Details of the Target
General Information of Target
| Target ID | LDTP12966 | |||||
|---|---|---|---|---|---|---|
| Target Name | Vesicle-associated membrane protein-associated protein A (VAPA) | |||||
| Gene Name | VAPA | |||||
| Gene ID | 9218 | |||||
| Synonyms |
VAP33; Vesicle-associated membrane protein-associated protein A; VAMP-A; VAMP-associated protein A; VAP-A; 33 kDa VAMP-associated protein; VAP-33 |
|||||
| 3D Structure | ||||||
| Sequence |
MARPPRQHPGVWASLLLLLLTGPAACAASPADDGAGPGGRGPRGRARGDTGADEAVPRHD
SSYGTFAGEFYDLRYLSEEGYPFPTAPPVDPFAKIKVDDCGKTKGCFRYGKPGCNAETCD YFLSYRMIGADVEFELSADTDGWVAVGFSSDKKMGGDDVMACVHDDNGRVRIQHFYNVGQ WAKEIQRNPARDEEGVFENNRVTCRFKRPVNVPRDETIVDLHLSWYYLFAWGPAIQGSIT RHDIDSPPASERVVSIYKYEDIFMPSAAYQTFSSPFCLLLIVALTFYLLMGTP |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
VAMP-associated protein (VAP) (TC 9.B.17) family
|
|||||
| Subcellular location |
Endoplasmic reticulum membrane
|
|||||
| Function |
Endoplasmic reticulum (ER)-anchored protein that mediates the formation of contact sites between the ER and endosomes via interaction with FFAT motif-containing proteins such as STARD3 or WDR44. STARD3-VAPA interaction enables cholesterol transfer from the ER to endosomes. Via interaction with WDR44 participates in neosynthesized protein export. In addition, recruited to the plasma membrane through OSBPL3 binding. The OSBPL3-VAPA complex stimulates RRAS signaling which in turn attenuates integrin beta-1 (ITGB1) activation at the cell surface. With OSBPL3, may regulate ER morphology. May play a role in vesicle trafficking.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| IGR1 | SNV: p.I67T | DBIA Probe Info | |||
| PF382 | SNV: p.M96V | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
Alkylaryl probe 1 Probe Info |
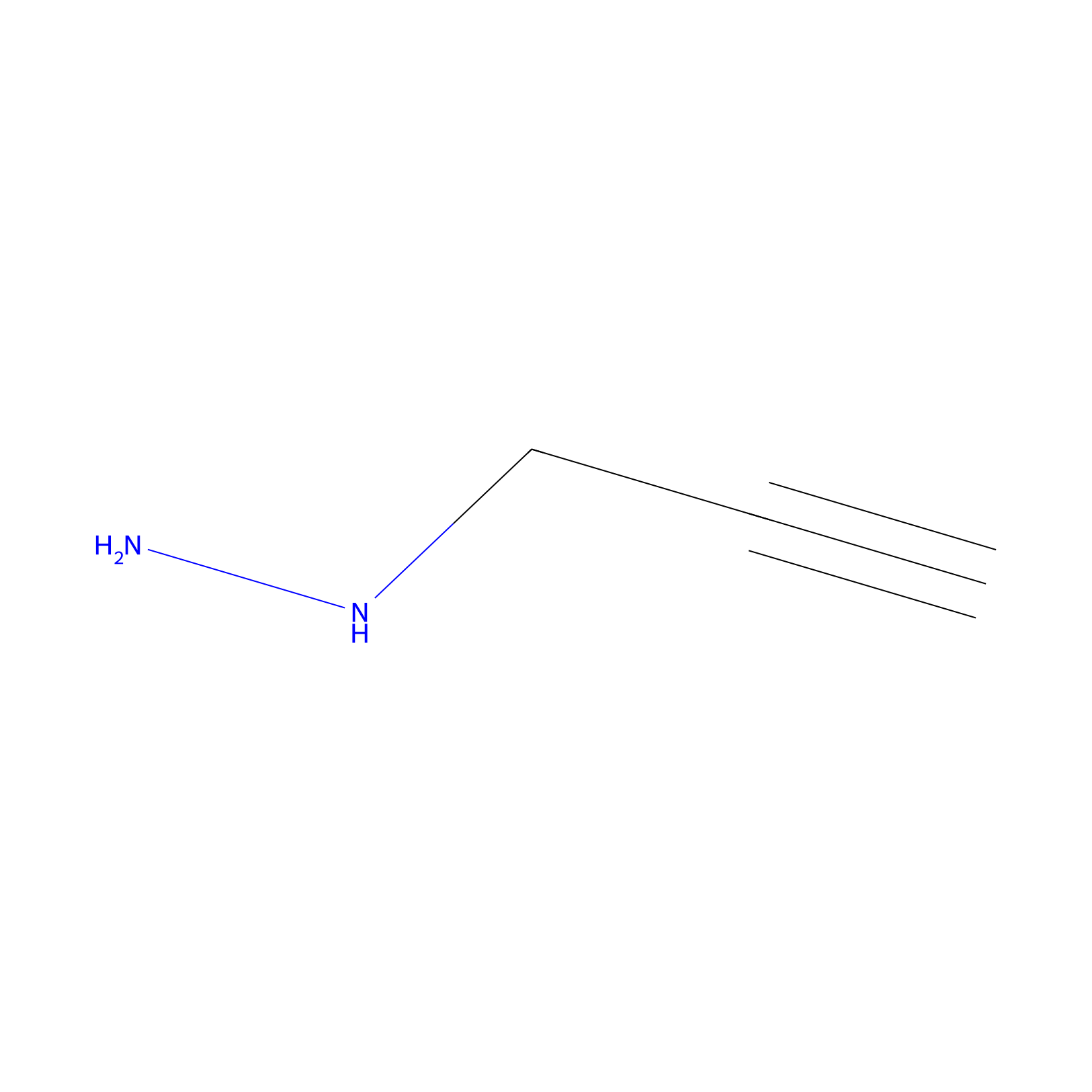 |
20.00 | LDD0387 | [2] | |
|
A-EBA Probe Info |
 |
4.95 | LDD0215 | [3] | |
|
CY-1 Probe Info |
 |
100.00 | LDD0243 | [4] | |
|
CY4 Probe Info |
 |
100.00 | LDD0244 | [4] | |
|
N1 Probe Info |
 |
100.00 | LDD0242 | [4] | |
|
W1 Probe Info |
 |
14.36 | LDD0235 | [5] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [6] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [6] | |
|
ONAyne Probe Info |
 |
N.A. | LDD0273 | [7] | |
|
OPA-S-S-alkyne Probe Info |
 |
K174(1.89); K180(1.96); K188(1.99); K211(4.31) | LDD3494 | [8] | |
|
DBIA Probe Info |
 |
C128(1.50) | LDD3311 | [9] | |
|
DA-P3 Probe Info |
 |
5.28 | LDD0180 | [10] | |
|
5E-2FA Probe Info |
 |
H11(0.00); H208(0.00); H163(0.00) | LDD2235 | [11] | |
|
ATP probe Probe Info |
 |
K125(0.00); K211(0.00); K26(0.00); K24(0.00) | LDD0199 | [12] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C128(0.00); C179(0.00); C60(0.00) | LDD0038 | [13] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0032 | [14] | |
|
Lodoacetamide azide Probe Info |
 |
C128(0.00); C60(0.00); C179(0.00) | LDD0037 | [13] | |
|
ATP probe Probe Info |
 |
K180(0.00); K52(0.00) | LDD0035 | [15] | |
|
BTD Probe Info |
 |
N.A. | LDD0004 | [16] | |
|
NAIA_4 Probe Info |
 |
C60(0.00); C179(0.00) | LDD2226 | [17] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [18] | |
|
WYneO Probe Info |
 |
N.A. | LDD0022 | [16] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [16] | |
|
NHS Probe Info |
 |
K205(0.00); K38(0.00); K10(0.00); K125(0.00) | LDD0010 | [16] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [19] | |
|
STPyne Probe Info |
 |
K125(0.00); K211(0.00) | LDD0009 | [16] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [16] | |
|
Phosphinate-6 Probe Info |
 |
C60(0.00); C179(0.00) | LDD0018 | [20] | |
|
1d-yne Probe Info |
 |
N.A. | LDD0357 | [21] | |
|
Acrolein Probe Info |
 |
