrane-associated small GTPase which cycles between an active GTP-bound and inactive GDP-bound state. In active state binds to a variety of effector proteins to regulate cellular responses, such as secretory processes, phagocytose of apoptotic cells and epithelial cell polarization. Augments the production of reactive oxygen species (ROS) by NADPH oxidase.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
N1 Probe Info |
 |
100.00 | LDD0242 | [1] | |
|
TH211 Probe Info |
 |
Y139(20.00) | LDD0260 | [2] | |
|
STPyne Probe Info |
 |
K133(10.00); K147(8.58); K153(7.18); K96(10.00) | LDD0277 | [3] | |
|
OPA-S-S-alkyne Probe Info |
 |
K166(1.37) | LDD3494 | [4] | |
|
JZ128-DTB Probe Info |
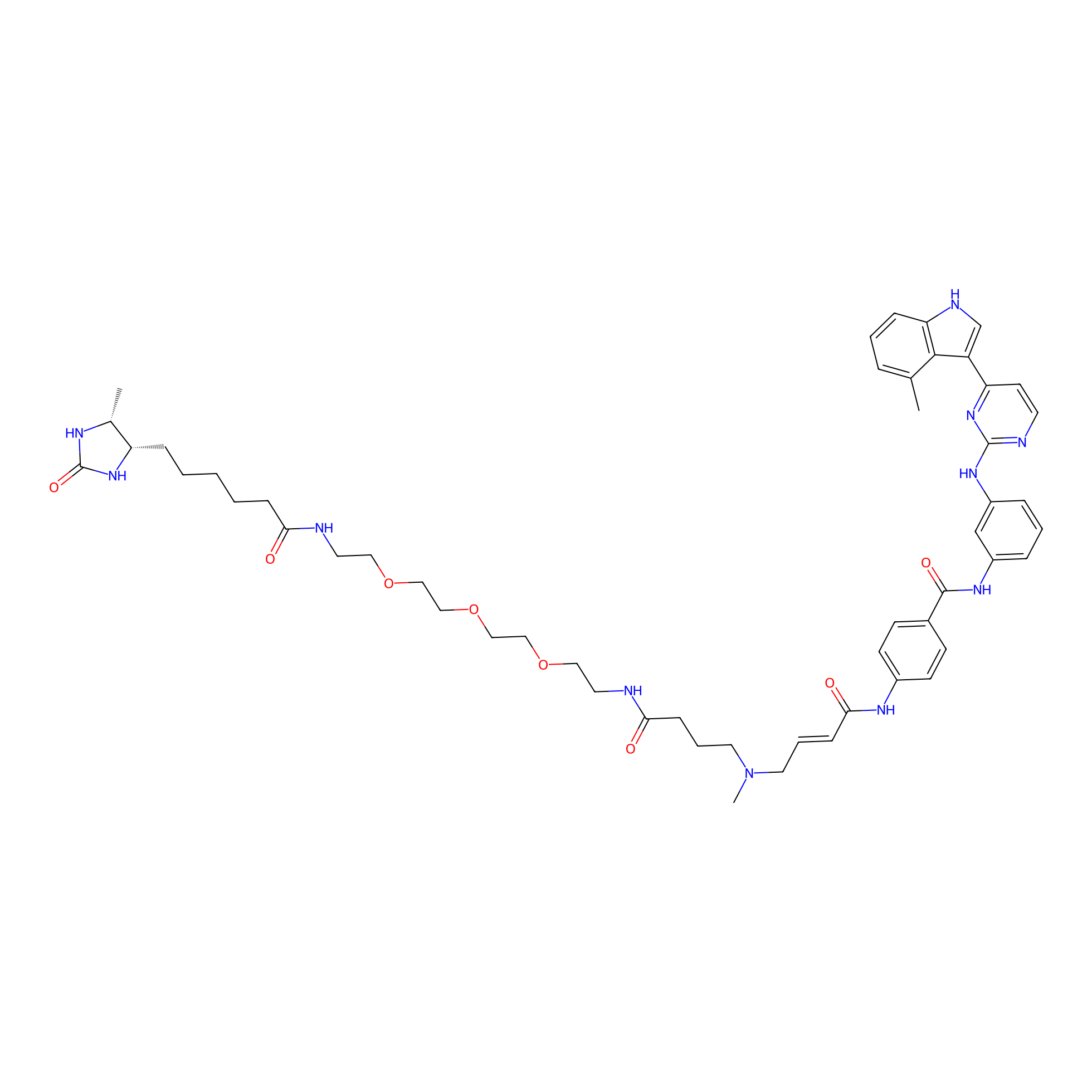 |
C157(0.00); C178(0.00) | LDD0462 | [5] | |
|
THZ1-DTB Probe Info |
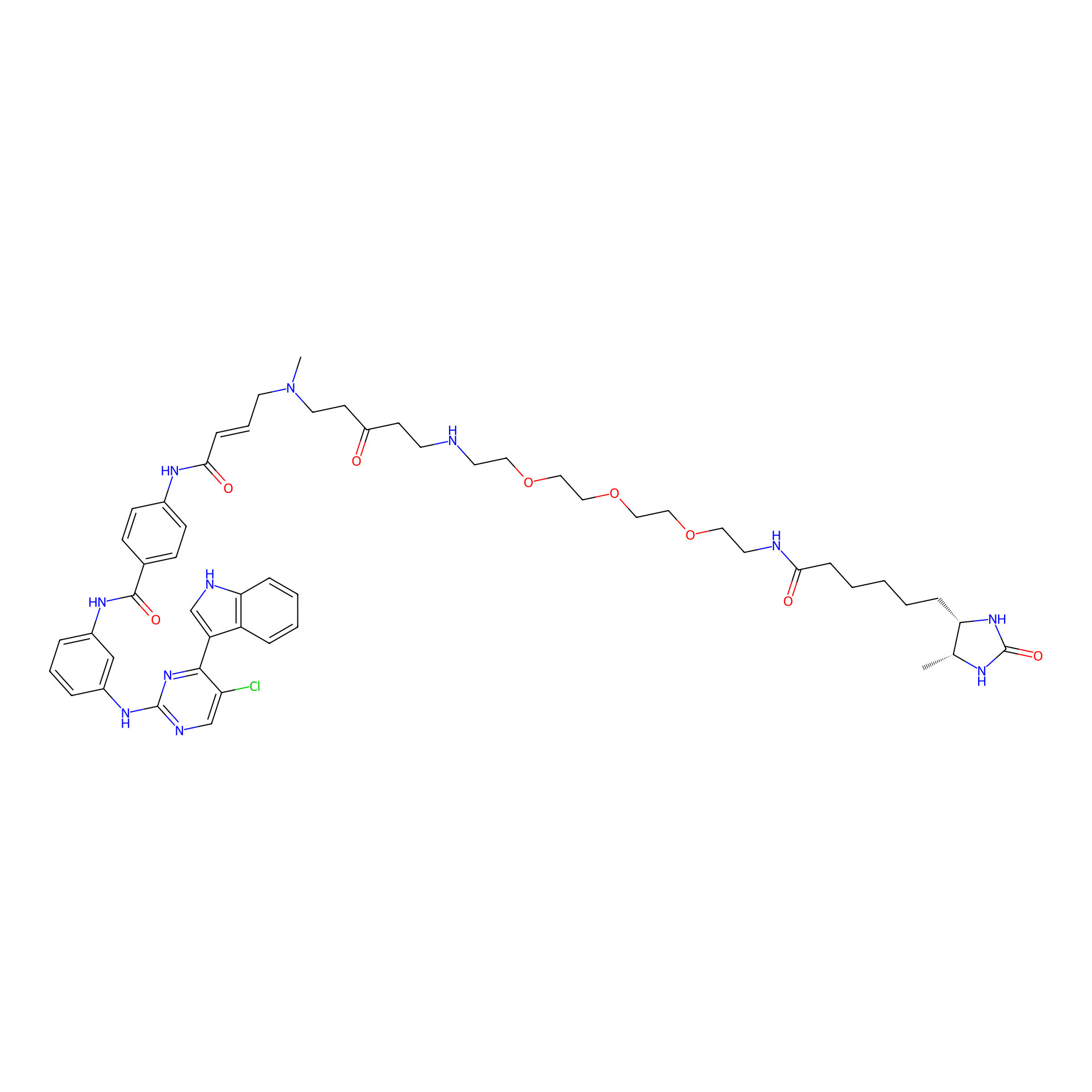 |
C157(1.10) | LDD0460 | [5] | |
|
BTD Probe Info |
 |
C178(1.35) | LDD1700 | [6] | |
|
AHL-Pu-1 Probe Info |
 |
C157(2.70) | LDD0172 | [7] | |
|
IPM Probe Info |
 |
C178(34.40) | LDD1701 | [6] | |
|
DBIA Probe Info |
 |
C178(1.40) | LDD0078 | [8] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C178(0.00); C6(0.00); C105(0.00); C81(0.00) | LDD0038 | [9] | |
|
IA-alkyne Probe Info |
 |
C178(0.00); C105(0.00); C81(0.00) | LDD0036 | [9] | |
|
IPIAA_H Probe Info |
 |
C6(0.00); C81(0.00) | LDD0030 | [10] | |
|
IPIAA_L Probe Info |
 |
C178(0.00); C6(0.00); C157(0.00); C81(0.00) | LDD0031 | [10] | |
|
Lodoacetamide azide Probe Info |
 |
C178(0.00); C6(0.00); C105(0.00); C81(0.00) | LDD0037 | [9] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [11] | |
|
TFBX Probe Info |
 |
C157(0.00); C178(0.00); C6(0.00) | LDD0027 | [12] | |
|
Compound 10 Probe Info |
 |
C105(0.00); C178(0.00) | LDD2216 | [13] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [13] | |
|
NHS Probe Info |
 |
K128(0.00); K166(0.00) | LDD0010 | [14] | |
|
PF-06672131 Probe Info |
 |
N.A. | LDD0152 | [15] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [16] | |
|
Cinnamaldehyde Probe Info |
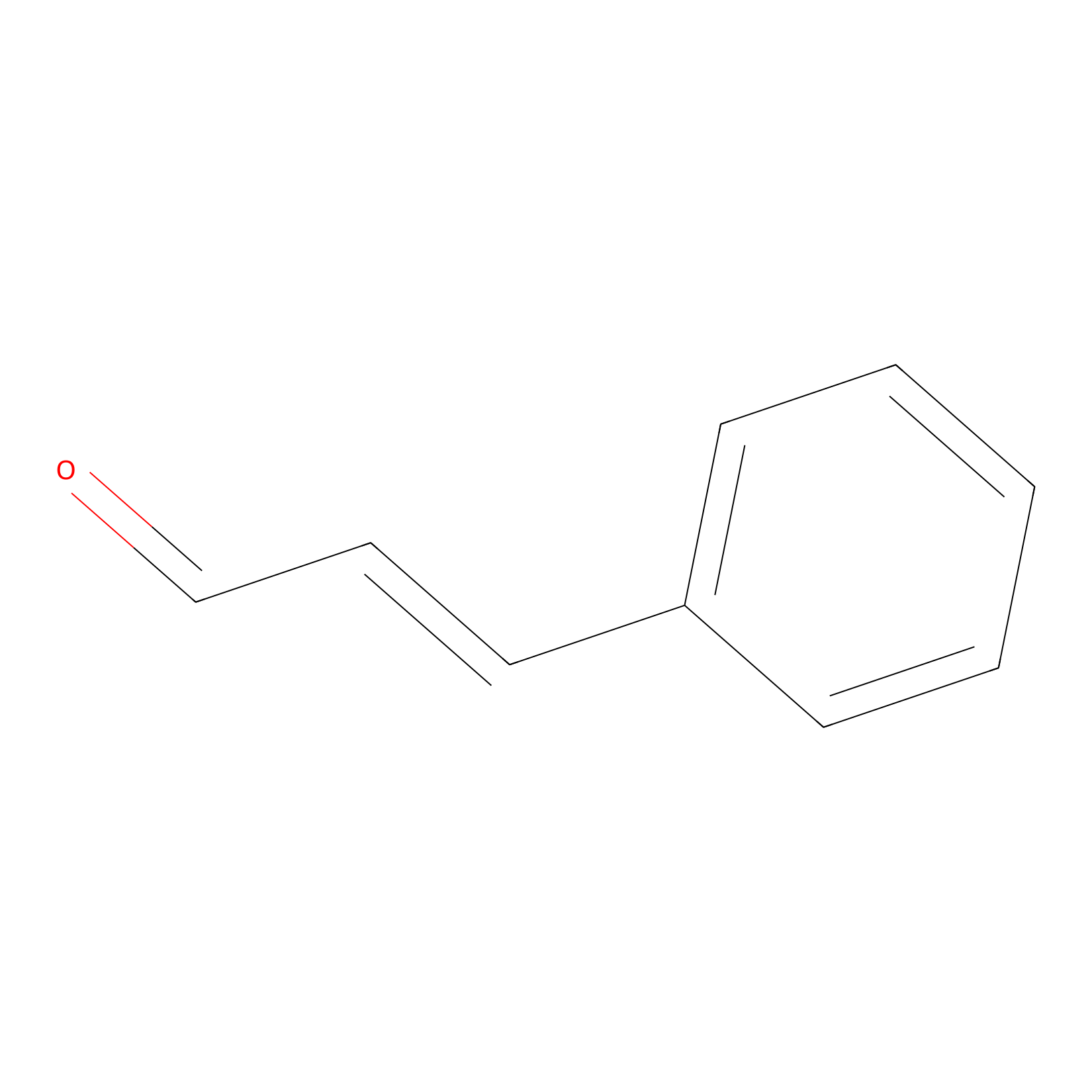 |
N.A. | LDD0220 | [16] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [16] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [16] | |
|
NAIA_5 Probe Info |
 |
C6(0.00); C157(0.00) | LDD2223 | [11] | |
|
TPP-AC Probe Info |
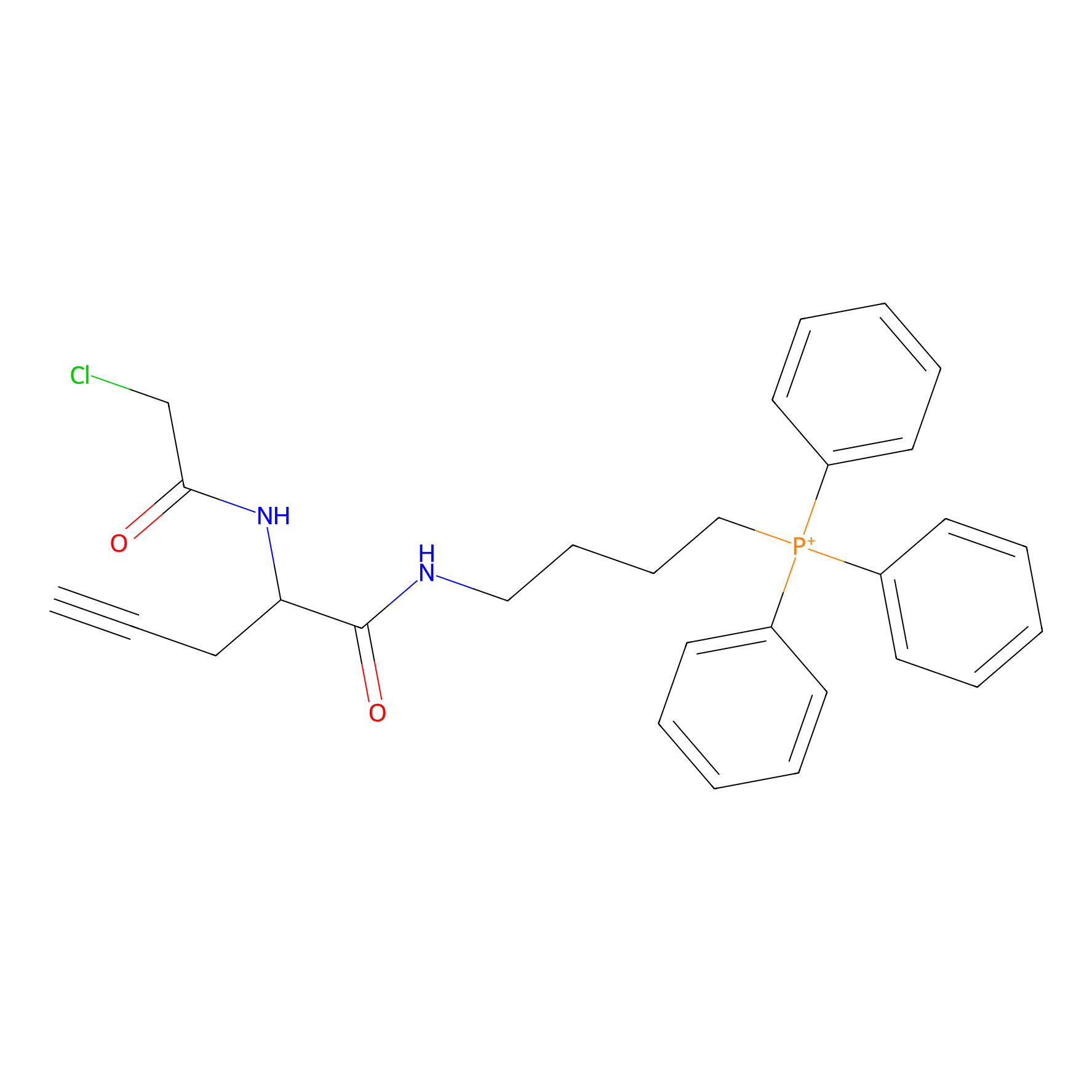 |
N.A. | LDD0427 | [17] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0537 | 2-Cyano-N,N-dimethylacetamide | MDA-MB-231 | C105(1.03) | LDD2130 | [6] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C105(1.17); C178(0.99) | LDD2117 | [6] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C178(0.93) | LDD2152 | [6] |
| LDCM0510 | 3-(4-(Hydroxydiphenylmethyl)piperidin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C178(1.39) | LDD2103 | [6] |
| LDCM0539 | 3-(4-Isopropylpiperazin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C105(0.55); C178(0.58) | LDD2132 | [6] |
| LDCM0538 | 4-(Cyanoacetyl)morpholine | MDA-MB-231 | C178(0.63) | LDD2131 | [6] |
| LDCM0025 | 4SU-RNA | HEK-293T | C157(2.70) | LDD0172 | [7] |
| LDCM0214 | AC1 | HCT 116 | C178(1.12) | LDD0531 | [8] |
