Details of the Target
General Information of Target
| Target ID | LDTP12190 | |||||
|---|---|---|---|---|---|---|
| Target Name | NmrA-like family domain-containing protein 1 (NMRAL1) | |||||
| Gene Name | NMRAL1 | |||||
| Gene ID | 57407 | |||||
| Synonyms |
HSCARG; NmrA-like family domain-containing protein 1 |
|||||
| 3D Structure | ||||||
| Sequence |
MTWICLSCMLWPEDLEAPKTHRFKVKTFKKVKPCGICRQVITQEGCTCKVCSFSCHRKCQ
AKVAAPCVPPSNHELVPITTENAPKNVVDKGEGASRGGNTRKSLEDNGSTRVTPSVQPHL QPIRNMSVSRTMEDSCELDLVYVTERIIAVSFPSTANEENFRSNLREVAQMLKSKHGGNY LLFNLSERRPDITKLHAKVLEFGWPDLHTPALEKICSICKAMDTWLNADPHNVVVLHNKG NRGRIGVVIAAYMHYSNISASADQALDRFAMKRFYEDKIVPIGQPSQRRYVHYFSGLLSG SIKMNNKPLFLHHVIMHGIPNFESKGGCRPFLRIYQAMQPVYTSGIYNIPGDSQTSVCIT IEPGLLLKGDILLKCYHKKFRSPARDVIFRVQFHTCAIHDLGVVFGKEDLDDAFKDDRFP EYGKVEFVFSYGPEKIQGMEHLENGPSVSVDYNTSDPLIRWDSYDNFSGHRDDGMEEVVG HTQGPLDGSLYAKVKKKDSLHGSTGAVNATRPTLSATPNHVEHTLSVSSDSGNSTASTKT DKTDEPVPGASSATAALSPQEKRELDRLLSGFGLEREKQGAMYHTQHLRSRPAGGSAVPS SGRHVVPAQVHVNGGALASERETDILDDELPNQDGHSAGSMGTLSSLDGVTNTSEGGYPE ALSPLTNGLDKSYPMEPMVNGGGYPYESASRAGPAHAGHTAPMRPSYSAQEGLAGYQREG PHPAWPQPVTTSHYAHDPSGMFRSQSFSEAEPQLPPAPVRGGSSREAVQRGLNSWQQQQQ QQQQPRPPPRQQERAHLESLVASRPSPQPLAETPIPSLPEFPRAASQQEIEQSIETLNML MLDLEPASAAAPLHKSQSVPGAWPGASPLSSQPLSGSSRQSHPLTQSRSGYIPSGHSLGT PEPAPRASLESVPPGRSYSPYDYQPCLAGPNQDFHSKSPASSSLPAFLPTTHSPPGPQQP PASLPGLTAQPLLSPKEATSDPSRTPEEEPLNLEGLVAHRVAGVQAREKQPAEPPAPLRR RAASDGQYENQSPEATSPRSPGVRSPVQCVSPELALTIALNPGGRPKEPHLHSYKEAFEE MEGTSPSSPPPSGVRSPPGLAKTPLSALGLKPHNPADILLHPTGEPRSYVESVARTAVAG PRAQDSEPKSFSAPATQAYGHEIPLRNGTLGGSFVSPSPLSTSSPILSADSTSVGSFPSG ESSDQGPRTPTQPLLESGFRSGSLGQPSPSAQRNYQSSSPLPTVGSSYSSPDYSLQHFSS SPESQARAQFSVAGVHTVPGSPQARHRTVGTNTPPSPGFGWRAINPSMAAPSSPSLSHHQ MMGPPGTGFHGSTVSSPQSSAATTPGSPSLCRHPAGVYQVSGLHNKVATTPGSPSLGRHP GAHQGNLASGLHSNAIASPGSPSLGRHLGGSGSVVPGSPCLDRHVAYGGYSTPEDRRPTL SRQSSASGYQAPSTPSFPVSPAYYPGLSSPATSPSPDSAAFRQGSPTPALPEKRRMSVGD RAGSLPNYATINGKVSSPVASGMSSPSGGSTVSFSHTLPDFSKYSMPDNSPETRAKVKFV QDTSKYWYKPEISREQAIALLKDQEPGAFIIRDSHSFRGAYGLAMKVSSPPPTIMQQNKK GDMTHELVRHFLIETGPRGVKLKGCPNEPNFGSLSALVYQHSIIPLALPCKLVIPNRDPT DESKDSSGPANSTADLLKQGAACNVLFVNSVDMESLTGPQAISKATSETLAADPTPAATI VHFKVSAQGITLTDNQRKLFFRRHYPLNTVTFCDLDPQERKWMKTEGGAPAKLFGFVARK QGSTTDNACHLFAELDPNQPASAIVNFVSKVMLNAGQKR |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
NmrA-type oxidoreductase family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Redox sensor protein. Undergoes restructuring and subcellular redistribution in response to changes in intracellular NADPH/NADP(+) levels. At low NADPH concentrations the protein is found mainly as a monomer, and binds argininosuccinate synthase (ASS1), the enzyme involved in nitric oxide synthesis. Association with ASS1 impairs its activity and reduces the production of nitric oxide, which subsecuently prevents apoptosis. Under normal NADPH concentrations, the protein is found as a dimer and hides the binding site for ASS1. The homodimer binds one molecule of NADPH. Has higher affinity for NADPH than for NADP(+). Binding to NADPH is necessary to form a stable dimer.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