H208(0.00); H11(0.00); C179(0.00); H195(0.00) | LDD0217 | [22] | |
|
Crotonaldehyde Probe Info |
 |
H208(0.00); C179(0.00); H195(0.00) | LDD0219 | [22] | |
|
Methacrolein Probe Info |
 |
H208(0.00); C179(0.00) | LDD0218 | [22] | |
|
AOyne Probe Info |
 |
14.50 | LDD0443 | [23] | |
|
MPP-AC Probe Info |
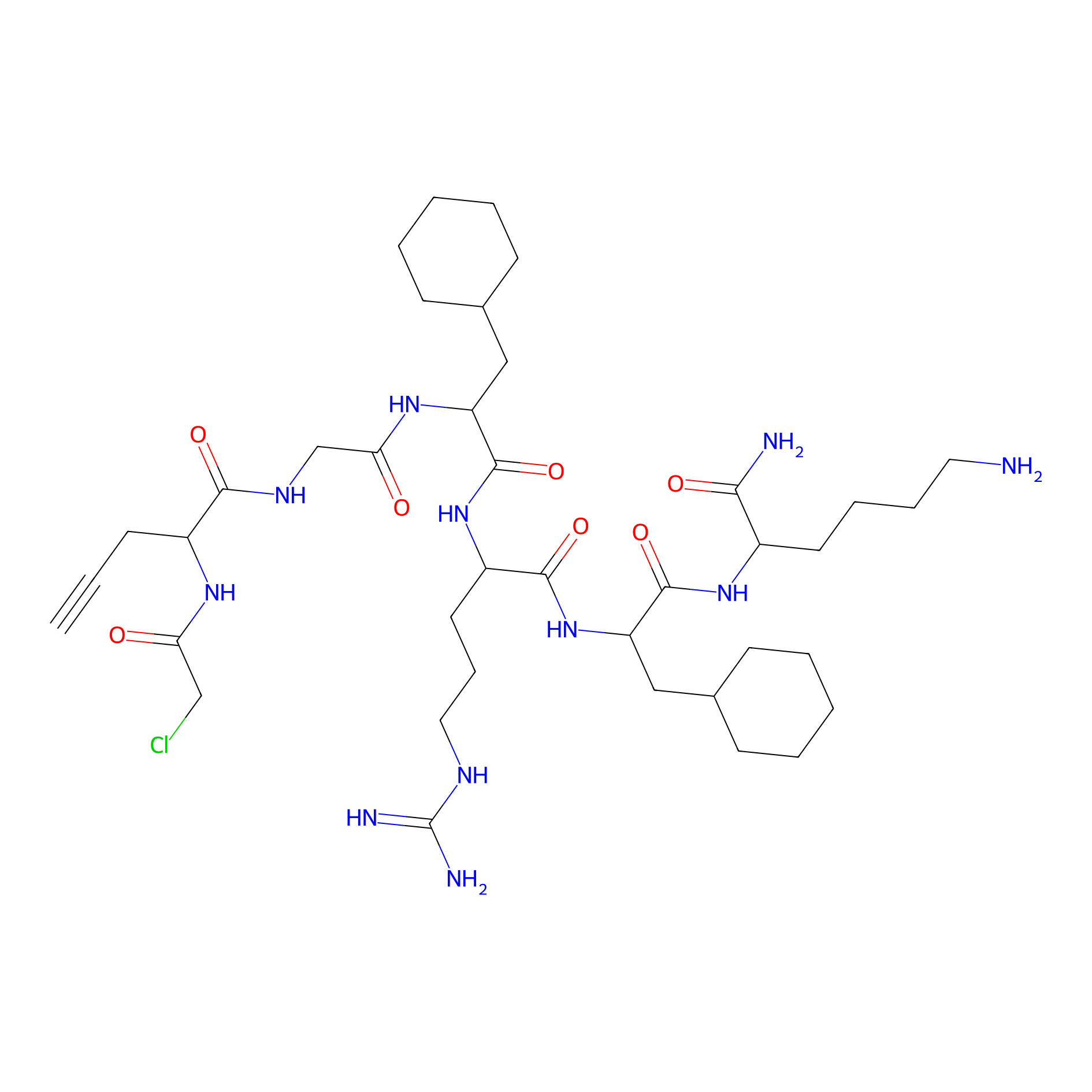 |
N.A. | LDD0428 | [24] | |
|
NAIA_5 Probe Info |
 |
C128(0.00); C179(0.00); C60(0.00) | LDD2223 | [17] | |
|
TER-AC Probe Info |
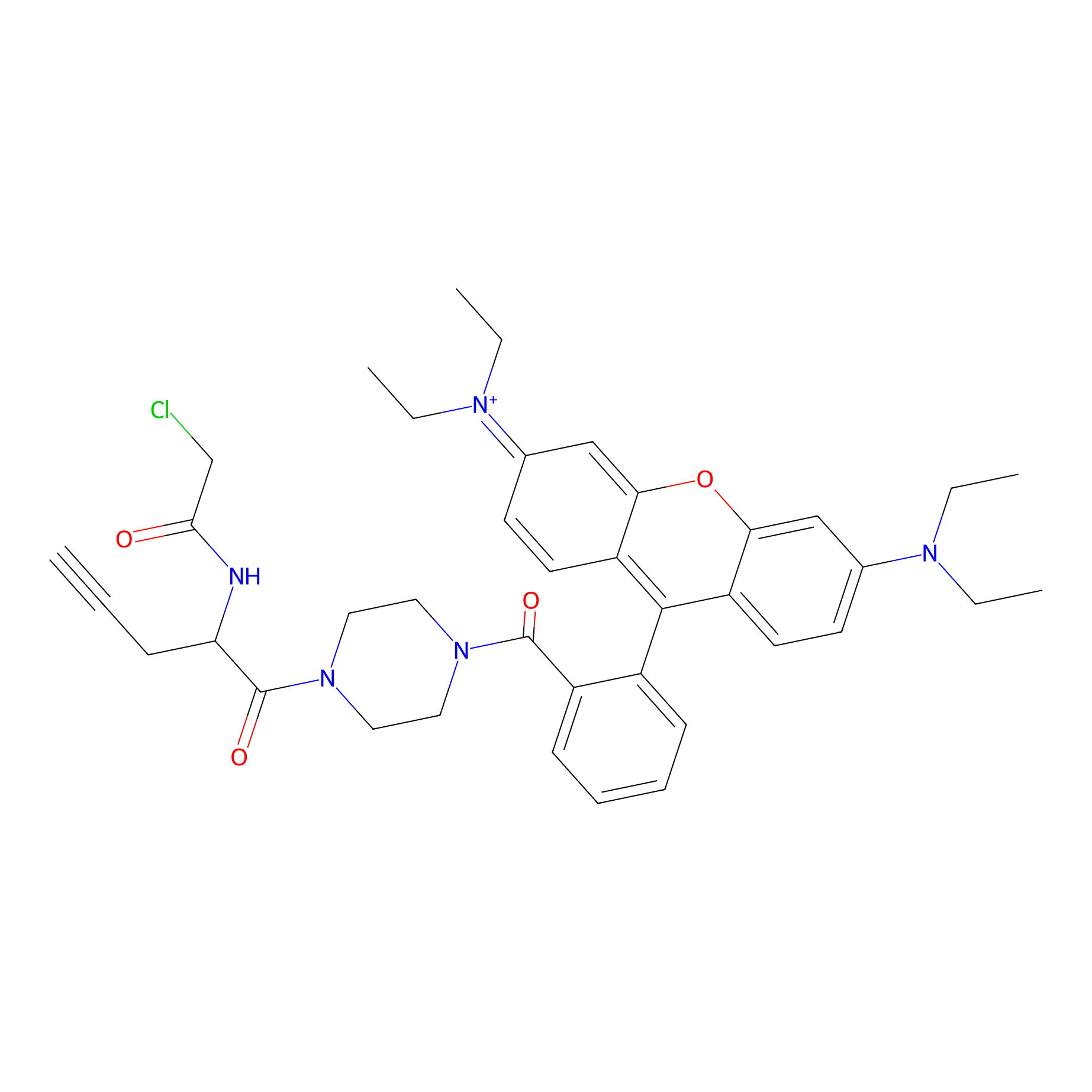 |
N.A. | LDD0426 | [24] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C191 Probe Info |
 |
9.92 | LDD1868 | [25] | |
|
C193 Probe Info |
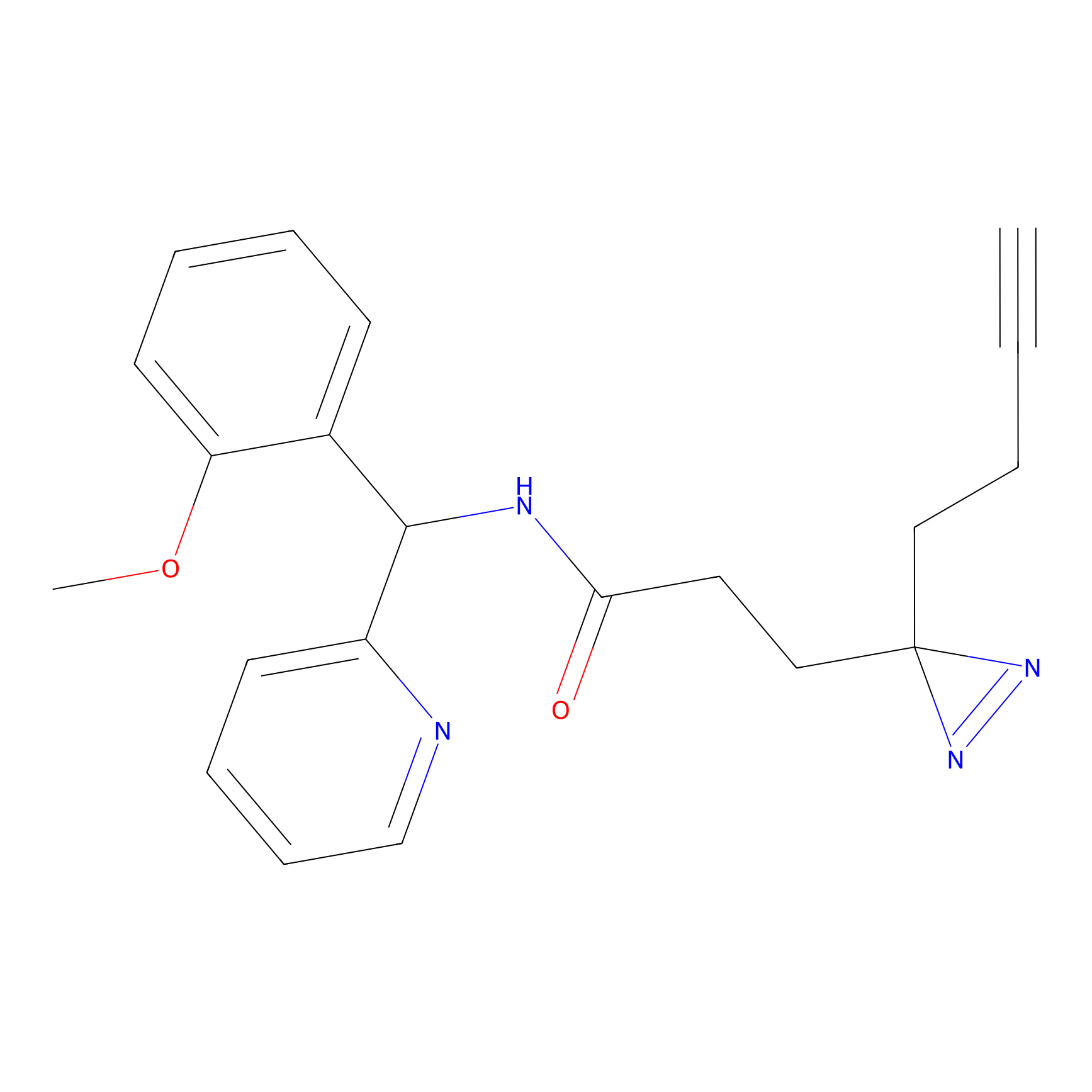 |
5.03 | LDD1869 | [25] | |
|
FFF probe13 Probe Info |
 |
20.00 | LDD0475 | [26] | |
|
FFF probe2 Probe Info |
 |
6.26 | LDD0463 | [26] | |
|
FFF probe3 Probe Info |
 |
8.30 | LDD0464 | [26] | |
|
DA-2 Probe Info |
 |
N.A. | LDD0072 | [27] | |
|
OEA-DA Probe Info |
 |
3.76 | LDD0046 | [28] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C179(0.53); C128(0.33) | LDD2142 | [29] |
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C128(0.75) | LDD2112 | [29] |
| LDCM0502 | 1-(Cyanoacetyl)piperidine | MDA-MB-231 | C179(0.60) | LDD2095 | [29] |
| LDCM0537 | 2-Cyano-N,N-dimethylacetamide | MDA-MB-231 | C179(1.00) | LDD2130 | [29] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C179(1.27); C128(1.24) | LDD2117 | [29] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C179(1.12); C128(1.33) | LDD2152 | [29] |
| LDCM0510 | 3-(4-(Hydroxydiphenylmethyl)piperidin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C179(0.95); C128(0.97) | LDD2103 | [29] |
| LDCM0539 | 3-(4-Isopropylpiperazin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C179(0.52) | LDD2132 | [29] |
| LDCM0538 | 4-(Cyanoacetyl)morpholine | MDA-MB-231 | C179(0.72) | LDD2131 | [29] |
| LDCM0214 | AC1 | HEK-293T | C179(1.28); C128(0.93); C60(1.33) | LDD1507 | [30] |
| LDCM0215 | AC10 | HEK-293T | C128(1.00); C60(1.18) | LDD1508 | [30] |
| LDCM0226 | AC11 | HEK-293T | C179(1.13); C128(1.06); C60(1.04) | LDD1509 | [30] |
| LDCM0237 | AC12 | HEK-293T | C179(1.39); C128(0.99); C60(1.05) | LDD1510 | [30] |
| LDCM0259 | AC14 | HEK-293T | C179(1.07); C128(1.10); C60(0.96) | LDD1512 | [30] |
| LDCM0270 | AC15 | HEK-293T | C179(1.07); C128(1.02); C60(0.93) | LDD1513 | [30] |
| LDCM0276 | AC17 | HEK-293T | C179(1.06); C128(0.93); C60(1.07) | LDD1515 | [30] |
| LDCM0277 | AC18 | HEK-293T | C128(0.97); C60(1.13) | LDD1516 | [30] |
| LDCM0278 | AC19 | HEK-293T | C179(0.92); C128(0.95); C60(1.11) | LDD1517 | [30] |
| LDCM0279 | AC2 | HEK-293T | C128(1.00); C60(1.24) | LDD1518 | [30] |
| LDCM0280 | AC20 | HEK-293T | C179(1.24); C128(1.06); C60(1.11) | LDD1519 | [30] |
| LDCM0281 | AC21 | HEK-293T | C179(1.06); C128(1.07); C60(1.12) | LDD1520 | [30] |
| LDCM0282 | AC22 | HEK-293T | C179(1.00); C128(1.10); C60(1.17) | LDD1521 | [30] |
| LDCM0283 | AC23 | HEK-293T | C179(1.10); C128(1.06); C60(1.08) | LDD1522 | [30] |
| LDCM0284 | AC24 | HEK-293T | C179(1.04); C128(1.19); C60(0.96) | LDD1523 | [30] |
| LDCM0285 | AC25 | HEK-293T | C179(1.13); C128(0.99); C60(1.10) | LDD1524 | [30] |
| LDCM0286 | AC26 | HEK-293T | C128(1.03); C60(1.13) | LDD1525 | [30] |
| LDCM0287 | AC27 | HEK-293T | C179(1.08); C128(1.11); C60(1.01) | LDD1526 | [30] |
| LDCM0288 | AC28 | HEK-293T | C179(1.31); C128(1.10); C60(1.13) | LDD1527 | [30] |
| LDCM0289 | AC29 | HEK-293T | C179(1.19); C128(1.09); C60(1.12) | LDD1528 | [30] |
| LDCM0290 | AC3 | HEK-293T | C179(1.14); C128(1.04); C60(1.15) | LDD1529 | [30] |
| LDCM0291 | AC30 | HEK-293T | C179(1.06); C128(1.12); C60(1.17) | LDD1530 | [30] |