| LDCM0215 | AC10 | HCT 116 | C178(0.98) | LDD0532 | [8] |
| LDCM0216 | AC100 | HCT 116 | C178(0.90) | LDD0533 | [8] |
| LDCM0217 | AC101 | HCT 116 | C178(0.91) | LDD0534 | [8] |
| LDCM0218 | AC102 | HCT 116 | C178(1.14) | LDD0535 | [8] |
| LDCM0219 | AC103 | HCT 116 | C178(0.88) | LDD0536 | [8] |
| LDCM0220 | AC104 | HCT 116 | C178(1.04) | LDD0537 | [8] |
| LDCM0221 | AC105 | HCT 116 | C178(0.86) | LDD0538 | [8] |
| LDCM0222 | AC106 | HCT 116 | C178(0.99) | LDD0539 | [8] |
| LDCM0223 | AC107 | HCT 116 | C178(0.76) | LDD0540 | [8] |
| LDCM0224 | AC108 | HCT 116 | C178(1.10) | LDD0541 | [8] |
| LDCM0225 | AC109 | HCT 116 | C178(0.95) | LDD0542 | [8] |
| LDCM0226 | AC11 | HCT 116 | C178(1.02) | LDD0543 | [8] |
| LDCM0227 | AC110 | HCT 116 | C178(1.17) | LDD0544 | [8] |
| LDCM0228 | AC111 | HCT 116 | C178(0.97) | LDD0545 | [8] |
| LDCM0229 | AC112 | HCT 116 | C178(0.80) | LDD0546 | [8] |
| LDCM0230 | AC113 | HCT 116 | C178(0.82) | LDD0547 | [8] |
| LDCM0231 | AC114 | HCT 116 | C178(0.75) | LDD0548 | [8] |
| LDCM0232 | AC115 | HCT 116 | C178(1.00) | LDD0549 | [8] |
| LDCM0233 | AC116 | HCT 116 | C178(0.82) | LDD0550 | [8] |
| LDCM0234 | AC117 | HCT 116 | C178(1.15) | LDD0551 | [8] |
| LDCM0235 | AC118 | HCT 116 | C178(0.84) | LDD0552 | [8] |
| LDCM0236 | AC119 | HCT 116 | C178(0.91) | LDD0553 | [8] |
| LDCM0237 | AC12 | HCT 116 | C178(0.84) | LDD0554 | [8] |
| LDCM0238 | AC120 | HCT 116 | C178(0.71) | LDD0555 | [8] |
| LDCM0239 | AC121 | HCT 116 | C178(0.74) | LDD0556 | [8] |
| LDCM0240 | AC122 | HCT 116 | C178(0.91) | LDD0557 | [8] |
| LDCM0241 | AC123 | HCT 116 | C178(0.68) | LDD0558 | [8] |
| LDCM0242 | AC124 | HCT 116 | C178(0.89) | LDD0559 | [8] |
| LDCM0243 | AC125 | HCT 116 | C178(0.84) | LDD0560 | [8] |
| LDCM0244 | AC126 | HCT 116 | C178(0.87) | LDD0561 | [8] |
| LDCM0245 | AC127 | HCT 116 | C178(0.78) | LDD0562 | [8] |
| LDCM0259 | AC14 | HCT 116 | C178(0.81) | LDD0576 | [8] |
| LDCM0263 | AC143 | HCT 116 | C178(1.34) | LDD0580 | [8] |
| LDCM0264 | AC144 | HCT 116 | C178(0.83) | LDD0581 | [8] |
| LDCM0265 | AC145 | HCT 116 | C178(0.94) | LDD0582 | [8] |
| LDCM0266 | AC146 | HCT 116 | C178(1.06) | LDD0583 | [8] |
| LDCM0267 | AC147 | HCT 116 | C178(0.85) | LDD0584 | [8] |
| LDCM0268 | AC148 | HCT 116 | C178(1.28) | LDD0585 | [8] |
| LDCM0269 | AC149 | HCT 116 | C178(1.26) | LDD0586 | [8] |
| LDCM0270 | AC15 | HCT 116 | C178(0.78) | LDD0587 | [8] |
| LDCM0271 | AC150 | HCT 116 | C178(0.97) | LDD0588 | [8] |
| LDCM0272 | AC151 | HCT 116 | C178(0.75) | LDD0589 | [8] |
| LDCM0273 | AC152 | HCT 116 | C178(0.95) | LDD0590 | [8] |
| LDCM0274 | AC153 | HCT 116 | C178(0.99) | LDD0591 | [8] |
| LDCM0621 | AC154 | HCT 116 | C178(0.82) | LDD2158 | [8] |
| LDCM0622 | AC155 | HCT 116 | C178(0.97) | LDD2159 | [8] |
| LDCM0623 | AC156 | HCT 116 | C178(1.01) | LDD2160 | [8] |
| LDCM0624 | AC157 | HCT 116 | C178(0.91) | LDD2161 | [8] |
| LDCM0276 | AC17 | HCT 116 | C178(0.91) | LDD0593 | [8] |
| LDCM0277 | AC18 | HCT 116 | C178(1.10) | LDD0594 | [8] |
| LDCM0278 | AC19 | HCT 116 | C178(0.93) | LDD0595 | [8] |
| LDCM0279 | AC2 | HCT 116 | C178(1.04) | LDD0596 | [8] |