DBIA Probe Info |
 |
C217(1.20) | LDD0078 | [2] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [3] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C154(0.00); C217(0.00) | LDD0038 | [4] | |
|
IA-alkyne Probe Info |
 |
C154(0.00); C217(0.00) | LDD0036 | [4] | |
|
Lodoacetamide azide Probe Info |
 |
C154(0.00); C217(0.00) | LDD0037 | [4] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [5] | |
|
Compound 10 Probe Info |
 |
N.A. | LDD2216 | [6] | |
|
IPM Probe Info |
 |
N.A. | LDD0147 | [5] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [7] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [8] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [9] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [10] | |
|
Kambe_22 Probe Info |
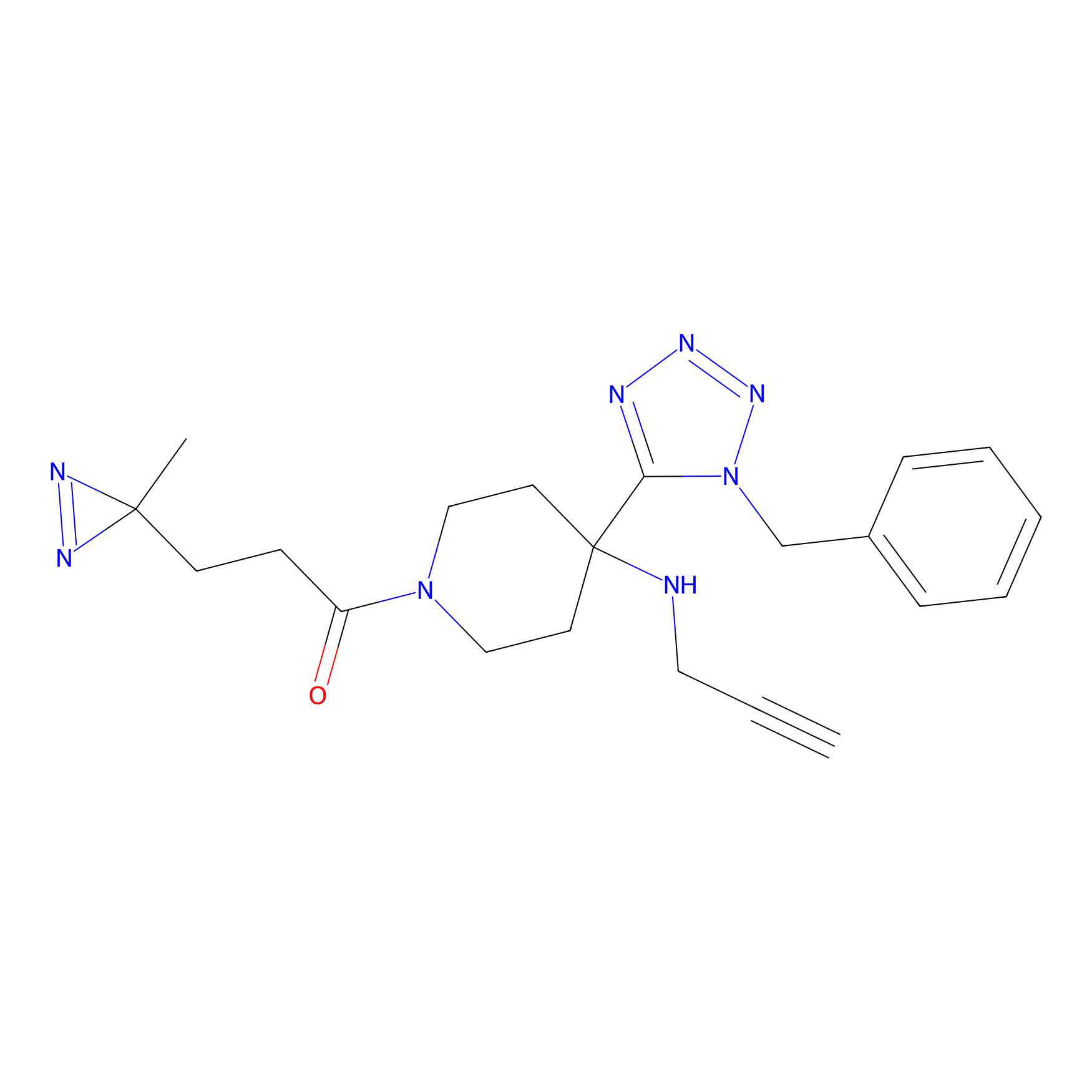 |
2.28 | LDD0132 | [11] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C154(2.03) | LDD0372 | [12] |
| LDCM0214 | AC1 | HCT 116 | C217(0.91) | LDD0531 | [2] |
| LDCM0215 | AC10 | HCT 116 | C217(1.14) | LDD0532 | [2] |
| LDCM0216 | AC100 | HCT 116 | C217(0.73) | LDD0533 | [2] |
| LDCM0217 | AC101 | HCT 116 | C217(0.70) | LDD0534 | [2] |
| LDCM0218 | AC102 | HCT 116 | C217(0.64) | LDD0535 | [2] |
| LDCM0219 | AC103 | HCT 116 | C217(0.59) | LDD0536 | [2] |
| LDCM0220 | AC104 | HCT 116 | C217(0.68) | LDD0537 | [2] |
| LDCM0221 | AC105 | HCT 116 | C217(0.60) | LDD0538 | [2] |
| LDCM0222 | AC106 | HCT 116 | C217(0.67) | LDD0539 | [2] |
| LDCM0223 | AC107 | HCT 116 | C217(0.67) | LDD0540 | [2] |
| LDCM0224 | AC108 | HCT 116 | C217(0.60) | LDD0541 | [2] |
| LDCM0225 | AC109 | HCT 116 | C217(0.61) | LDD0542 | [2] |
| LDCM0226 | AC11 | HCT 116 | C217(1.37) | LDD0543 | [2] |
| LDCM0227 | AC110 | HCT 116 | C217(0.64) | LDD0544 | [2] |
| LDCM0228 | AC111 | HCT 116 | C217(0.61) | LDD0545 | [2] |
| LDCM0229 | AC112 | HCT 116 | C217(0.61) | LDD0546 | [2] |
| LDCM0230 | AC113 | HCT 116 | C217(1.41) | LDD0547 | [2] |
| LDCM0231 | AC114 | HCT 116 | C217(0.93) | LDD0548 | [2] |
| LDCM0232 | AC115 | HCT 116 | C217(0.71) | LDD0549 | [2] |
| LDCM0233 | AC116 | HCT 116 | C217(0.74) | LDD0550 | [2] |
| LDCM0234 | AC117 | HCT 116 | C217(0.99) | LDD0551 | [2] |
| LDCM0235 | AC118 | HCT 116 | C217(1.05) | LDD0552 | [2] |
| LDCM0236 | AC119 | HCT 116 | C217(0.79) | LDD0553 | [2] |
| LDCM0237 | AC12 | HCT 116 | C217(1.30) | LDD0554 | [2] |
| LDCM0238 | AC120 | HCT 116 | C217(1.04) | LDD0555 | [2] |
| LDCM0239 | AC121 | HCT 116 | C217(1.02) | LDD0556 | [2] |
| LDCM0240 | AC122 | HCT 116 | C217(0.91) | LDD0557 | [2] |
| LDCM0241 | AC123 | HCT 116 | C217(0.99) | LDD0558 | [2] |
| LDCM0242 | AC124 | HCT 116 | C217(1.34) | LDD0559 | [2] |
| LDCM0243 | AC125 | HCT 116 | C217(0.87) | LDD0560 | [2] |
| LDCM0244 | AC126 | HCT 116 | C217(0.74) | LDD0561 | [2] |