| LDCM0292 | AC31 | HEK-293T | C179(1.01); C128(1.02); C60(1.10) | LDD1531 | [30] |
| LDCM0293 | AC32 | HEK-293T | C179(1.05); C128(1.03); C60(0.99) | LDD1532 | [30] |
| LDCM0294 | AC33 | HEK-293T | C179(1.14); C128(0.95); C60(1.14) | LDD1533 | [30] |
| LDCM0295 | AC34 | HEK-293T | C128(0.98); C60(1.04) | LDD1534 | [30] |
| LDCM0296 | AC35 | HEK-293T | C179(1.29); C128(1.06); C60(1.08) | LDD1535 | [30] |
| LDCM0297 | AC36 | HEK-293T | C179(1.50); C128(1.08); C60(1.12) | LDD1536 | [30] |
| LDCM0298 | AC37 | HEK-293T | C179(1.33); C128(1.17); C60(1.01) | LDD1537 | [30] |
| LDCM0299 | AC38 | HEK-293T | C179(1.08); C128(1.15); C60(1.00) | LDD1538 | [30] |
| LDCM0300 | AC39 | HEK-293T | C179(1.09); C128(1.00); C60(1.00) | LDD1539 | [30] |
| LDCM0301 | AC4 | HEK-293T | C179(1.26); C128(1.05); C60(1.19) | LDD1540 | [30] |
| LDCM0302 | AC40 | HEK-293T | C179(1.06); C128(1.11); C60(0.98) | LDD1541 | [30] |
| LDCM0303 | AC41 | HEK-293T | C179(1.10); C128(0.89); C60(1.10) | LDD1542 | [30] |
| LDCM0304 | AC42 | HEK-293T | C128(0.92); C60(1.34) | LDD1543 | [30] |
| LDCM0305 | AC43 | HEK-293T | C179(1.15); C128(1.04); C60(1.08) | LDD1544 | [30] |
| LDCM0306 | AC44 | HEK-293T | C179(1.33); C128(1.00); C60(1.15) | LDD1545 | [30] |
| LDCM0307 | AC45 | HEK-293T | C179(1.16); C128(1.10); C60(1.01) | LDD1546 | [30] |
| LDCM0308 | AC46 | HEK-293T | C179(1.04); C128(1.10); C60(1.02) | LDD1547 | [30] |
| LDCM0309 | AC47 | HEK-293T | C179(1.03); C128(1.01); C60(1.12) | LDD1548 | [30] |
| LDCM0310 | AC48 | HEK-293T | C179(1.09); C128(1.10); C60(1.03) | LDD1549 | [30] |
| LDCM0311 | AC49 | HEK-293T | C179(1.12); C128(0.94); C60(1.07) | LDD1550 | [30] |
| LDCM0312 | AC5 | HEK-293T | C179(1.30); C128(1.11); C60(1.09) | LDD1551 | [30] |
| LDCM0313 | AC50 | HEK-293T | C128(1.05); C60(1.16) | LDD1552 | [30] |
| LDCM0314 | AC51 | HEK-293T | C179(1.27); C128(1.10); C60(1.09) | LDD1553 | [30] |
| LDCM0315 | AC52 | HEK-293T | C179(1.16); C128(0.99); C60(1.06) | LDD1554 | [30] |
| LDCM0316 | AC53 | HEK-293T | C179(1.22); C128(1.06); C60(1.06) | LDD1555 | [30] |
| LDCM0317 | AC54 | HEK-293T | C179(1.08); C128(1.14); C60(1.13) | LDD1556 | [30] |
| LDCM0318 | AC55 | HEK-293T | C179(1.03); C128(1.00); C60(1.00) | LDD1557 | [30] |
| LDCM0319 | AC56 | HEK-293T | C179(1.00); C128(1.11); C60(1.00) | LDD1558 | [30] |
| LDCM0320 | AC57 | HEK-293T | C179(1.19); C128(0.93); C60(1.19) | LDD1559 | [30] |
| LDCM0321 | AC58 | HEK-293T | C128(1.10); C60(1.06) | LDD1560 | [30] |
| LDCM0322 | AC59 | HEK-293T | C179(1.16); C128(1.14); C60(1.12) | LDD1561 | [30] |
| LDCM0323 | AC6 | HEK-293T | C179(1.20); C128(1.07); C60(1.18) | LDD1562 | [30] |
| LDCM0324 | AC60 | HEK-293T | C179(1.35); C128(1.06); C60(1.13) | LDD1563 | [30] |
| LDCM0325 | AC61 | HEK-293T | C179(1.22); C128(1.06); C60(1.15) | LDD1564 | [30] |
| LDCM0326 | AC62 | HEK-293T | C179(1.19); C128(1.07); C60(1.08) | LDD1565 | [30] |
| LDCM0327 | AC63 | HEK-293T | C179(1.17); C128(0.95); C60(1.04) | LDD1566 | [30] |
| LDCM0328 | AC64 | HEK-293T | C179(1.12); C128(1.23); C60(1.13) | LDD1567 | [30] |
| LDCM0334 | AC7 | HEK-293T | C179(1.20); C128(1.00); C60(1.08) | LDD1568 | [30] |
| LDCM0345 | AC8 | HEK-293T | C179(1.03); C128(1.09); C60(1.08) | LDD1569 | [30] |
| LDCM0545 | Acetamide | MDA-MB-231 | C179(0.58) | LDD2138 | [29] |
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C179(0.80) | LDD2113 | [29] |
| LDCM0248 | AKOS034007472 | HEK-293T | C179(1.15); C128(1.04); C60(1.05) | LDD1511 | [30] |
| LDCM0356 | AKOS034007680 | HEK-293T | C179(1.10); C128(0.91); C60(1.16) | LDD1570 | [30] |
| LDCM0275 | AKOS034007705 | HEK-293T | C179(1.01); C128(1.14); C60(0.94) | LDD1514 | [30] |