| LDCM0280 | AC20 | HCT 116 | C178(1.02) | LDD0597 | [8] |
| LDCM0281 | AC21 | HCT 116 | C178(0.89) | LDD0598 | [8] |
| LDCM0282 | AC22 | HCT 116 | C178(0.98) | LDD0599 | [8] |
| LDCM0283 | AC23 | HCT 116 | C178(0.93) | LDD0600 | [8] |
| LDCM0284 | AC24 | HCT 116 | C178(0.95) | LDD0601 | [8] |
| LDCM0285 | AC25 | HCT 116 | C178(0.95) | LDD0602 | [8] |
| LDCM0286 | AC26 | HCT 116 | C178(1.05) | LDD0603 | [8] |
| LDCM0287 | AC27 | HCT 116 | C178(1.36) | LDD0604 | [8] |
| LDCM0288 | AC28 | HCT 116 | C178(1.01) | LDD0605 | [8] |
| LDCM0289 | AC29 | HCT 116 | C178(0.97) | LDD0606 | [8] |
| LDCM0290 | AC3 | HCT 116 | C178(1.06) | LDD0607 | [8] |
| LDCM0291 | AC30 | HCT 116 | C178(1.22) | LDD0608 | [8] |
| LDCM0292 | AC31 | HCT 116 | C178(1.21) | LDD0609 | [8] |
| LDCM0293 | AC32 | HCT 116 | C178(1.31) | LDD0610 | [8] |
| LDCM0294 | AC33 | HCT 116 | C178(1.20) | LDD0611 | [8] |
| LDCM0295 | AC34 | HCT 116 | C178(1.83) | LDD0612 | [8] |
| LDCM0296 | AC35 | HCT 116 | C178(0.68) | LDD0613 | [8] |
| LDCM0297 | AC36 | HCT 116 | C178(0.84) | LDD0614 | [8] |
| LDCM0298 | AC37 | HCT 116 | C178(0.97) | LDD0615 | [8] |
| LDCM0299 | AC38 | HCT 116 | C178(0.96) | LDD0616 | [8] |
| LDCM0300 | AC39 | HCT 116 | C178(0.83) | LDD0617 | [8] |
| LDCM0301 | AC4 | HCT 116 | C178(1.08) | LDD0618 | [8] |
| LDCM0302 | AC40 | HCT 116 | C178(0.83) | LDD0619 | [8] |
| LDCM0303 | AC41 | HCT 116 | C178(0.86) | LDD0620 | [8] |
| LDCM0304 | AC42 | HCT 116 | C178(0.96) | LDD0621 | [8] |
| LDCM0305 | AC43 | HCT 116 | C178(1.04) | LDD0622 | [8] |
| LDCM0306 | AC44 | HCT 116 | C178(1.03) | LDD0623 | [8] |
| LDCM0307 | AC45 | HCT 116 | C178(0.78) | LDD0624 | [8] |
| LDCM0308 | AC46 | HCT 116 | C178(1.04) | LDD0625 | [8] |
| LDCM0309 | AC47 | HCT 116 | C178(0.98) | LDD0626 | [8] |
| LDCM0310 | AC48 | HCT 116 | C178(1.00) | LDD0627 | [8] |
| LDCM0311 | AC49 | HCT 116 | C178(1.02) | LDD0628 | [8] |
| LDCM0312 | AC5 | HCT 116 | C178(1.26) | LDD0629 | [8] |
| LDCM0313 | AC50 | HCT 116 | C178(0.97) | LDD0630 | [8] |
| LDCM0314 | AC51 | HCT 116 | C178(0.93) | LDD0631 | [8] |
| LDCM0315 | AC52 | HCT 116 | C178(1.26) | LDD0632 | [8] |
| LDCM0316 | AC53 | HCT 116 | C178(0.99) | LDD0633 | [8] |
| LDCM0317 | AC54 | HCT 116 | C178(0.87) | LDD0634 | [8] |
| LDCM0318 | AC55 | HCT 116 | C178(0.98) | LDD0635 | [8] |
| LDCM0319 | AC56 | HCT 116 | C178(1.05) | LDD0636 | [8] |
| LDCM0320 | AC57 | HCT 116 | C178(1.01) | LDD0637 | [8] |
| LDCM0321 | AC58 | HCT 116 | C178(1.04) | LDD0638 | [8] |
| LDCM0322 | AC59 | HCT 116 | C178(1.13) | LDD0639 | [8] |
| LDCM0323 | AC6 | HCT 116 | C178(1.02) | LDD0640 | [8] |
| LDCM0324 | AC60 | HCT 116 | C178(1.26) | LDD0641 | [8] |
| LDCM0325 | AC61 | HCT 116 | C178(1.13) | LDD0642 | [8] |
| LDCM0326 | AC62 | HCT 116 | C178(1.18) | LDD0643 | [8] |
| LDCM0327 | AC63 | HCT 116 | C178(1.30) | LDD0644 | [8] |
| LDCM0328 | AC64 | HCT 116 | C178(1.36) | LDD0645 | [8] |
| LDCM0329 | AC65 | HCT 116 | C178(1.28) | LDD0646 | [8] |
| LDCM0330 | AC66 | HCT 116 | C178(1.09) | LDD0647 | [8] |
| LDCM0331 | AC67 | HCT 116 | C178(1.14) | LDD0648 | [8] |
| LDCM0332 | AC68 | HCT 116 | C178(0.83) | LDD0649 | [8] |