| LDCM0245 | AC127 | HCT 116 | C217(0.76) | LDD0562 | [2] |
| LDCM0246 | AC128 | HCT 116 | C217(0.60) | LDD0563 | [2] |
| LDCM0247 | AC129 | HCT 116 | C217(1.17) | LDD0564 | [2] |
| LDCM0249 | AC130 | HCT 116 | C217(0.79) | LDD0566 | [2] |
| LDCM0250 | AC131 | HCT 116 | C217(1.00) | LDD0567 | [2] |
| LDCM0251 | AC132 | HCT 116 | C217(1.17) | LDD0568 | [2] |
| LDCM0252 | AC133 | HCT 116 | C217(0.77) | LDD0569 | [2] |
| LDCM0253 | AC134 | HCT 116 | C217(0.72) | LDD0570 | [2] |
| LDCM0254 | AC135 | HCT 116 | C217(0.73) | LDD0571 | [2] |
| LDCM0255 | AC136 | HCT 116 | C217(0.80) | LDD0572 | [2] |
| LDCM0256 | AC137 | HCT 116 | C217(1.10) | LDD0573 | [2] |
| LDCM0257 | AC138 | HCT 116 | C217(0.81) | LDD0574 | [2] |
| LDCM0258 | AC139 | HCT 116 | C217(1.22) | LDD0575 | [2] |
| LDCM0259 | AC14 | HCT 116 | C217(1.23) | LDD0576 | [2] |
| LDCM0260 | AC140 | HCT 116 | C217(0.80) | LDD0577 | [2] |
| LDCM0261 | AC141 | HCT 116 | C217(0.91) | LDD0578 | [2] |
| LDCM0262 | AC142 | HCT 116 | C217(1.15) | LDD0579 | [2] |
| LDCM0263 | AC143 | HCT 116 | C217(1.08) | LDD0580 | [2] |
| LDCM0264 | AC144 | HCT 116 | C217(0.52) | LDD0581 | [2] |
| LDCM0265 | AC145 | HCT 116 | C217(0.60) | LDD0582 | [2] |
| LDCM0266 | AC146 | HCT 116 | C217(0.51) | LDD0583 | [2] |
| LDCM0267 | AC147 | HCT 116 | C217(0.79) | LDD0584 | [2] |
| LDCM0268 | AC148 | HCT 116 | C217(0.49) | LDD0585 | [2] |
| LDCM0269 | AC149 | HCT 116 | C217(0.56) | LDD0586 | [2] |
| LDCM0270 | AC15 | HCT 116 | C217(1.03) | LDD0587 | [2] |
| LDCM0271 | AC150 | HCT 116 | C217(0.74) | LDD0588 | [2] |
| LDCM0272 | AC151 | HCT 116 | C217(0.78) | LDD0589 | [2] |
| LDCM0273 | AC152 | HCT 116 | C217(0.57) | LDD0590 | [2] |
| LDCM0274 | AC153 | HCT 116 | C217(0.51) | LDD0591 | [2] |
| LDCM0621 | AC154 | HCT 116 | C217(0.65) | LDD2158 | [2] |
| LDCM0622 | AC155 | HCT 116 | C217(0.59) | LDD2159 | [2] |
| LDCM0623 | AC156 | HCT 116 | C217(1.09) | LDD2160 | [2] |
| LDCM0624 | AC157 | HCT 116 | C217(0.90) | LDD2161 | [2] |
| LDCM0276 | AC17 | HCT 116 | C217(0.90) | LDD0593 | [2] |
| LDCM0277 | AC18 | HCT 116 | C217(0.81) | LDD0594 | [2] |
| LDCM0278 | AC19 | HCT 116 | C217(0.83) | LDD0595 | [2] |
| LDCM0279 | AC2 | HCT 116 | C217(0.84) | LDD0596 | [2] |
| LDCM0280 | AC20 | HCT 116 | C217(0.89) | LDD0597 | [2] |
| LDCM0281 | AC21 | HCT 116 | C217(0.74) | LDD0598 | [2] |
| LDCM0282 | AC22 | HCT 116 | C217(1.14) | LDD0599 | [2] |
| LDCM0283 | AC23 | HCT 116 | C217(1.20) | LDD0600 | [2] |
| LDCM0284 | AC24 | HCT 116 | C217(1.08) | LDD0601 | [2] |
| LDCM0285 | AC25 | PaTu 8988t | C217(0.77) | LDD1164 | [2] |
| LDCM0286 | AC26 | PaTu 8988t | C217(0.55) | LDD1165 | [2] |
| LDCM0287 | AC27 | PaTu 8988t | C217(0.59) | LDD1166 | [2] |
| LDCM0288 | AC28 | PaTu 8988t | C217(0.52) | LDD1167 | [2] |
| LDCM0289 | AC29 | PaTu 8988t | C217(0.71) | LDD1168 | [2] |
| LDCM0290 | AC3 | HCT 116 | C217(0.91) | LDD0607 | [2] |
| LDCM0291 | AC30 | PaTu 8988t | C217(0.75) | LDD1170 | [2] |