| LDCM0156 | Aniline | NCI-H1299 | 11.76 | LDD0403 | [1] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C179(0.71) | LDD2091 | [29] |
| LDCM0630 | CCW28-3 | 231MFP | C60(1.36) | LDD2214 | [31] |
| LDCM0108 | Chloroacetamide | HeLa | H208(0.00); H11(0.00); H195(0.00); C179(0.00) | LDD0222 | [22] |
| LDCM0632 | CL-Sc | Hep-G2 | C179(20.00); C179(2.46); C128(2.02); C179(0.78) | LDD2227 | [17] |
| LDCM0367 | CL1 | HEK-293T | C179(1.47); C128(0.91); C60(1.05) | LDD1571 | [30] |
| LDCM0368 | CL10 | HEK-293T | C179(0.34); C128(0.84); C60(0.61) | LDD1572 | [30] |
| LDCM0369 | CL100 | HEK-293T | C128(1.01); C60(1.43) | LDD1573 | [30] |
| LDCM0370 | CL101 | HEK-293T | C179(1.27); C128(0.94); C60(1.22) | LDD1574 | [30] |
| LDCM0371 | CL102 | HEK-293T | C179(0.97); C128(0.94); C60(1.09) | LDD1575 | [30] |
| LDCM0372 | CL103 | HEK-293T | C179(1.14); C128(1.03); C60(0.98) | LDD1576 | [30] |
| LDCM0373 | CL104 | HEK-293T | C128(1.04); C60(1.10) | LDD1577 | [30] |
| LDCM0374 | CL105 | HEK-293T | C179(1.21); C128(0.96); C60(1.07) | LDD1578 | [30] |
| LDCM0375 | CL106 | HEK-293T | C179(0.91); C128(1.01); C60(0.97) | LDD1579 | [30] |
| LDCM0376 | CL107 | HEK-293T | C179(0.95); C128(1.04); C60(1.02) | LDD1580 | [30] |
| LDCM0377 | CL108 | HEK-293T | C128(1.07); C60(1.20) | LDD1581 | [30] |
| LDCM0378 | CL109 | HEK-293T | C179(1.16); C128(1.01); C60(1.07) | LDD1582 | [30] |
| LDCM0379 | CL11 | HEK-293T | C179(0.47); C128(1.03); C60(0.68) | LDD1583 | [30] |
| LDCM0380 | CL110 | HEK-293T | C179(0.91); C128(0.94); C60(0.92) | LDD1584 | [30] |
| LDCM0381 | CL111 | HEK-293T | C179(0.95); C128(0.96); C60(1.02) | LDD1585 | [30] |
| LDCM0382 | CL112 | HEK-293T | C128(1.08); C60(1.23) | LDD1586 | [30] |
| LDCM0383 | CL113 | HEK-293T | C179(1.32); C128(1.01); C60(1.23) | LDD1587 | [30] |
| LDCM0384 | CL114 | HEK-293T | C179(1.00); C128(0.95); C60(0.96) | LDD1588 | [30] |
| LDCM0385 | CL115 | HEK-293T | C179(1.04); C128(1.08); C60(1.03) | LDD1589 | [30] |
| LDCM0386 | CL116 | HEK-293T | C128(1.06); C60(1.18) | LDD1590 | [30] |
| LDCM0387 | CL117 | HEK-293T | C179(1.24); C128(1.01); C60(1.24) | LDD1591 | [30] |
| LDCM0388 | CL118 | HEK-293T | C179(1.05); C128(1.08); C60(0.98) | LDD1592 | [30] |
| LDCM0389 | CL119 | HEK-293T | C179(1.04); C128(1.05); C60(1.03) | LDD1593 | [30] |
| LDCM0390 | CL12 | HEK-293T | C179(0.38); C128(1.03); C60(0.82) | LDD1594 | [30] |
| LDCM0391 | CL120 | HEK-293T | C128(1.11); C60(1.23) | LDD1595 | [30] |
| LDCM0392 | CL121 | HEK-293T | C179(1.33); C128(1.04); C60(1.14) | LDD1596 | [30] |
| LDCM0393 | CL122 | HEK-293T | C179(1.11); C128(1.03); C60(0.97) | LDD1597 | [30] |
| LDCM0394 | CL123 | HEK-293T | C179(0.92); C128(0.91); C60(1.02) | LDD1598 | [30] |
| LDCM0395 | CL124 | HEK-293T | C128(1.16); C60(1.17) | LDD1599 | [30] |
| LDCM0396 | CL125 | HEK-293T | C179(1.51); C128(1.04); C60(1.28) | LDD1600 | [30] |
| LDCM0397 | CL126 | HEK-293T | C179(1.38); C128(0.99); C60(1.04) | LDD1601 | [30] |
| LDCM0398 | CL127 | HEK-293T | C179(1.24); C128(1.07); C60(1.15) | LDD1602 | [30] |
| LDCM0399 | CL128 | HEK-293T | C128(1.15); C60(1.24) | LDD1603 | [30] |
| LDCM0400 | CL13 | HEK-293T | C179(1.54); C128(0.85); C60(1.07) | LDD1604 | [30] |
| LDCM0401 | CL14 | HEK-293T | C179(1.04); C128(1.01); C60(0.94) | LDD1605 | [30] |
| LDCM0402 | CL15 | HEK-293T | C179(0.90); C128(0.80); C60(0.83) | LDD1606 | [30] |
| LDCM0403 | CL16 | HEK-293T | C128(1.11); C60(0.99) | LDD1607 | [30] |
| LDCM0404 | CL17 | HEK-293T | C179(0.84); C128(0.69); C60(0.86) | LDD1608 | [30] |
| LDCM0405 | CL18 | HEK-293T | C128(0.97); C60(0.88) | LDD1609 | [30] |
| LDCM0406 | CL19 | HEK-293T | C179(0.85); C128(0.99); C60(0.88) | LDD1610 | [30] |
| LDCM0407 | CL2 | HEK-293T | C179(1.09); C128(0.89); C60(0.90) | LDD1611 | [30] |