| LDCM0333 | AC69 | HCT 116 | C178(0.70) | LDD0650 | [8] |
| LDCM0334 | AC7 | HCT 116 | C178(0.87) | LDD0651 | [8] |
| LDCM0335 | AC70 | HCT 116 | C178(0.95) | LDD0652 | [8] |
| LDCM0336 | AC71 | HCT 116 | C178(0.97) | LDD0653 | [8] |
| LDCM0337 | AC72 | HCT 116 | C178(1.20) | LDD0654 | [8] |
| LDCM0338 | AC73 | HCT 116 | C178(0.97) | LDD0655 | [8] |
| LDCM0339 | AC74 | HCT 116 | C178(0.91) | LDD0656 | [8] |
| LDCM0340 | AC75 | HCT 116 | C178(1.17) | LDD0657 | [8] |
| LDCM0341 | AC76 | HCT 116 | C178(1.00) | LDD0658 | [8] |
| LDCM0342 | AC77 | HCT 116 | C178(0.82) | LDD0659 | [8] |
| LDCM0343 | AC78 | HCT 116 | C178(0.98) | LDD0660 | [8] |
| LDCM0344 | AC79 | HCT 116 | C178(1.05) | LDD0661 | [8] |
| LDCM0345 | AC8 | HCT 116 | C178(0.99) | LDD0662 | [8] |
| LDCM0346 | AC80 | HCT 116 | C178(0.94) | LDD0663 | [8] |
| LDCM0347 | AC81 | HCT 116 | C178(1.02) | LDD0664 | [8] |
| LDCM0348 | AC82 | HCT 116 | C178(1.03) | LDD0665 | [8] |
| LDCM0349 | AC83 | HCT 116 | C178(1.33) | LDD0666 | [8] |
| LDCM0350 | AC84 | HCT 116 | C178(1.30) | LDD0667 | [8] |
| LDCM0351 | AC85 | HCT 116 | C178(1.35) | LDD0668 | [8] |
| LDCM0352 | AC86 | HCT 116 | C178(1.27) | LDD0669 | [8] |
| LDCM0353 | AC87 | HCT 116 | C178(1.46) | LDD0670 | [8] |
| LDCM0354 | AC88 | HCT 116 | C178(1.38) | LDD0671 | [8] |
| LDCM0355 | AC89 | HCT 116 | C178(1.27) | LDD0672 | [8] |
| LDCM0357 | AC90 | HCT 116 | C178(1.43) | LDD0674 | [8] |
| LDCM0358 | AC91 | HCT 116 | C178(1.39) | LDD0675 | [8] |
| LDCM0359 | AC92 | HCT 116 | C178(1.50) | LDD0676 | [8] |
| LDCM0360 | AC93 | HCT 116 | C178(1.32) | LDD0677 | [8] |
| LDCM0361 | AC94 | HCT 116 | C178(1.36) | LDD0678 | [8] |
| LDCM0362 | AC95 | HCT 116 | C178(1.48) | LDD0679 | [8] |
| LDCM0363 | AC96 | HCT 116 | C178(1.44) | LDD0680 | [8] |
| LDCM0364 | AC97 | HCT 116 | C178(1.55) | LDD0681 | [8] |
| LDCM0365 | AC98 | HCT 116 | C178(1.00) | LDD0682 | [8] |
| LDCM0366 | AC99 | HCT 116 | C178(0.81) | LDD0683 | [8] |
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C178(1.02) | LDD2113 | [6] |
| LDCM0248 | AKOS034007472 | HCT 116 | C178(0.95) | LDD0565 | [8] |
| LDCM0356 | AKOS034007680 | HCT 116 | C178(1.03) | LDD0673 | [8] |
| LDCM0275 | AKOS034007705 | HCT 116 | C178(0.93) | LDD0592 | [8] |
| LDCM0020 | ARS-1620 | HCC44 | C178(1.40) | LDD0078 | [8] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C105(1.03); C178(0.43) | LDD2091 | [6] |
| LDCM0630 | CCW28-3 | 231MFP | C157(2.36) | LDD2214 | [18] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [16] |
| LDCM0367 | CL1 | HCT 116 | C178(1.12) | LDD0684 | [8] |
| LDCM0368 | CL10 | HCT 116 | C178(1.27) | LDD0685 | [8] |
| LDCM0369 | CL100 | HCT 116 | C178(1.06) | LDD0686 | [8] |
| LDCM0370 | CL101 | HCT 116 | C178(0.78) | LDD0687 | [8] |
| LDCM0371 | CL102 | HCT 116 | C178(0.71) | LDD0688 | [8] |
| LDCM0372 | CL103 | HCT 116 | C178(0.89) | LDD0689 | [8] |
| LDCM0373 | CL104 | HCT 116 | C178(0.87) | LDD0690 | [8] |
| LDCM0374 | CL105 | HCT 116 | C178(1.01) | LDD0691 | [8] |
| LDCM0375 | CL106 | HCT 116 | C178(1.06) | LDD0692 | [8] |
| LDCM0376 | CL107 | HCT 116 | C178(0.88) | LDD0693 | [8] |