| LDCM0292 | AC31 | PaTu 8988t | C217(0.83) | LDD1171 | [2] |
| LDCM0293 | AC32 | PaTu 8988t | C217(0.82) | LDD1172 | [2] |
| LDCM0294 | AC33 | PaTu 8988t | C217(0.64) | LDD1173 | [2] |
| LDCM0295 | AC34 | PaTu 8988t | C217(0.63) | LDD1174 | [2] |
| LDCM0296 | AC35 | HCT 116 | C217(0.90) | LDD0613 | [2] |
| LDCM0297 | AC36 | HCT 116 | C217(0.81) | LDD0614 | [2] |
| LDCM0298 | AC37 | HCT 116 | C217(0.79) | LDD0615 | [2] |
| LDCM0299 | AC38 | HCT 116 | C217(1.09) | LDD0616 | [2] |
| LDCM0300 | AC39 | HCT 116 | C217(1.07) | LDD0617 | [2] |
| LDCM0301 | AC4 | HCT 116 | C217(0.95) | LDD0618 | [2] |
| LDCM0302 | AC40 | HCT 116 | C217(0.93) | LDD0619 | [2] |
| LDCM0303 | AC41 | HCT 116 | C217(0.90) | LDD0620 | [2] |
| LDCM0304 | AC42 | HCT 116 | C217(1.04) | LDD0621 | [2] |
| LDCM0305 | AC43 | HCT 116 | C217(1.24) | LDD0622 | [2] |
| LDCM0306 | AC44 | HCT 116 | C217(0.82) | LDD0623 | [2] |
| LDCM0307 | AC45 | HCT 116 | C217(0.76) | LDD0624 | [2] |
| LDCM0308 | AC46 | HCT 116 | C217(0.81) | LDD0625 | [2] |
| LDCM0309 | AC47 | HCT 116 | C217(0.72) | LDD0626 | [2] |
| LDCM0310 | AC48 | HCT 116 | C217(0.74) | LDD0627 | [2] |
| LDCM0311 | AC49 | HCT 116 | C217(0.68) | LDD0628 | [2] |
| LDCM0312 | AC5 | HCT 116 | C217(1.59) | LDD0629 | [2] |
| LDCM0313 | AC50 | HCT 116 | C217(0.62) | LDD0630 | [2] |
| LDCM0314 | AC51 | HCT 116 | C217(1.34) | LDD0631 | [2] |
| LDCM0315 | AC52 | HCT 116 | C217(1.12) | LDD0632 | [2] |
| LDCM0316 | AC53 | HCT 116 | C217(0.77) | LDD0633 | [2] |
| LDCM0317 | AC54 | HCT 116 | C217(0.79) | LDD0634 | [2] |
| LDCM0318 | AC55 | HCT 116 | C217(0.98) | LDD0635 | [2] |
| LDCM0319 | AC56 | HCT 116 | C217(0.97) | LDD0636 | [2] |
| LDCM0320 | AC57 | HCT 116 | C217(0.26) | LDD0637 | [2] |
| LDCM0321 | AC58 | HCT 116 | C217(0.27) | LDD0638 | [2] |
| LDCM0322 | AC59 | HCT 116 | C217(0.26) | LDD0639 | [2] |
| LDCM0323 | AC6 | HCT 116 | C217(0.93) | LDD0640 | [2] |
| LDCM0324 | AC60 | HCT 116 | C217(0.38) | LDD0641 | [2] |
| LDCM0325 | AC61 | HCT 116 | C217(0.29) | LDD0642 | [2] |
| LDCM0326 | AC62 | HCT 116 | C217(0.22) | LDD0643 | [2] |
| LDCM0327 | AC63 | HCT 116 | C217(0.38) | LDD0644 | [2] |
| LDCM0328 | AC64 | HCT 116 | C217(0.28) | LDD0645 | [2] |
| LDCM0329 | AC65 | HCT 116 | C217(0.38) | LDD0646 | [2] |
| LDCM0330 | AC66 | HCT 116 | C217(0.36) | LDD0647 | [2] |
| LDCM0331 | AC67 | HCT 116 | C217(0.34) | LDD0648 | [2] |
| LDCM0332 | AC68 | HCT 116 | C217(0.92) | LDD0649 | [2] |
| LDCM0333 | AC69 | HCT 116 | C217(0.95) | LDD0650 | [2] |
| LDCM0334 | AC7 | HCT 116 | C217(0.96) | LDD0651 | [2] |
| LDCM0335 | AC70 | HCT 116 | C217(0.96) | LDD0652 | [2] |
| LDCM0336 | AC71 | HCT 116 | C217(0.93) | LDD0653 | [2] |
| LDCM0337 | AC72 | HCT 116 | C217(1.02) | LDD0654 | [2] |
| LDCM0338 | AC73 | HCT 116 | C217(0.99) | LDD0655 | [2] |
| LDCM0339 | AC74 | HCT 116 | C217(1.22) | LDD0656 | [2] |
| LDCM0340 | AC75 | HCT 116 | C217(1.16) | LDD0657 | [2] |
| LDCM0341 | AC76 | HCT 116 | C217(0.85) | LDD0658 | [2] |
