Details of the Target
General Information of Target
| Target ID | LDTP13352 | |||||
|---|---|---|---|---|---|---|
| Target Name | GTP:AMP phosphotransferase AK3, mitochondrial (AK3) | |||||
| Gene Name | AK3 | |||||
| Gene ID | 50808 | |||||
| Synonyms |
AK3L1; AK6; AKL3L; GTP:AMP phosphotransferase AK3, mitochondrial; EC 2.7.4.10; Adenylate kinase 3; AK 3; Adenylate kinase 3 alpha-like 1 |
|||||
| 3D Structure | ||||||
| Sequence |
MAPSGPGSSARRRCRRVLYWIPVVFITLLLGWSYYAYAIQLCIVSMENTGEQVVCLMAYH
LLFAMFVWSYWKTIFTLPMNPSKEFHLSYAEKDLLEREPRGEAHQEVLRRAAKDLPIYTR TMSGAIRYCDRCQLIKPDRCHHCSVCDKCILKMDHHCPWVNNCVGFSNYKFFLLFLAYSL LYCLFIAATDLQYFIKFWTNGLPDTQAKFHIMFLFFAAAMFSVSLSSLFGYHCWLVSKNK STLEAFRSPVFRHGTDKNGFSLGFSKNMRQVFGDEKKYWLLPIFSSLGDGCSFPTCLVNQ DPEQASTPAGLNSTAKNLENHQFPAKPLRESQSHLLTDSQSWTESSINPGKCKAGMSNPA LTMENET |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Adenylate kinase family, AK3 subfamily
|
|||||
| Subcellular location |