| LDCM0377 | CL108 | HCT 116 | C178(0.96) | LDD0694 | [8] |
| LDCM0378 | CL109 | HCT 116 | C178(1.07) | LDD0695 | [8] |
| LDCM0379 | CL11 | HCT 116 | C178(0.88) | LDD0696 | [8] |
| LDCM0380 | CL110 | HCT 116 | C178(0.89) | LDD0697 | [8] |
| LDCM0381 | CL111 | HCT 116 | C178(1.05) | LDD0698 | [8] |
| LDCM0382 | CL112 | HCT 116 | C178(1.01) | LDD0699 | [8] |
| LDCM0383 | CL113 | HCT 116 | C178(1.04) | LDD0700 | [8] |
| LDCM0384 | CL114 | HCT 116 | C178(1.19) | LDD0701 | [8] |
| LDCM0385 | CL115 | HCT 116 | C178(1.01) | LDD0702 | [8] |
| LDCM0386 | CL116 | HCT 116 | C178(1.41) | LDD0703 | [8] |
| LDCM0387 | CL117 | HCT 116 | C178(0.74) | LDD0704 | [8] |
| LDCM0388 | CL118 | HCT 116 | C178(0.73) | LDD0705 | [8] |
| LDCM0389 | CL119 | HCT 116 | C178(1.10) | LDD0706 | [8] |
| LDCM0390 | CL12 | HCT 116 | C178(1.13) | LDD0707 | [8] |
| LDCM0391 | CL120 | HCT 116 | C178(0.93) | LDD0708 | [8] |
| LDCM0392 | CL121 | HCT 116 | C178(1.16) | LDD0709 | [8] |
| LDCM0393 | CL122 | HCT 116 | C178(1.03) | LDD0710 | [8] |
| LDCM0394 | CL123 | HCT 116 | C178(1.00) | LDD0711 | [8] |
| LDCM0395 | CL124 | HCT 116 | C178(1.08) | LDD0712 | [8] |
| LDCM0396 | CL125 | HCT 116 | C178(1.12) | LDD0713 | [8] |
| LDCM0397 | CL126 | HCT 116 | C178(1.33) | LDD0714 | [8] |
| LDCM0398 | CL127 | HCT 116 | C178(1.35) | LDD0715 | [8] |
| LDCM0399 | CL128 | HCT 116 | C178(1.14) | LDD0716 | [8] |
| LDCM0400 | CL13 | HCT 116 | C178(1.53) | LDD0717 | [8] |
| LDCM0401 | CL14 | HCT 116 | C178(1.47) | LDD0718 | [8] |
| LDCM0402 | CL15 | HCT 116 | C178(1.05) | LDD0719 | [8] |
| LDCM0403 | CL16 | HCT 116 | C178(1.47) | LDD0720 | [8] |
| LDCM0407 | CL2 | HCT 116 | C178(1.19) | LDD0724 | [8] |
| LDCM0418 | CL3 | HCT 116 | C178(1.15) | LDD0735 | [8] |
| LDCM0420 | CL31 | HCT 116 | C178(1.06) | LDD0737 | [8] |
| LDCM0421 | CL32 | HCT 116 | C178(1.26) | LDD0738 | [8] |
| LDCM0422 | CL33 | HCT 116 | C178(1.26) | LDD0739 | [8] |
| LDCM0423 | CL34 | HCT 116 | C178(1.06) | LDD0740 | [8] |
| LDCM0424 | CL35 | HCT 116 | C178(1.21) | LDD0741 | [8] |
| LDCM0425 | CL36 | HCT 116 | C178(1.00) | LDD0742 | [8] |
| LDCM0426 | CL37 | HCT 116 | C178(1.33) | LDD0743 | [8] |
| LDCM0428 | CL39 | HCT 116 | C178(1.13) | LDD0745 | [8] |
| LDCM0429 | CL4 | HCT 116 | C178(0.97) | LDD0746 | [8] |
| LDCM0430 | CL40 | HCT 116 | C178(1.09) | LDD0747 | [8] |
| LDCM0431 | CL41 | HCT 116 | C178(1.08) | LDD0748 | [8] |
| LDCM0432 | CL42 | HCT 116 | C178(1.07) | LDD0749 | [8] |
| LDCM0433 | CL43 | HCT 116 | C178(1.21) | LDD0750 | [8] |
| LDCM0434 | CL44 | HCT 116 | C178(1.25) | LDD0751 | [8] |
| LDCM0435 | CL45 | HCT 116 | C178(1.20) | LDD0752 | [8] |
| LDCM0436 | CL46 | HCT 116 | C178(1.02) | LDD0753 | [8] |
| LDCM0437 | CL47 | HCT 116 | C178(1.51) | LDD0754 | [8] |
| LDCM0438 | CL48 | HCT 116 | C178(1.14) | LDD0755 | [8] |
| LDCM0439 | CL49 | HCT 116 | C178(1.14) | LDD0756 | [8] |
| LDCM0440 | CL5 | HCT 116 | C178(1.21) | LDD0757 | [8] |
| LDCM0441 | CL50 | HCT 116 | C178(1.15) | LDD0758 | [8] |
| LDCM0442 | CL51 | HCT 116 | C178(1.10) | LDD0759 | [8] |
| LDCM0443 | CL52 | HCT 116 | C178(1.39) | LDD0760 | [8] |