| LDCM0408 | CL20 | HEK-293T | C179(0.87); C128(1.00); C60(0.84) | LDD1612 | [30] |
| LDCM0409 | CL21 | HEK-293T | C179(0.43); C128(0.96); C60(0.70) | LDD1613 | [30] |
| LDCM0410 | CL22 | HEK-293T | C179(0.37); C128(1.01); C60(0.72) | LDD1614 | [30] |
| LDCM0411 | CL23 | HEK-293T | C179(0.33); C128(1.00); C60(0.76) | LDD1615 | [30] |
| LDCM0412 | CL24 | HEK-293T | C179(0.25); C128(1.02); C60(0.78) | LDD1616 | [30] |
| LDCM0413 | CL25 | HEK-293T | C179(1.34); C128(0.91); C60(1.09) | LDD1617 | [30] |
| LDCM0414 | CL26 | HEK-293T | C179(1.06); C128(0.98); C60(0.92) | LDD1618 | [30] |
| LDCM0415 | CL27 | HEK-293T | C179(0.99); C128(1.05); C60(1.02) | LDD1619 | [30] |
| LDCM0416 | CL28 | HEK-293T | C128(1.00); C60(1.03) | LDD1620 | [30] |
| LDCM0417 | CL29 | HEK-293T | C179(0.89); C128(0.97); C60(0.98) | LDD1621 | [30] |
| LDCM0418 | CL3 | HEK-293T | C179(1.00); C128(0.96); C60(0.92) | LDD1622 | [30] |
| LDCM0419 | CL30 | HEK-293T | C128(0.98); C60(0.88) | LDD1623 | [30] |
| LDCM0420 | CL31 | HEK-293T | C179(0.82); C128(0.90); C60(0.80) | LDD1624 | [30] |
| LDCM0421 | CL32 | HEK-293T | C179(0.81); C128(1.02); C60(0.83) | LDD1625 | [30] |
| LDCM0422 | CL33 | HEK-293T | C179(0.36); C128(0.93); C60(0.61) | LDD1626 | [30] |
| LDCM0423 | CL34 | HEK-293T | C179(0.32); C128(1.13); C60(0.68) | LDD1627 | [30] |
| LDCM0424 | CL35 | HEK-293T | C179(0.31); C128(1.00); C60(0.75) | LDD1628 | [30] |
| LDCM0425 | CL36 | HEK-293T | C179(0.22); C128(1.14); C60(0.74) | LDD1629 | [30] |
| LDCM0426 | CL37 | HEK-293T | C179(1.36); C128(0.95); C60(1.24) | LDD1630 | [30] |
| LDCM0428 | CL39 | HEK-293T | C179(0.97); C128(1.04); C60(0.97) | LDD1632 | [30] |
| LDCM0429 | CL4 | HEK-293T | C128(0.87); C60(1.02) | LDD1633 | [30] |
| LDCM0430 | CL40 | HEK-293T | C128(1.07); C60(1.06) | LDD1634 | [30] |
| LDCM0431 | CL41 | HEK-293T | C179(0.86); C128(0.98); C60(1.00) | LDD1635 | [30] |
| LDCM0432 | CL42 | HEK-293T | C128(1.03); C60(0.90) | LDD1636 | [30] |
| LDCM0433 | CL43 | HEK-293T | C179(0.76); C128(1.18); C60(0.82) | LDD1637 | [30] |
| LDCM0434 | CL44 | HEK-293T | C179(0.89); C128(1.06); C60(0.87) | LDD1638 | [30] |
| LDCM0435 | CL45 | HEK-293T | C179(0.49); C128(0.98); C60(0.74) | LDD1639 | [30] |
| LDCM0436 | CL46 | HEK-293T | C179(0.30); C128(1.16); C60(0.74) | LDD1640 | [30] |
| LDCM0437 | CL47 | HEK-293T | C179(0.29); C128(1.01); C60(0.76) | LDD1641 | [30] |
| LDCM0438 | CL48 | HEK-293T | C179(0.16); C128(1.07); C60(0.75) | LDD1642 | [30] |
| LDCM0439 | CL49 | HEK-293T | C179(1.43); C128(0.97); C60(1.28) | LDD1643 | [30] |
| LDCM0440 | CL5 | HEK-293T | C179(1.09); C128(0.84); C60(1.00) | LDD1644 | [30] |
| LDCM0441 | CL50 | HEK-293T | C179(1.13); C128(0.97); C60(0.89) | LDD1645 | [30] |
| LDCM0443 | CL52 | HEK-293T | C128(1.02); C60(1.03) | LDD1646 | [30] |
| LDCM0444 | CL53 | HEK-293T | C179(0.82); C128(0.81); C60(0.89) | LDD1647 | [30] |
| LDCM0445 | CL54 | HEK-293T | C128(0.94); C60(0.78) | LDD1648 | [30] |
| LDCM0446 | CL55 | HEK-293T | C179(0.77); C128(1.03); C60(0.78) | LDD1649 | [30] |
| LDCM0447 | CL56 | HEK-293T | C179(0.82); C128(1.03); C60(0.83) | LDD1650 | [30] |
| LDCM0448 | CL57 | HEK-293T | C179(0.43); C128(0.91); C60(0.66) | LDD1651 | [30] |
| LDCM0449 | CL58 | HEK-293T | C179(0.39); C128(1.12); C60(0.70) | LDD1652 | [30] |
| LDCM0450 | CL59 | HEK-293T | C179(0.32); C128(0.97); C60(0.74) | LDD1653 | [30] |
| LDCM0451 | CL6 | HEK-293T | C128(0.79); C60(0.87) | LDD1654 | [30] |
| LDCM0452 | CL60 | HEK-293T | C179(0.22); C128(1.07); C60(0.78) | LDD1655 | [30] |
| LDCM0453 | CL61 | HEK-293T | C179(1.59); C128(0.98); C60(1.28) | LDD1656 | [30] |
| LDCM0454 | CL62 | HEK-293T | C179(1.14); C128(1.00); C60(1.02) | LDD1657 | [30] |