| LDCM0342 | AC77 | HCT 116 | C217(0.74) | LDD0659 | [2] |
| LDCM0343 | AC78 | HCT 116 | C217(0.84) | LDD0660 | [2] |
| LDCM0344 | AC79 | HCT 116 | C217(1.02) | LDD0661 | [2] |
| LDCM0345 | AC8 | HCT 116 | C217(1.03) | LDD0662 | [2] |
| LDCM0346 | AC80 | HCT 116 | C217(1.00) | LDD0663 | [2] |
| LDCM0347 | AC81 | HCT 116 | C217(0.84) | LDD0664 | [2] |
| LDCM0348 | AC82 | HCT 116 | C217(1.02) | LDD0665 | [2] |
| LDCM0349 | AC83 | HCT 116 | C217(0.64) | LDD0666 | [2] |
| LDCM0350 | AC84 | HCT 116 | C217(0.65) | LDD0667 | [2] |
| LDCM0351 | AC85 | HCT 116 | C217(0.74) | LDD0668 | [2] |
| LDCM0352 | AC86 | HCT 116 | C217(0.92) | LDD0669 | [2] |
| LDCM0353 | AC87 | HCT 116 | C217(1.00) | LDD0670 | [2] |
| LDCM0354 | AC88 | HCT 116 | C217(0.84) | LDD0671 | [2] |
| LDCM0355 | AC89 | HCT 116 | C217(0.76) | LDD0672 | [2] |
| LDCM0357 | AC90 | HCT 116 | C217(1.48) | LDD0674 | [2] |
| LDCM0358 | AC91 | HCT 116 | C217(0.74) | LDD0675 | [2] |
| LDCM0359 | AC92 | HCT 116 | C217(0.62) | LDD0676 | [2] |
| LDCM0360 | AC93 | HCT 116 | C217(0.85) | LDD0677 | [2] |
| LDCM0361 | AC94 | HCT 116 | C217(0.87) | LDD0678 | [2] |
| LDCM0362 | AC95 | HCT 116 | C217(0.77) | LDD0679 | [2] |
| LDCM0363 | AC96 | HCT 116 | C217(0.77) | LDD0680 | [2] |
| LDCM0364 | AC97 | HCT 116 | C217(0.64) | LDD0681 | [2] |
| LDCM0365 | AC98 | HCT 116 | C217(0.58) | LDD0682 | [2] |
| LDCM0366 | AC99 | HCT 116 | C217(0.81) | LDD0683 | [2] |
| LDCM0248 | AKOS034007472 | HCT 116 | C217(1.24) | LDD0565 | [2] |
| LDCM0356 | AKOS034007680 | HCT 116 | C217(1.23) | LDD0673 | [2] |
| LDCM0275 | AKOS034007705 | HCT 116 | C217(0.88) | LDD0592 | [2] |
| LDCM0156 | Aniline | NCI-H1299 | 14.99 | LDD0403 | [1] |
| LDCM0020 | ARS-1620 | HCC44 | C217(1.20) | LDD0078 | [2] |
| LDCM0632 | CL-Sc | Hep-G2 | C154(0.68) | LDD2227 | [9] |
| LDCM0367 | CL1 | HCT 116 | C217(0.96) | LDD0684 | [2] |
| LDCM0368 | CL10 | HCT 116 | C217(0.73) | LDD0685 | [2] |
| LDCM0369 | CL100 | HCT 116 | C217(0.87) | LDD0686 | [2] |
| LDCM0370 | CL101 | HCT 116 | C217(1.10) | LDD0687 | [2] |
| LDCM0371 | CL102 | HCT 116 | C217(1.17) | LDD0688 | [2] |
| LDCM0372 | CL103 | HCT 116 | C217(1.13) | LDD0689 | [2] |
| LDCM0373 | CL104 | HCT 116 | C217(1.11) | LDD0690 | [2] |
| LDCM0374 | CL105 | HCT 116 | C217(0.71) | LDD0691 | [2] |
| LDCM0375 | CL106 | HCT 116 | C217(0.50) | LDD0692 | [2] |
| LDCM0376 | CL107 | HCT 116 | C217(0.59) | LDD0693 | [2] |
| LDCM0377 | CL108 | HCT 116 | C217(0.68) | LDD0694 | [2] |
| LDCM0378 | CL109 | HCT 116 | C217(0.75) | LDD0695 | [2] |
| LDCM0379 | CL11 | HCT 116 | C217(0.83) | LDD0696 | [2] |
| LDCM0380 | CL110 | HCT 116 | C217(0.73) | LDD0697 | [2] |
| LDCM0381 | CL111 | HCT 116 | C217(0.98) | LDD0698 | [2] |
| LDCM0382 | CL112 | PaTu 8988t | C217(0.89) | LDD1261 | [2] |
| LDCM0383 | CL113 | PaTu 8988t | C217(0.67) | LDD1262 | [2] |
| LDCM0384 | CL114 | PaTu 8988t | C217(0.72) | LDD1263 | [2] |
| LDCM0385 | CL115 | PaTu 8988t | C217(0.80) | LDD1264 | [2] |