Mitochondrion matrix
|
|||||
| Function |
Involved in maintaining the homeostasis of cellular nucleotides by catalyzing the interconversion of nucleoside phosphates. Has GTP:AMP phosphotransferase and ITP:AMP phosphotransferase activities. {|HAMAP-Rule:MF_03169}.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
TH211 Probe Info |
 |
Y85(20.00) | LDD0260 | [1] | |
|
C-Sul Probe Info |
 |
21.24 | LDD0066 | [2] | |
|
STPyne Probe Info |
 |
K64(1.41) | LDD0277 | [3] | |
|
DBIA Probe Info |
 |
C81(1.92) | LDD3312 | [4] | |
|
Acrolein Probe Info |
 |
H35(0.00); H30(0.00) | LDD0221 | [5] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [6] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [5] | |
|
HHS-465 Probe Info |
 |
N.A. | LDD2240 | [7] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C056 Probe Info |
 |
23.59 | LDD1753 | [8] | |
|
C071 Probe Info |
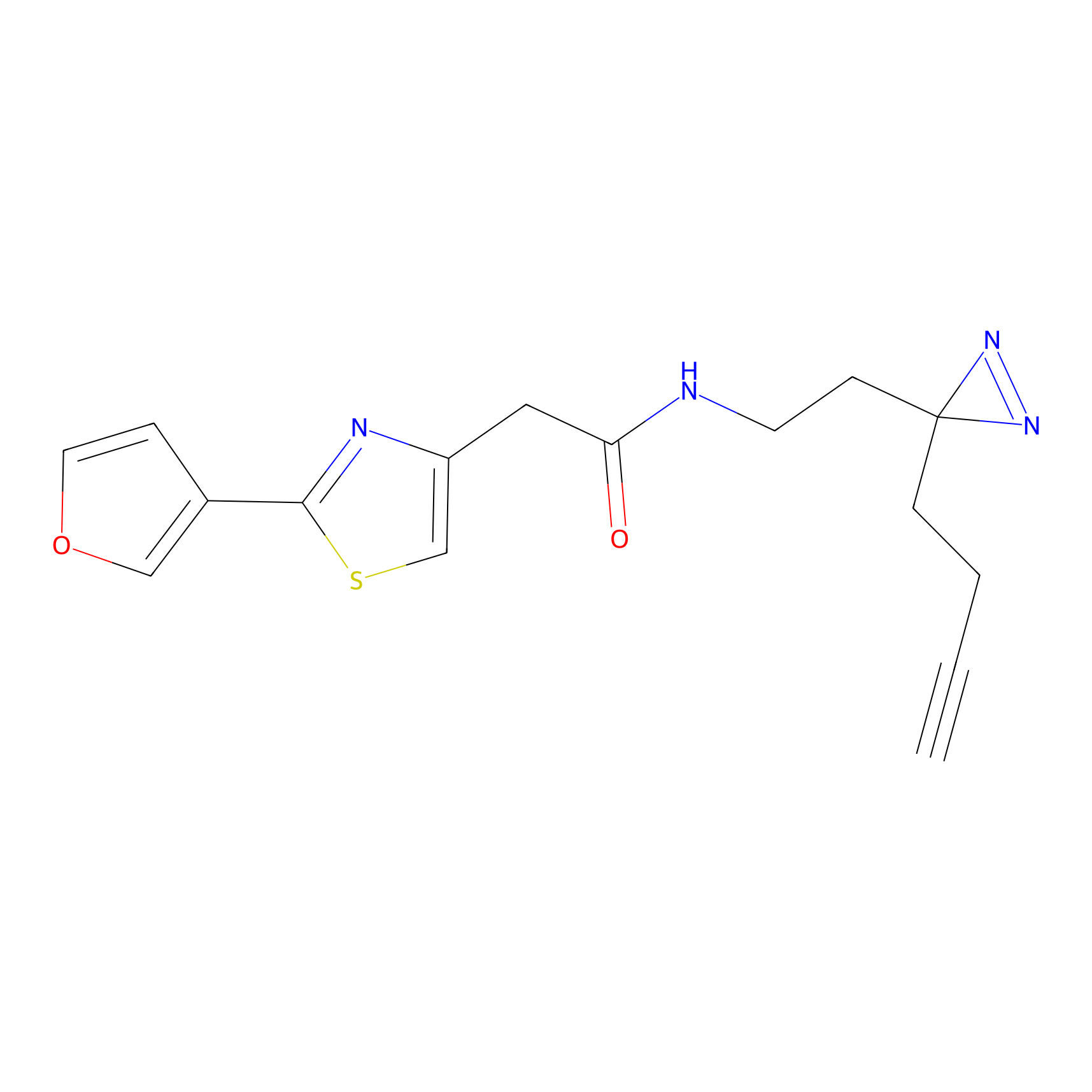 |