| LDCM0455 | CL63 | HEK-293T | C179(1.04); C128(1.06); C60(1.04) | LDD1658 | [30] |
| LDCM0456 | CL64 | HEK-293T | C128(0.98); C60(1.01) | LDD1659 | [30] |
| LDCM0457 | CL65 | HEK-293T | C179(1.00); C128(0.94); C60(1.05) | LDD1660 | [30] |
| LDCM0458 | CL66 | HEK-293T | C128(0.96); C60(0.93) | LDD1661 | [30] |
| LDCM0459 | CL67 | HEK-293T | C179(0.87); C128(1.04); C60(0.81) | LDD1662 | [30] |
| LDCM0460 | CL68 | HEK-293T | C179(0.83); C128(1.08); C60(0.82) | LDD1663 | [30] |
| LDCM0461 | CL69 | HEK-293T | C179(0.47); C128(1.04); C60(0.72) | LDD1664 | [30] |
| LDCM0462 | CL7 | HEK-293T | C179(1.00); C128(0.94); C60(0.86) | LDD1665 | [30] |
| LDCM0463 | CL70 | HEK-293T | C179(0.38); C128(1.10); C60(0.71) | LDD1666 | [30] |
| LDCM0464 | CL71 | HEK-293T | C179(0.34); C128(0.99); C60(0.80) | LDD1667 | [30] |
| LDCM0465 | CL72 | HEK-293T | C179(0.32); C128(1.02); C60(0.76) | LDD1668 | [30] |
| LDCM0466 | CL73 | HEK-293T | C179(1.58); C128(1.02); C60(1.19) | LDD1669 | [30] |
| LDCM0467 | CL74 | HEK-293T | C179(1.28); C128(1.07); C60(1.01) | LDD1670 | [30] |
| LDCM0469 | CL76 | HEK-293T | C128(1.04); C60(1.10) | LDD1672 | [30] |
| LDCM0470 | CL77 | HEK-293T | C179(0.87); C128(0.82); C60(0.95) | LDD1673 | [30] |
| LDCM0471 | CL78 | HEK-293T | C128(1.10); C60(0.96) | LDD1674 | [30] |
| LDCM0472 | CL79 | HEK-293T | C179(0.98); C128(1.12); C60(0.87) | LDD1675 | [30] |
| LDCM0473 | CL8 | HEK-293T | C179(0.53); C128(0.82); C60(0.73) | LDD1676 | [30] |
| LDCM0474 | CL80 | HEK-293T | C179(0.87); C128(1.17); C60(0.88) | LDD1677 | [30] |
| LDCM0475 | CL81 | HEK-293T | C179(0.58); C128(1.06); C60(0.77) | LDD1678 | [30] |
| LDCM0476 | CL82 | HEK-293T | C179(0.46); C128(1.27); C60(0.77) | LDD1679 | [30] |
| LDCM0477 | CL83 | HEK-293T | C179(0.38); C128(1.10); C60(0.74) | LDD1680 | [30] |
| LDCM0478 | CL84 | HEK-293T | C179(0.38); C128(1.10); C60(0.76) | LDD1681 | [30] |
| LDCM0479 | CL85 | HEK-293T | C179(1.59); C128(0.95); C60(1.49) | LDD1682 | [30] |
| LDCM0480 | CL86 | HEK-293T | C179(1.78); C128(1.01); C60(1.29) | LDD1683 | [30] |
| LDCM0481 | CL87 | HEK-293T | C179(1.40); C128(1.02); C60(1.25) | LDD1684 | [30] |
| LDCM0482 | CL88 | HEK-293T | C128(1.04); C60(1.31) | LDD1685 | [30] |
| LDCM0483 | CL89 | HEK-293T | C179(1.59); C128(0.92); C60(1.08) | LDD1686 | [30] |
| LDCM0484 | CL9 | HEK-293T | C179(0.51); C128(0.93); C60(0.74) | LDD1687 | [30] |
| LDCM0485 | CL90 | HEK-293T | C128(0.77); C60(0.77) | LDD1688 | [30] |
| LDCM0486 | CL91 | HEK-293T | C179(0.97); C128(1.15); C60(0.92) | LDD1689 | [30] |
| LDCM0487 | CL92 | HEK-293T | C179(0.98); C128(0.93); C60(0.85) | LDD1690 | [30] |
| LDCM0488 | CL93 | HEK-293T | C179(0.56); C128(1.03); C60(0.71) | LDD1691 | [30] |
| LDCM0489 | CL94 | HEK-293T | C179(0.53); C128(1.12); C60(0.82) | LDD1692 | [30] |
| LDCM0490 | CL95 | HEK-293T | C179(0.45); C128(0.86); C60(0.84) | LDD1693 | [30] |
| LDCM0491 | CL96 | HEK-293T | C179(0.55); C128(1.02); C60(0.82) | LDD1694 | [30] |
| LDCM0492 | CL97 | HEK-293T | C179(1.36); C128(0.95); C60(1.19) | LDD1695 | [30] |
| LDCM0493 | CL98 | HEK-293T | C179(1.06); C128(0.97); C60(1.00) | LDD1696 | [30] |
| LDCM0494 | CL99 | HEK-293T | C179(1.02); C128(0.96); C60(1.16) | LDD1697 | [30] |
| LDCM0028 | Dobutamine | HEK-293T | 5.28 | LDD0180 | [10] |
| LDCM0495 | E2913 | HEK-293T | C179(1.04); C128(0.91); C60(1.08) | LDD1698 | [30] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C179(0.50) | LDD1702 | [29] |
| LDCM0625 | F8 | Ramos | C179(2.86); C128(1.12) | LDD2187 | [32] |
| LDCM0572 | Fragment10 | Ramos | C179(1.01); C128(0.60) | LDD2189 | [32] |
| LDCM0573 | Fragment11 | Ramos | C179(0.13); C128(0.20) | LDD2190 | [32] |