| LDCM0386 | CL116 | PaTu 8988t | C217(0.50) | LDD1265 | [2] |
| LDCM0387 | CL117 | HCT 116 | C217(1.33) | LDD0704 | [2] |
| LDCM0388 | CL118 | HCT 116 | C217(1.17) | LDD0705 | [2] |
| LDCM0389 | CL119 | HCT 116 | C217(1.31) | LDD0706 | [2] |
| LDCM0390 | CL12 | HCT 116 | C217(0.84) | LDD0707 | [2] |
| LDCM0391 | CL120 | HCT 116 | C217(1.07) | LDD0708 | [2] |
| LDCM0392 | CL121 | HCT 116 | C217(0.83) | LDD0709 | [2] |
| LDCM0393 | CL122 | HCT 116 | C217(0.80) | LDD0710 | [2] |
| LDCM0394 | CL123 | HCT 116 | C217(0.74) | LDD0711 | [2] |
| LDCM0395 | CL124 | HCT 116 | C217(0.73) | LDD0712 | [2] |
| LDCM0396 | CL125 | HCT 116 | C217(1.28) | LDD0713 | [2] |
| LDCM0397 | CL126 | HCT 116 | C217(0.42) | LDD0714 | [2] |
| LDCM0398 | CL127 | HCT 116 | C217(0.47) | LDD0715 | [2] |
| LDCM0399 | CL128 | HCT 116 | C217(0.31) | LDD0716 | [2] |
| LDCM0400 | CL13 | HCT 116 | C217(0.82) | LDD0717 | [2] |
| LDCM0401 | CL14 | HCT 116 | C217(0.81) | LDD0718 | [2] |
| LDCM0402 | CL15 | HCT 116 | C217(0.82) | LDD0719 | [2] |
| LDCM0403 | CL16 | HCT 116 | C217(0.53) | LDD0720 | [2] |
| LDCM0404 | CL17 | HCT 116 | C217(0.41) | LDD0721 | [2] |
| LDCM0405 | CL18 | HCT 116 | C217(0.27) | LDD0722 | [2] |
| LDCM0406 | CL19 | HCT 116 | C217(0.42) | LDD0723 | [2] |
| LDCM0407 | CL2 | HCT 116 | C217(0.84) | LDD0724 | [2] |
| LDCM0408 | CL20 | HCT 116 | C217(0.31) | LDD0725 | [2] |
| LDCM0409 | CL21 | HCT 116 | C217(0.28) | LDD0726 | [2] |
| LDCM0410 | CL22 | HCT 116 | C217(0.34) | LDD0727 | [2] |
| LDCM0411 | CL23 | HCT 116 | C217(0.36) | LDD0728 | [2] |
| LDCM0412 | CL24 | HCT 116 | C217(0.26) | LDD0729 | [2] |
| LDCM0413 | CL25 | HCT 116 | C217(0.29) | LDD0730 | [2] |
| LDCM0414 | CL26 | HCT 116 | C217(0.34) | LDD0731 | [2] |
| LDCM0415 | CL27 | HCT 116 | C217(0.42) | LDD0732 | [2] |
| LDCM0416 | CL28 | HCT 116 | C217(0.31) | LDD0733 | [2] |
| LDCM0417 | CL29 | HCT 116 | C217(0.36) | LDD0734 | [2] |
| LDCM0418 | CL3 | HCT 116 | C217(0.85) | LDD0735 | [2] |
| LDCM0419 | CL30 | HCT 116 | C217(0.49) | LDD0736 | [2] |
| LDCM0420 | CL31 | HCT 116 | C217(0.84) | LDD0737 | [2] |
| LDCM0421 | CL32 | HCT 116 | C217(0.76) | LDD0738 | [2] |
| LDCM0422 | CL33 | HCT 116 | C217(0.73) | LDD0739 | [2] |
| LDCM0423 | CL34 | HCT 116 | C217(0.57) | LDD0740 | [2] |
| LDCM0424 | CL35 | HCT 116 | C217(0.58) | LDD0741 | [2] |
| LDCM0425 | CL36 | HCT 116 | C217(0.67) | LDD0742 | [2] |
| LDCM0426 | CL37 | HCT 116 | C217(0.64) | LDD0743 | [2] |
| LDCM0428 | CL39 | HCT 116 | C217(0.72) | LDD0745 | [2] |
| LDCM0429 | CL4 | HCT 116 | C217(0.86) | LDD0746 | [2] |
| LDCM0430 | CL40 | HCT 116 | C217(0.81) | LDD0747 | [2] |
| LDCM0431 | CL41 | HCT 116 | C217(0.89) | LDD0748 | [2] |
| LDCM0432 | CL42 | HCT 116 | C217(0.73) | LDD0749 | [2] |
| LDCM0433 | CL43 | HCT 116 | C217(0.76) | LDD0750 | [2] |
| LDCM0434 | CL44 | HCT 116 | C217(0.67) | LDD0751 | [2] |
| LDCM0435 | CL45 | HCT 116 | C217(0.75) | LDD0752 | [2] |
| LDCM0436 | CL46 | HCT 116 | C217(0.91) | LDD0753 | [2] |