8.34 | LDD1767 | [8] | |
|
C094 Probe Info |
 |
46.85 | LDD1785 | [8] | |
|
C158 Probe Info |
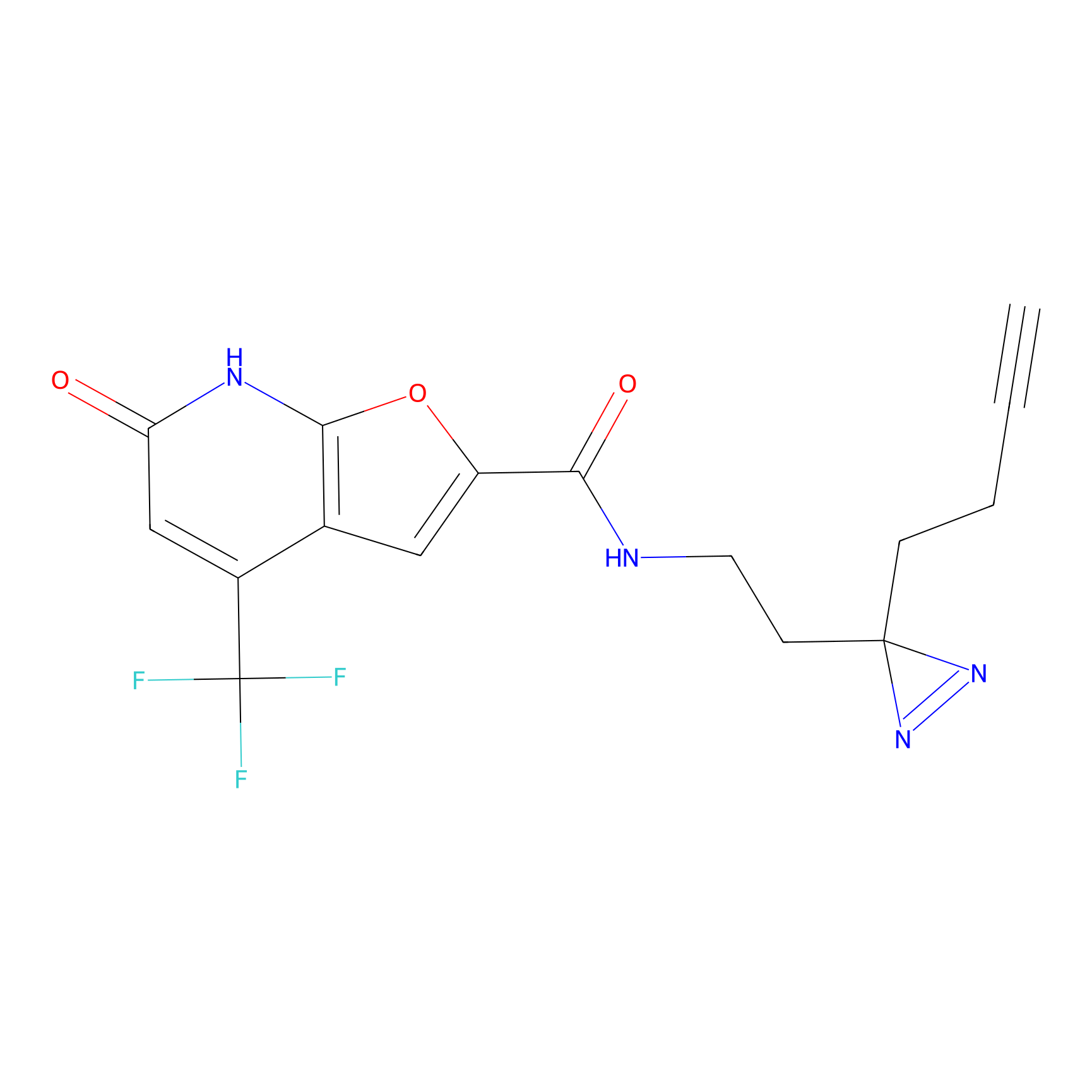 |
14.72 | LDD1838 | [8] | |
|
C161 Probe Info |
 |
10.78 | LDD1841 | [8] | |
|
C201 Probe Info |
 |
42.52 | LDD1877 | [8] | |
|
C206 Probe Info |
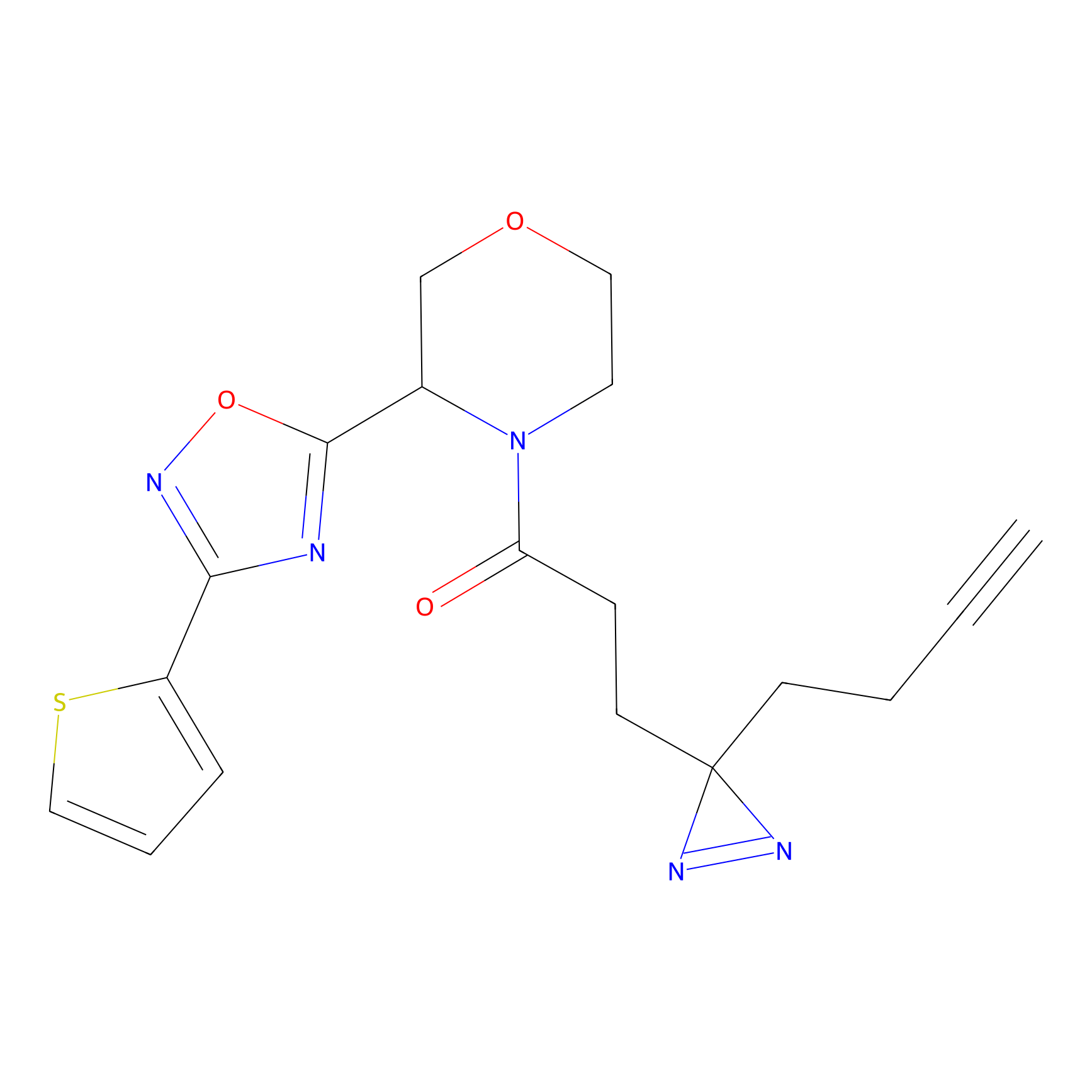 |
23.43 | LDD1881 | [8] | |
|
C210 Probe Info |
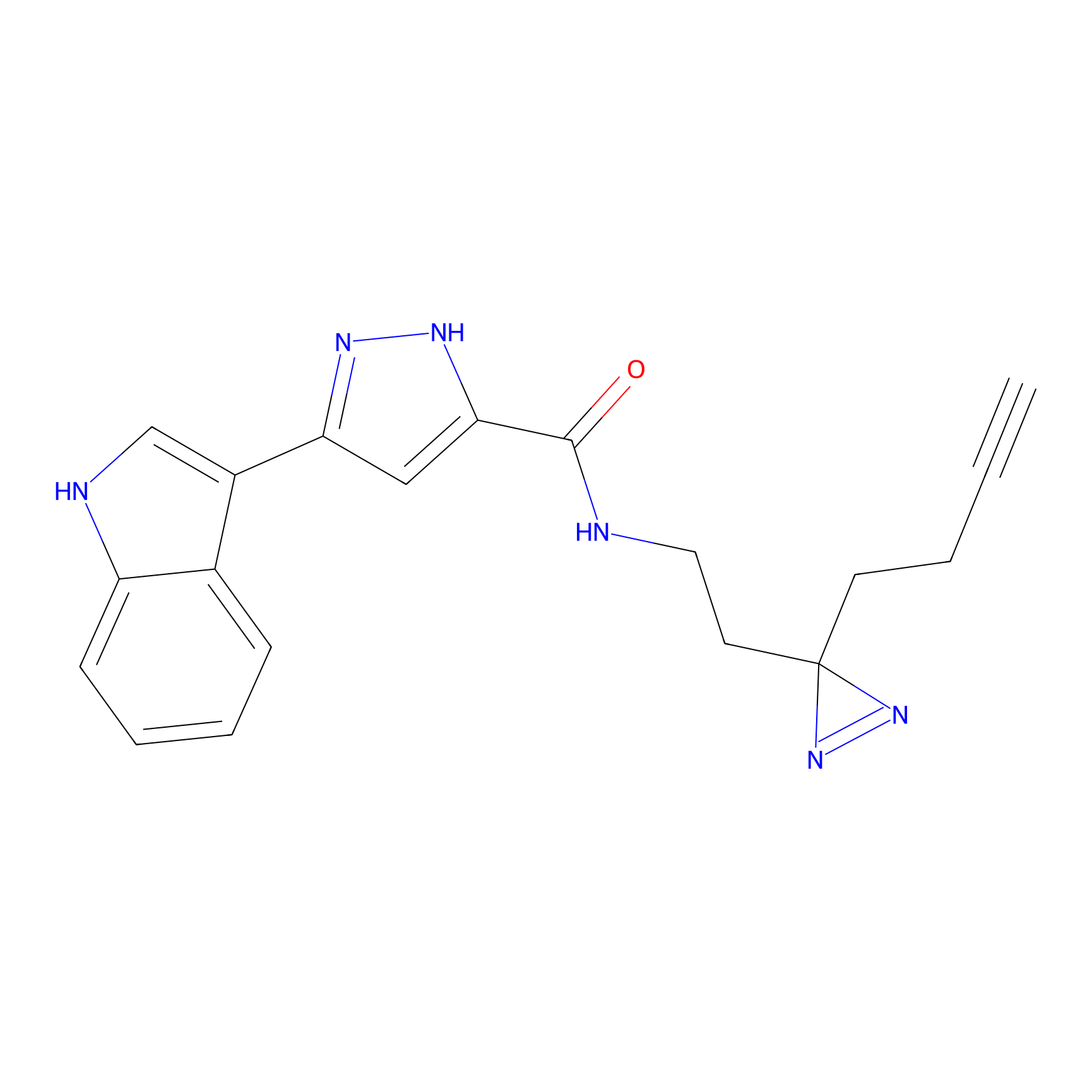 |
68.12 | LDD1884 | [8] | |
|
C232 Probe Info |
 |