| LDCM0574 | Fragment12 | Ramos | C179(0.55); C128(0.96) | LDD2191 | [32] |
| LDCM0575 | Fragment13 | Ramos | C179(1.28); C128(1.33) | LDD2192 | [32] |
| LDCM0576 | Fragment14 | Ramos | C179(0.54); C128(0.66) | LDD2193 | [32] |
| LDCM0579 | Fragment20 | Ramos | C179(0.60); C128(0.72) | LDD2194 | [32] |
| LDCM0580 | Fragment21 | Ramos | C179(0.65); C128(0.87) | LDD2195 | [32] |
| LDCM0582 | Fragment23 | Ramos | C179(1.36); C128(1.05) | LDD2196 | [32] |
| LDCM0578 | Fragment27 | Ramos | C179(1.74); C128(0.98) | LDD2197 | [32] |
| LDCM0586 | Fragment28 | Ramos | C179(0.70); C128(1.17) | LDD2198 | [32] |
| LDCM0588 | Fragment30 | Ramos | C179(0.79); C128(1.53) | LDD2199 | [32] |
| LDCM0589 | Fragment31 | Ramos | C179(1.02); C128(1.45) | LDD2200 | [32] |
| LDCM0590 | Fragment32 | Ramos | C179(1.07); C128(0.67) | LDD2201 | [32] |
| LDCM0468 | Fragment33 | HEK-293T | C179(1.10); C128(1.08); C60(1.02) | LDD1671 | [30] |
| LDCM0596 | Fragment38 | Ramos | C179(1.72); C128(1.24) | LDD2203 | [32] |
| LDCM0566 | Fragment4 | Ramos | C179(0.79); C128(0.73) | LDD2184 | [32] |
| LDCM0427 | Fragment51 | HEK-293T | C179(1.08); C128(1.04); C60(0.96) | LDD1631 | [30] |
| LDCM0610 | Fragment52 | Ramos | C179(0.92); C128(1.90) | LDD2204 | [32] |
| LDCM0614 | Fragment56 | Ramos | C179(1.09); C128(1.57) | LDD2205 | [32] |
| LDCM0569 | Fragment7 | Ramos | C179(0.70); C128(0.69) | LDD2186 | [32] |
| LDCM0571 | Fragment9 | Ramos | C179(0.39); C128(0.59) | LDD2188 | [32] |
| LDCM0107 | IAA | HeLa | H208(0.00); H11(0.00); H163(0.00); C179(0.00) | LDD0221 | [22] |
| LDCM0022 | KB02 | HEK-293T | C179(1.03); C128(0.97); C60(1.00) | LDD1492 | [30] |
| LDCM0023 | KB03 | HEK-293T | C179(0.94); C128(0.94); C60(1.04) | LDD1497 | [30] |
| LDCM0024 | KB05 | G361 | C128(1.50) | LDD3311 | [9] |
| LDCM0509 | N-(4-bromo-3,5-dimethylphenyl)-2-nitroacetamide | MDA-MB-231 | C179(0.97); C128(0.91) | LDD2102 | [29] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C179(0.72) | LDD2121 | [29] |
| LDCM0109 | NEM | HeLa | H208(0.00); H163(0.00); H11(0.00); H195(0.00) | LDD0223 | [22] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C179(0.70) | LDD2089 | [29] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C179(1.37) | LDD2090 | [29] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C179(0.89); C128(1.17) | LDD2092 | [29] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C179(1.54); C128(1.23) | LDD2093 | [29] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C179(1.77) | LDD2094 | [29] |
| LDCM0503 | Nucleophilic fragment 14b | MDA-MB-231 | C179(2.39); C128(1.23) | LDD2096 | [29] |
| LDCM0504 | Nucleophilic fragment 15a | MDA-MB-231 | C179(0.72); C128(0.89) | LDD2097 | [29] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C179(0.55); C128(0.68) | LDD2098 | [29] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C128(1.17) | LDD2099 | [29] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C128(0.43) | LDD2100 | [29] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C179(1.06); C128(0.87) | LDD2101 | [29] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C179(0.55); C128(0.51) | LDD2104 | [29] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C128(1.61) | LDD2105 | [29] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C179(0.53) | LDD2106 | [29] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C179(1.02); C128(0.98) | LDD2107 | [29] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C179(0.62); C128(0.48) | LDD2108 | [29] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C179(0.66); C128(0.73) | LDD2109 | [29] |