| LDCM0444 | CL53 | HCT 116 | C178(1.01) | LDD0761 | [8] |
| LDCM0445 | CL54 | HCT 116 | C178(1.04) | LDD0762 | [8] |
| LDCM0446 | CL55 | HCT 116 | C178(1.15) | LDD0763 | [8] |
| LDCM0447 | CL56 | HCT 116 | C178(1.41) | LDD0764 | [8] |
| LDCM0448 | CL57 | HCT 116 | C178(1.05) | LDD0765 | [8] |
| LDCM0449 | CL58 | HCT 116 | C178(1.21) | LDD0766 | [8] |
| LDCM0450 | CL59 | HCT 116 | C178(1.22) | LDD0767 | [8] |
| LDCM0451 | CL6 | HCT 116 | C178(1.49) | LDD0768 | [8] |
| LDCM0452 | CL60 | HCT 116 | C178(1.16) | LDD0769 | [8] |
| LDCM0453 | CL61 | HCT 116 | C178(1.14) | LDD0770 | [8] |
| LDCM0454 | CL62 | HCT 116 | C178(1.15) | LDD0771 | [8] |
| LDCM0455 | CL63 | HCT 116 | C178(1.12) | LDD0772 | [8] |
| LDCM0456 | CL64 | HCT 116 | C178(1.11) | LDD0773 | [8] |
| LDCM0457 | CL65 | HCT 116 | C178(1.06) | LDD0774 | [8] |
| LDCM0458 | CL66 | HCT 116 | C178(1.12) | LDD0775 | [8] |
| LDCM0459 | CL67 | HCT 116 | C178(1.12) | LDD0776 | [8] |
| LDCM0460 | CL68 | HCT 116 | C178(1.12) | LDD0777 | [8] |
| LDCM0461 | CL69 | HCT 116 | C178(1.25) | LDD0778 | [8] |
| LDCM0462 | CL7 | HCT 116 | C178(1.27) | LDD0779 | [8] |
| LDCM0463 | CL70 | HCT 116 | C178(1.34) | LDD0780 | [8] |
| LDCM0464 | CL71 | HCT 116 | C178(1.15) | LDD0781 | [8] |
| LDCM0465 | CL72 | HCT 116 | C178(1.27) | LDD0782 | [8] |
| LDCM0466 | CL73 | HCT 116 | C178(1.27) | LDD0783 | [8] |
| LDCM0467 | CL74 | HCT 116 | C178(1.17) | LDD0784 | [8] |
| LDCM0469 | CL76 | HCT 116 | C178(1.18) | LDD0786 | [8] |
| LDCM0470 | CL77 | HCT 116 | C178(1.10) | LDD0787 | [8] |
| LDCM0471 | CL78 | HCT 116 | C178(1.12) | LDD0788 | [8] |
| LDCM0472 | CL79 | HCT 116 | C178(1.32) | LDD0789 | [8] |
| LDCM0473 | CL8 | HCT 116 | C178(1.15) | LDD0790 | [8] |
| LDCM0474 | CL80 | HCT 116 | C178(0.99) | LDD0791 | [8] |
| LDCM0475 | CL81 | HCT 116 | C178(1.07) | LDD0792 | [8] |
| LDCM0476 | CL82 | HCT 116 | C178(1.29) | LDD0793 | [8] |
| LDCM0477 | CL83 | HCT 116 | C178(1.08) | LDD0794 | [8] |
| LDCM0478 | CL84 | HCT 116 | C178(1.15) | LDD0795 | [8] |
| LDCM0479 | CL85 | HCT 116 | C178(1.12) | LDD0796 | [8] |
| LDCM0480 | CL86 | HCT 116 | C178(1.17) | LDD0797 | [8] |
| LDCM0481 | CL87 | HCT 116 | C178(1.00) | LDD0798 | [8] |
| LDCM0482 | CL88 | HCT 116 | C178(1.03) | LDD0799 | [8] |
| LDCM0483 | CL89 | HCT 116 | C178(1.15) | LDD0800 | [8] |
| LDCM0484 | CL9 | HCT 116 | C178(1.25) | LDD0801 | [8] |
| LDCM0485 | CL90 | HCT 116 | C178(0.98) | LDD0802 | [8] |
| LDCM0486 | CL91 | HCT 116 | C178(1.03) | LDD0803 | [8] |
| LDCM0487 | CL92 | HCT 116 | C178(0.81) | LDD0804 | [8] |
| LDCM0488 | CL93 | HCT 116 | C178(1.32) | LDD0805 | [8] |
| LDCM0489 | CL94 | HCT 116 | C178(1.44) | LDD0806 | [8] |
| LDCM0490 | CL95 | HCT 116 | C178(0.91) | LDD0807 | [8] |
| LDCM0491 | CL96 | HCT 116 | C178(1.12) | LDD0808 | [8] |
| LDCM0492 | CL97 | HCT 116 | C178(1.48) | LDD0809 | [8] |
| LDCM0493 | CL98 | HCT 116 | C178(1.20) | LDD0810 | [8] |
| LDCM0494 | CL99 | HCT 116 | C178(0.97) | LDD0811 | [8] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C178(10.26); C105(6.69) | LDD1702 | [6] |
| LDCM0625 | F8 | Ramos | C178(0.57); C157(0.59); C105(8.56) | LDD2187 | [19] |