50.56 | LDD1905 | [8] | |
|
C244 Probe Info |
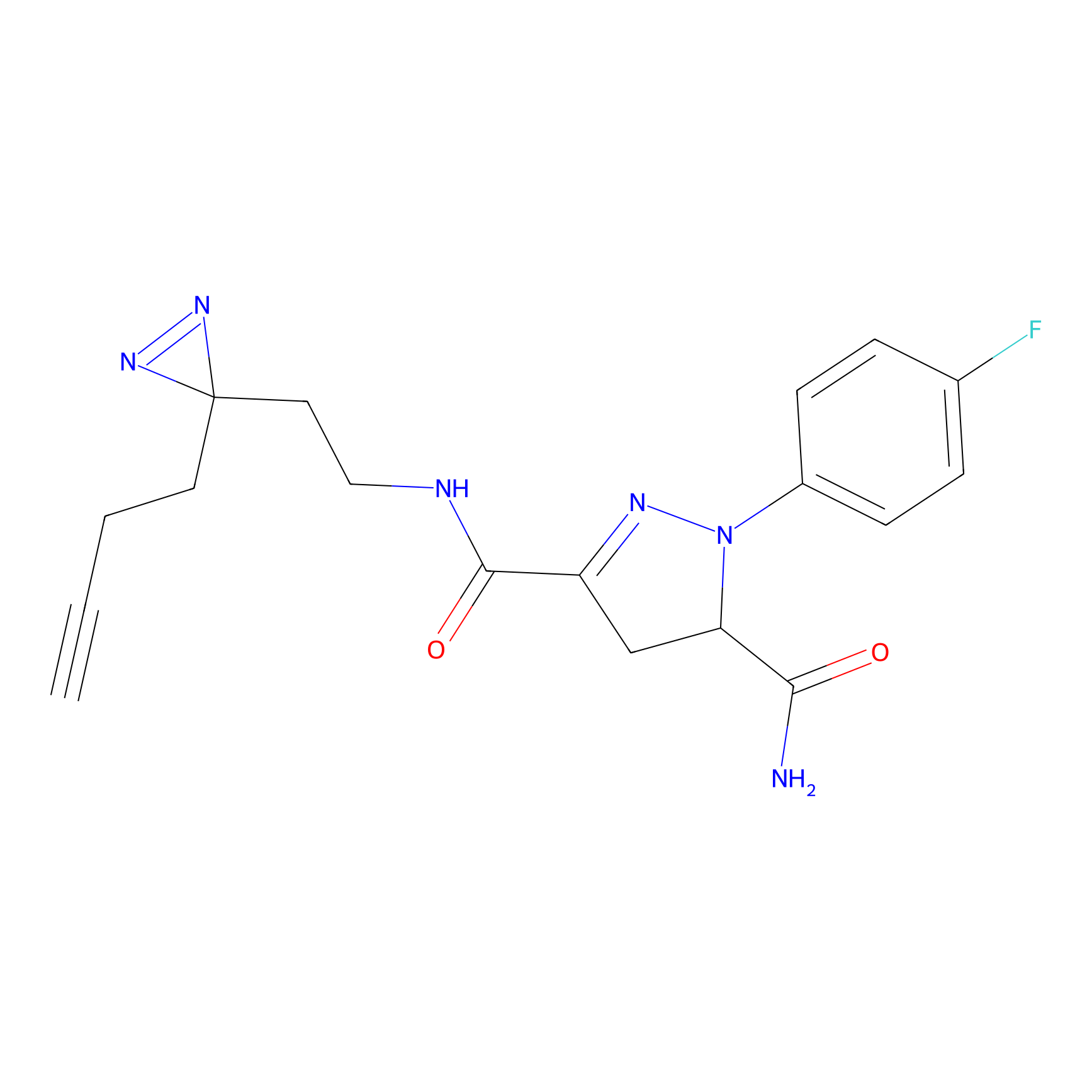 |
25.63 | LDD1917 | [8] | |
|
C246 Probe Info |
 |
17.51 | LDD1919 | [8] | |
|
C278 Probe Info |
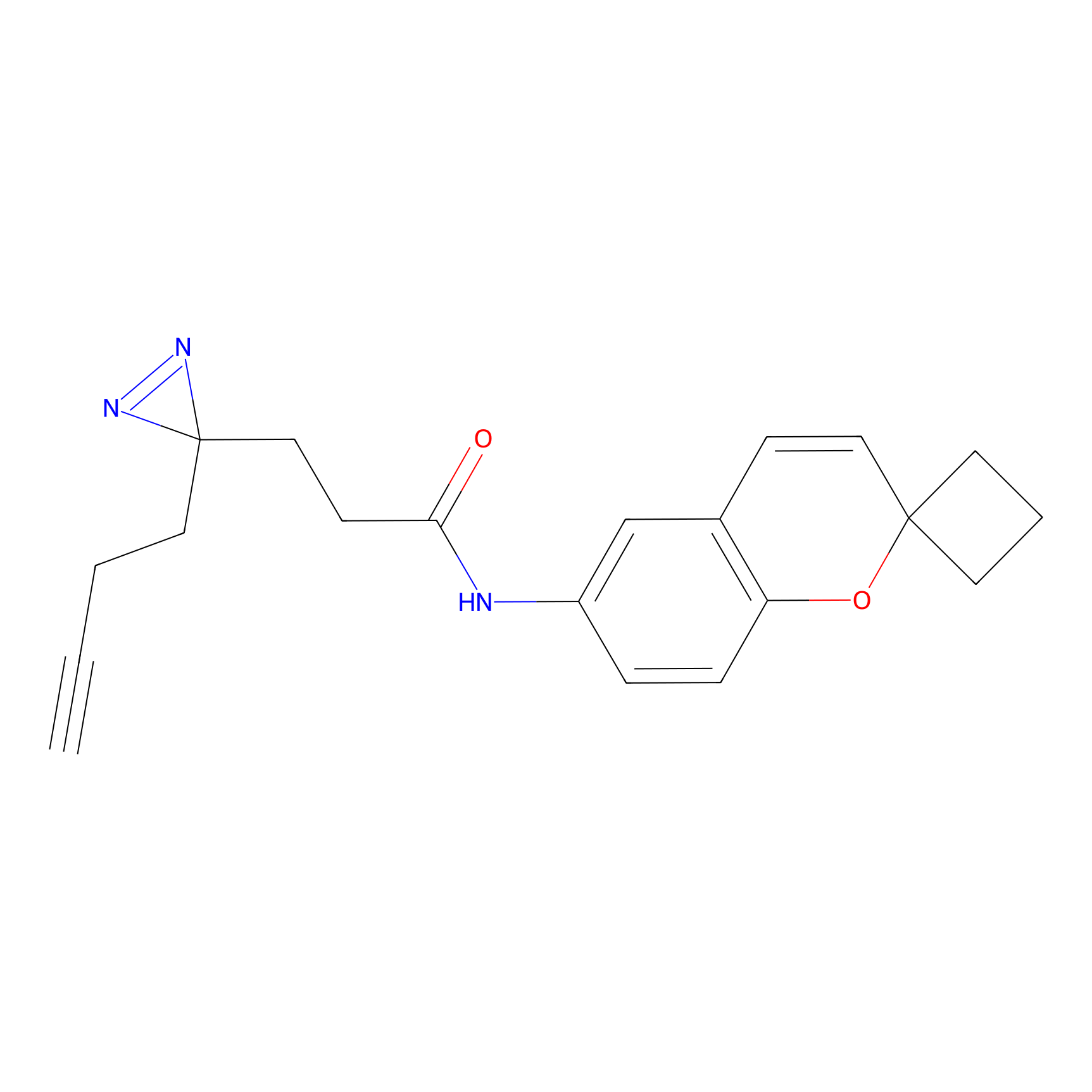 |
99.73 | LDD1948 | [8] | |
|
C280 Probe Info |
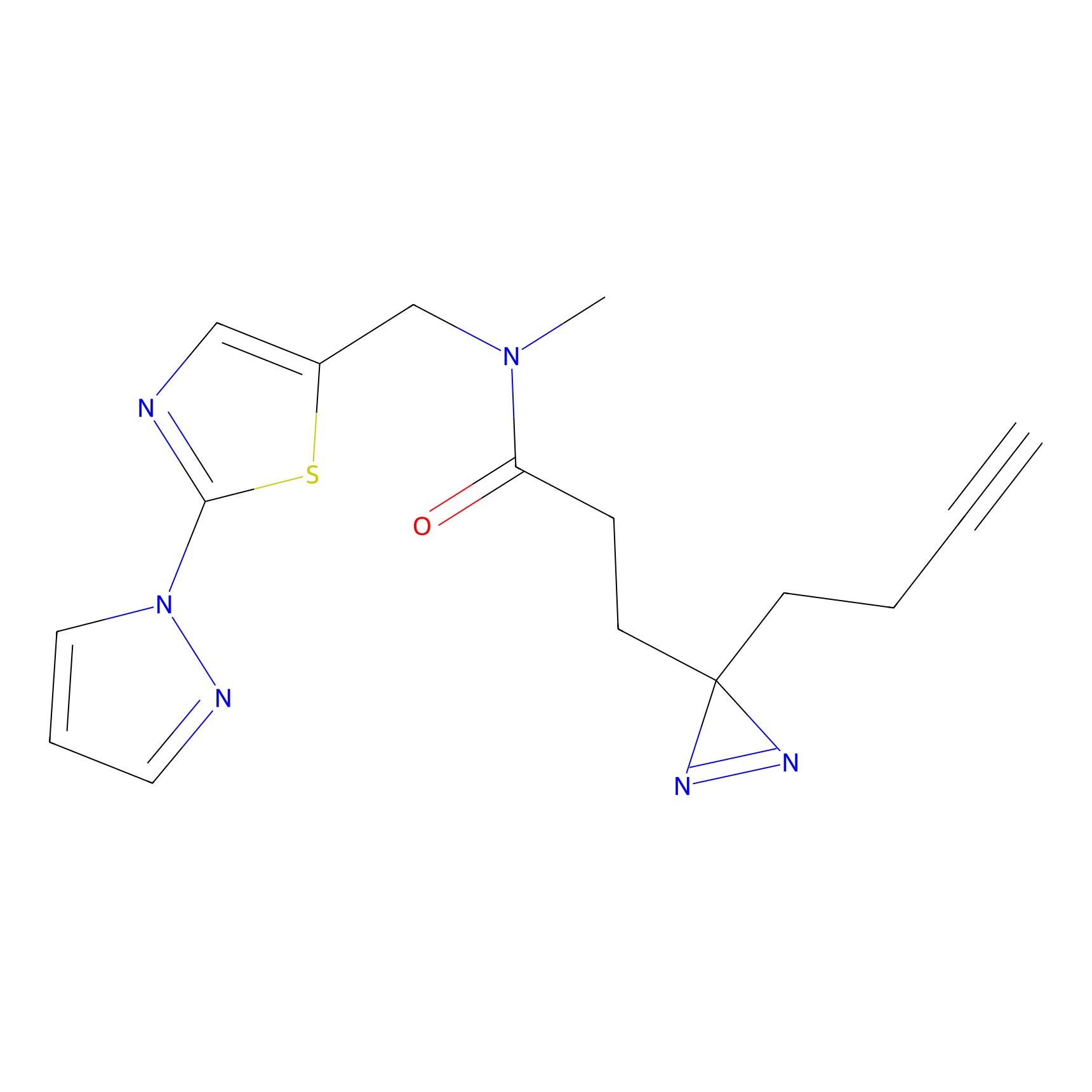 |
12.91 | LDD1950 | [8] | |
|
C293 Probe Info |
 |
63.12 | LDD1963 | [8] | |
|
C349 Probe Info |
 |
9.92 | LDD2010 | [8] | |
|
C362 Probe Info |
 |
99.73 | LDD2023 | [8] | |
|
C364 Probe Info |
 |
22.01 | LDD2025 | [8] | |
|
C366 Probe Info |
 |
7.36 | LDD2027 | [8] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0108 | Chloroacetamide | HeLa | H30(0.00); H35(0.00) | LDD0222 | [5] |
| LDCM0107 | IAA | HeLa | H35(0.00); H30(0.00) | LDD0221 | [5] |
| LDCM0022 | KB02 | 42-MG-BA | C81(2.01) | LDD2244 | [4] |
| LDCM0023 | KB03 | 22RV1 | C81(2.11) | LDD2660 | [4] |
| LDCM0024 | KB05 | HMCB | C81(1.92) | LDD3312 | [4] |
| LDCM0109 | NEM | HeLa | H35(0.00); H30(0.00) | LDD0223 | [5] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | Glyceraldehyde-3-phosphate dehydrogenase family | P04406 | |||
Transporter and channel
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Progranulin (GRN) | Granulin family | P28799 | |||
| Keratin-associated protein 10-8 (KRTAP10-8) | KRTAP type 10 family | P60410 | |||
| Amyloid protein-binding protein 2 (APPBP2) | . | Q92624 | |||
References