| LDCM0517 | Nucleophilic fragment 21b | MDA-MB-231 | C179(0.98) | LDD2110 | [29] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C179(1.81); C128(1.26) | LDD2111 | [29] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C179(0.59); C128(0.55) | LDD2114 | [29] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C179(0.44); C128(0.63) | LDD2115 | [29] |
| LDCM0523 | Nucleophilic fragment 24b | MDA-MB-231 | C179(0.80); C128(1.07) | LDD2116 | [29] |
| LDCM0525 | Nucleophilic fragment 25b | MDA-MB-231 | C128(1.72) | LDD2118 | [29] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C179(2.13); C128(1.56) | LDD2119 | [29] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C179(0.66); C128(0.85) | LDD2120 | [29] |
| LDCM0529 | Nucleophilic fragment 27b | MDA-MB-231 | C128(1.06) | LDD2122 | [29] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C179(0.90); C128(0.93) | LDD2123 | [29] |
| LDCM0531 | Nucleophilic fragment 28b | MDA-MB-231 | C128(1.06) | LDD2124 | [29] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C179(0.95); C128(0.95) | LDD2125 | [29] |
| LDCM0533 | Nucleophilic fragment 29b | MDA-MB-231 | C179(1.27); C128(1.09) | LDD2126 | [29] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C179(1.43); C128(1.24) | LDD2127 | [29] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C128(1.15) | LDD2128 | [29] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C179(1.04); C128(1.05) | LDD2129 | [29] |
| LDCM0540 | Nucleophilic fragment 35 | MDA-MB-231 | C179(0.56); C128(0.50) | LDD2133 | [29] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C179(0.38); C128(0.65) | LDD2134 | [29] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C179(1.24); C128(1.19) | LDD2135 | [29] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C179(1.58); C128(1.28) | LDD2136 | [29] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C179(0.95); C128(1.08) | LDD2137 | [29] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C179(3.19); C128(1.41) | LDD1700 | [29] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C179(0.91); C128(0.60) | LDD2141 | [29] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C128(1.12) | LDD2143 | [29] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C179(2.47); C128(2.77) | LDD2144 | [29] |
| LDCM0551 | Nucleophilic fragment 5b | MDA-MB-231 | C179(3.90); C128(0.81) | LDD2145 | [29] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C179(0.76); C128(0.96) | LDD2146 | [29] |
| LDCM0553 | Nucleophilic fragment 6b | MDA-MB-231 | C128(3.36) | LDD2147 | [29] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C179(0.42); C128(0.53) | LDD2148 | [29] |
| LDCM0555 | Nucleophilic fragment 7b | MDA-MB-231 | C179(4.32) | LDD2149 | [29] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C179(0.56) | LDD2150 | [29] |
| LDCM0557 | Nucleophilic fragment 8b | MDA-MB-231 | C179(1.67); C128(0.97) | LDD2151 | [29] |
| LDCM0559 | Nucleophilic fragment 9b | MDA-MB-231 | C179(2.17) | LDD2153 | [29] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C60(1.21) | LDD2206 | [33] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C60(0.92); C179(0.77); C179(0.68) | LDD2207 | [33] |
| LDCM0131 | RA190 | MM1.R | C128(1.08) | LDD0304 | [34] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Zinc finger and BTB domain-containing protein 22 (ZBTB22) | Krueppel C2H2-type zinc-finger protein family | O15209 | |||
GPCR
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Mu-type opioid receptor (OPRM1) | G-protein coupled receptor 1 family | P35372 | |||
| Adhesion G-protein coupled receptor G7 (ADGRG7) | G-protein coupled receptor 2 family | Q96K78 | |||
Immunoglobulin
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| B-cell antigen receptor complex-associated protein alpha chain (CD79A) | . | P11912 | |||
Other
References