| LDCM0437 | CL47 | HCT 116 | C217(0.86) | LDD0754 | [2] |
| LDCM0438 | CL48 | HCT 116 | C217(0.79) | LDD0755 | [2] |
| LDCM0439 | CL49 | HCT 116 | C217(1.09) | LDD0756 | [2] |
| LDCM0440 | CL5 | HCT 116 | C217(0.84) | LDD0757 | [2] |
| LDCM0441 | CL50 | HCT 116 | C217(1.12) | LDD0758 | [2] |
| LDCM0442 | CL51 | HCT 116 | C217(1.04) | LDD0759 | [2] |
| LDCM0443 | CL52 | HCT 116 | C217(0.90) | LDD0760 | [2] |
| LDCM0444 | CL53 | HCT 116 | C217(0.90) | LDD0761 | [2] |
| LDCM0445 | CL54 | HCT 116 | C217(0.81) | LDD0762 | [2] |
| LDCM0446 | CL55 | HCT 116 | C217(1.07) | LDD0763 | [2] |
| LDCM0447 | CL56 | HCT 116 | C217(0.88) | LDD0764 | [2] |
| LDCM0448 | CL57 | HCT 116 | C217(0.99) | LDD0765 | [2] |
| LDCM0449 | CL58 | HCT 116 | C217(0.98) | LDD0766 | [2] |
| LDCM0450 | CL59 | HCT 116 | C217(0.98) | LDD0767 | [2] |
| LDCM0451 | CL6 | HCT 116 | C217(0.74) | LDD0768 | [2] |
| LDCM0452 | CL60 | HCT 116 | C217(0.85) | LDD0769 | [2] |
| LDCM0453 | CL61 | HCT 116 | C217(1.18) | LDD0770 | [2] |
| LDCM0454 | CL62 | HCT 116 | C217(1.00) | LDD0771 | [2] |
| LDCM0455 | CL63 | HCT 116 | C217(1.01) | LDD0772 | [2] |
| LDCM0456 | CL64 | HCT 116 | C217(1.35) | LDD0773 | [2] |
| LDCM0457 | CL65 | HCT 116 | C217(1.19) | LDD0774 | [2] |
| LDCM0458 | CL66 | HCT 116 | C217(0.93) | LDD0775 | [2] |
| LDCM0459 | CL67 | HCT 116 | C217(1.05) | LDD0776 | [2] |
| LDCM0460 | CL68 | HCT 116 | C217(0.93) | LDD0777 | [2] |
| LDCM0461 | CL69 | HCT 116 | C217(0.93) | LDD0778 | [2] |
| LDCM0462 | CL7 | HCT 116 | C217(0.85) | LDD0779 | [2] |
| LDCM0463 | CL70 | HCT 116 | C217(0.89) | LDD0780 | [2] |
| LDCM0464 | CL71 | HCT 116 | C217(0.96) | LDD0781 | [2] |
| LDCM0465 | CL72 | HCT 116 | C217(1.09) | LDD0782 | [2] |
| LDCM0466 | CL73 | HCT 116 | C217(1.18) | LDD0783 | [2] |
| LDCM0467 | CL74 | HCT 116 | C217(1.00) | LDD0784 | [2] |
| LDCM0469 | CL76 | HCT 116 | C217(0.58) | LDD0786 | [2] |
| LDCM0470 | CL77 | HCT 116 | C217(0.68) | LDD0787 | [2] |
| LDCM0471 | CL78 | HCT 116 | C217(0.64) | LDD0788 | [2] |
| LDCM0472 | CL79 | HCT 116 | C217(0.43) | LDD0789 | [2] |
| LDCM0473 | CL8 | HCT 116 | C217(0.79) | LDD0790 | [2] |
| LDCM0474 | CL80 | HCT 116 | C217(0.53) | LDD0791 | [2] |
| LDCM0475 | CL81 | HCT 116 | C217(0.58) | LDD0792 | [2] |
| LDCM0476 | CL82 | HCT 116 | C217(0.41) | LDD0793 | [2] |
| LDCM0477 | CL83 | HCT 116 | C217(0.41) | LDD0794 | [2] |
| LDCM0478 | CL84 | HCT 116 | C217(0.41) | LDD0795 | [2] |
| LDCM0479 | CL85 | HCT 116 | C217(0.59) | LDD0796 | [2] |
| LDCM0480 | CL86 | HCT 116 | C217(0.83) | LDD0797 | [2] |
| LDCM0481 | CL87 | HCT 116 | C217(0.56) | LDD0798 | [2] |
| LDCM0482 | CL88 | HCT 116 | C217(0.42) | LDD0799 | [2] |
| LDCM0483 | CL89 | HCT 116 | C217(0.48) | LDD0800 | [2] |
| LDCM0484 | CL9 | HCT 116 | C217(0.66) | LDD0801 | [2] |
| LDCM0485 | CL90 | HCT 116 | C217(1.37) | LDD0802 | [2] |
| LDCM0486 | CL91 | HCT 116 | C217(0.78) | LDD0803 | [2] |
| LDCM0487 | CL92 | HCT 116 | C217(0.91) | LDD0804 | [2] |