| LDCM0572 | Fragment10 | Ramos | C178(1.18); C157(0.57); C105(1.44) | LDD2189 | [19] |
| LDCM0573 | Fragment11 | Ramos | C178(0.02); C157(0.02); C105(0.06) | LDD2190 | [19] |
| LDCM0574 | Fragment12 | Ramos | C178(1.12); C157(0.80); C105(1.05) | LDD2191 | [19] |
| LDCM0575 | Fragment13 | Ramos | C178(0.85); C157(0.53); C105(0.46) | LDD2192 | [19] |
| LDCM0576 | Fragment14 | Ramos | C178(0.77); C157(0.73); C105(1.10) | LDD2193 | [19] |
| LDCM0579 | Fragment20 | Ramos | C178(1.05); C157(0.85); C105(3.38) | LDD2194 | [19] |
| LDCM0580 | Fragment21 | Ramos | C178(0.72); C157(0.69); C105(0.40) | LDD2195 | [19] |
| LDCM0582 | Fragment23 | Ramos | C178(1.61); C157(0.49); C105(0.94) | LDD2196 | [19] |
| LDCM0578 | Fragment27 | Ramos | C178(0.67); C157(0.61); C105(1.13) | LDD2197 | [19] |
| LDCM0586 | Fragment28 | Ramos | C178(1.34); C157(7.44); C105(0.54) | LDD2198 | [19] |
| LDCM0588 | Fragment30 | Ramos | C178(1.09); C157(0.55); C105(0.46) | LDD2199 | [19] |
| LDCM0589 | Fragment31 | Ramos | C178(0.92); C157(0.76); C105(0.89) | LDD2200 | [19] |
| LDCM0590 | Fragment32 | Ramos | C178(1.15); C157(0.46); C105(1.23) | LDD2201 | [19] |
| LDCM0468 | Fragment33 | HCT 116 | C178(1.31) | LDD0785 | [8] |
| LDCM0596 | Fragment38 | Ramos | C178(1.08); C157(0.40); C105(2.00) | LDD2203 | [19] |
| LDCM0566 | Fragment4 | Ramos | C178(1.05); C157(0.90); C105(1.10) | LDD2184 | [19] |
| LDCM0427 | Fragment51 | HCT 116 | C178(1.25) | LDD0744 | [8] |
| LDCM0610 | Fragment52 | Ramos | C178(1.16); C157(0.61); C105(0.73) | LDD2204 | [19] |
| LDCM0614 | Fragment56 | Ramos | C178(1.06); C157(0.85); C105(0.17) | LDD2205 | [19] |
| LDCM0569 | Fragment7 | Ramos | C178(0.92); C157(0.79); C105(0.85) | LDD2186 | [19] |
| LDCM0571 | Fragment9 | Ramos | C178(1.28); C157(1.09); C105(0.58) | LDD2188 | [19] |
| LDCM0015 | HNE | MDA-MB-231 | C178(1.00); C157(0.91) | LDD0346 | [19] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [16] |
| LDCM0179 | JZ128 | PC-3 | C157(0.00); C178(0.00) | LDD0462 | [5] |
| LDCM0022 | KB02 | Ramos | C178(1.09); C157(0.50); C105(0.73) | LDD2182 | [19] |
| LDCM0023 | KB03 | MDA-MB-231 | C178(34.40) | LDD1701 | [6] |
| LDCM0024 | KB05 | G361 | C178(3.99); C176(1.43) | LDD3311 | [20] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C178(0.60) | LDD2121 | [6] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0224 | [16] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C178(0.53) | LDD2089 | [6] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C178(1.86) | LDD2090 | [6] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C178(0.98) | LDD2093 | [6] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C105(0.89) | LDD2094 | [6] |
| LDCM0503 | Nucleophilic fragment 14b | MDA-MB-231 | C105(0.45) | LDD2096 | [6] |