| LDCM0488 | CL93 | HCT 116 | C217(0.89) | LDD0805 | [2] |
| LDCM0489 | CL94 | HCT 116 | C217(1.07) | LDD0806 | [2] |
| LDCM0490 | CL95 | HCT 116 | C217(0.87) | LDD0807 | [2] |
| LDCM0491 | CL96 | HCT 116 | C217(0.79) | LDD0808 | [2] |
| LDCM0492 | CL97 | HCT 116 | C217(0.77) | LDD0809 | [2] |
| LDCM0493 | CL98 | HCT 116 | C217(0.82) | LDD0810 | [2] |
| LDCM0494 | CL99 | HCT 116 | C217(0.83) | LDD0811 | [2] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C217(2.07) | LDD1702 | [13] |
| LDCM0625 | F8 | Ramos | C217(1.41); C154(1.69) | LDD2187 | [14] |
| LDCM0572 | Fragment10 | MDA-MB-231 | C154(20.00) | LDD1389 | [15] |
| LDCM0573 | Fragment11 | Ramos | C154(3.43) | LDD2190 | [14] |
| LDCM0574 | Fragment12 | Ramos | C217(1.12) | LDD2191 | [14] |
| LDCM0575 | Fragment13 | Ramos | C217(1.01) | LDD2192 | [14] |
| LDCM0576 | Fragment14 | MDA-MB-231 | C154(1.53) | LDD1397 | [15] |
| LDCM0579 | Fragment20 | Ramos | C154(1.14) | LDD1403 | [15] |
| LDCM0580 | Fragment21 | Ramos | C217(0.85) | LDD2195 | [14] |
| LDCM0582 | Fragment23 | Ramos | C154(1.06) | LDD1409 | [15] |
| LDCM0578 | Fragment27 | Ramos | C154(1.32) | LDD1414 | [15] |
| LDCM0586 | Fragment28 | Ramos | C154(1.04) | LDD1416 | [15] |
| LDCM0588 | Fragment30 | Ramos | C217(0.94) | LDD2199 | [14] |
| LDCM0589 | Fragment31 | Ramos | C217(1.09) | LDD2200 | [14] |
| LDCM0590 | Fragment32 | Ramos | C154(1.28) | LDD1424 | [15] |
| LDCM0468 | Fragment33 | HCT 116 | C217(0.92) | LDD0785 | [2] |
| LDCM0596 | Fragment38 | MDA-MB-231 | C154(1.21) | LDD1433 | [15] |
| LDCM0566 | Fragment4 | Ramos | C154(0.70) | LDD2184 | [14] |
| LDCM0599 | Fragment41 | MDA-MB-231 | C154(1.32) | LDD1438 | [15] |
| LDCM0600 | Fragment42 | Ramos | C154(0.78) | LDD1440 | [15] |
| LDCM0427 | Fragment51 | HCT 116 | C217(0.65) | LDD0744 | [2] |
| LDCM0610 | Fragment52 | Ramos | C217(1.21) | LDD2204 | [14] |
| LDCM0611 | Fragment53 | Ramos | C154(1.19) | LDD1455 | [15] |
| LDCM0614 | Fragment56 | Ramos | C154(1.58) | LDD1459 | [15] |
| LDCM0568 | Fragment6 | MDA-MB-231 | C154(1.01) | LDD1382 | [15] |
| LDCM0569 | Fragment7 | Ramos | C217(1.18); C154(1.07) | LDD2186 | [14] |
| LDCM0571 | Fragment9 | Ramos | C154(20.00) | LDD1388 | [15] |
| LDCM0076 | kambe_cp71 | PC-3 | 2.28 | LDD0132 | [11] |
| LDCM0022 | KB02 | MDA-MB-231 | C154(1.26) | LDD1374 | [15] |
| LDCM0023 | KB03 | Jurkat | C217(7.36) | LDD0209 | [16] |
| LDCM0024 | KB05 | COLO792 | C217(2.16) | LDD3310 | [17] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0224 | [8] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C154(0.42) | LDD2207 | [18] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Argininosuccinate synthase (ASS1) | Argininosuccinate synthase family | P00966 | |||
| NmrA-like family domain-containing protein 1 (NMRAL1) | NmrA-type oxidoreductase family | Q9HBL8 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Optineurin (OPTN) | . | Q96CV9 | |||
References