| LDCM0504 | Nucleophilic fragment 15a | MDA-MB-231 | C105(1.02); C178(1.05) | LDD2097 | [6] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C105(1.10) | LDD2098 | [6] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C105(1.12); C178(1.21) | LDD2099 | [6] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C178(1.36) | LDD2101 | [6] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C105(0.97) | LDD2104 | [6] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C105(1.18); C178(1.36) | LDD2105 | [6] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C105(1.07); C178(0.86) | LDD2107 | [6] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C105(0.97); C178(0.80) | LDD2109 | [6] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C178(0.89) | LDD2111 | [6] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C105(0.75) | LDD2114 | [6] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C105(0.65) | LDD2115 | [6] |
| LDCM0523 | Nucleophilic fragment 24b | MDA-MB-231 | C105(0.79) | LDD2116 | [6] |
| LDCM0525 | Nucleophilic fragment 25b | MDA-MB-231 | C105(0.91); C178(0.38) | LDD2118 | [6] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C178(2.25) | LDD2119 | [6] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C105(0.70); C178(0.93) | LDD2120 | [6] |
| LDCM0529 | Nucleophilic fragment 27b | MDA-MB-231 | C105(1.00); C178(0.23) | LDD2122 | [6] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C105(1.11); C178(0.87) | LDD2123 | [6] |
| LDCM0531 | Nucleophilic fragment 28b | MDA-MB-231 | C105(0.92); C178(0.31) | LDD2124 | [6] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C105(0.98); C178(0.74) | LDD2125 | [6] |
| LDCM0533 | Nucleophilic fragment 29b | MDA-MB-231 | C105(1.00); C178(0.45) | LDD2126 | [6] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C178(0.90) | LDD2127 | [6] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C105(0.78); C178(0.98) | LDD2128 | [6] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C105(1.21); C178(0.81) | LDD2129 | [6] |
| LDCM0540 | Nucleophilic fragment 35 | MDA-MB-231 | C178(0.72) | LDD2133 | [6] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C105(0.43); C178(0.65) | LDD2134 | [6] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C178(0.95) | LDD2135 | [6] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C178(1.11) | LDD2136 | [6] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C105(1.02); C178(0.83) | LDD2137 | [6] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C178(1.35) | LDD1700 | [6] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C105(0.80); C178(0.62) | LDD2140 | [6] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C105(0.86); C178(1.04) | LDD2143 | [6] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C105(0.95); C178(0.89) | LDD2146 | [6] |
| LDCM0553 | Nucleophilic fragment 6b | MDA-MB-231 | C178(3.90) | LDD2147 | [6] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C105(0.63); C178(0.60) | LDD2148 | [6] |
| LDCM0555 | Nucleophilic fragment 7b | MDA-MB-231 | C105(0.84); C178(0.38) | LDD2149 | [6] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C105(0.99); C178(0.41) | LDD2150 | [6] |
| LDCM0557 | Nucleophilic fragment 8b | MDA-MB-231 | C105(0.80) | LDD2151 | [6] |
| LDCM0559 | Nucleophilic fragment 9b | MDA-MB-231 | C178(1.27) | LDD2153 | [6] |
| LDCM0131 | RA190 | MM1.R | C178(1.12) | LDD0304 | [21] |
| LDCM0021 | THZ1 | HeLa S3 | C157(1.10) | LDD0460 | [5] |
The Interaction Atlas With This Target
References
