Details of the Target
General Information of Target
| Target ID | LDTP01206 | |||||
|---|---|---|---|---|---|---|
| Target Name | Vesicle-trafficking protein SEC22b (SEC22B) | |||||
| Gene Name | SEC22B | |||||
| Gene ID | 9554 | |||||
| Synonyms |
SEC22L1; Vesicle-trafficking protein SEC22b; ER-Golgi SNARE of 24 kDa; ERS-24; ERS24; SEC22 vesicle-trafficking protein homolog B; SEC22 vesicle-trafficking protein-like 1 |
|||||
| 3D Structure | ||||||
| Sequence |
MVLLTMIARVADGLPLAASMQEDEQSGRDLQQYQSQAKQLFRKLNEQSPTRCTLEAGAMT
FHYIIEQGVCYLVLCEAAFPKKLAFAYLEDLHSEFDEQHGKKVPTVSRPYSFIEFDTFIQ KTKKLYIDSRARRNLGSINTELQDVQRIMVANIEEVLQRGEALSALDSKANNLSSLSKKY RQDAKYLNMRSTYAKLAAVAVFFIMLIVYVRFWWL |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
Synaptobrevin family
|
|||||
| Subcellular location |
Endoplasmic reticulum membrane
|
|||||
| Function | SNARE involved in targeting and fusion of ER-derived transport vesicles with the Golgi complex as well as Golgi-derived retrograde transport vesicles with the ER. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
CY-1 Probe Info |
 |
100.00 | LDD0243 | [1] | |
|
CY4 Probe Info |
 |
100.00 | LDD0244 | [1] | |
|
C-Sul Probe Info |
 |
3.02 | LDD0066 | [2] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [3] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [3] | |
|
1oxF11yne Probe Info |
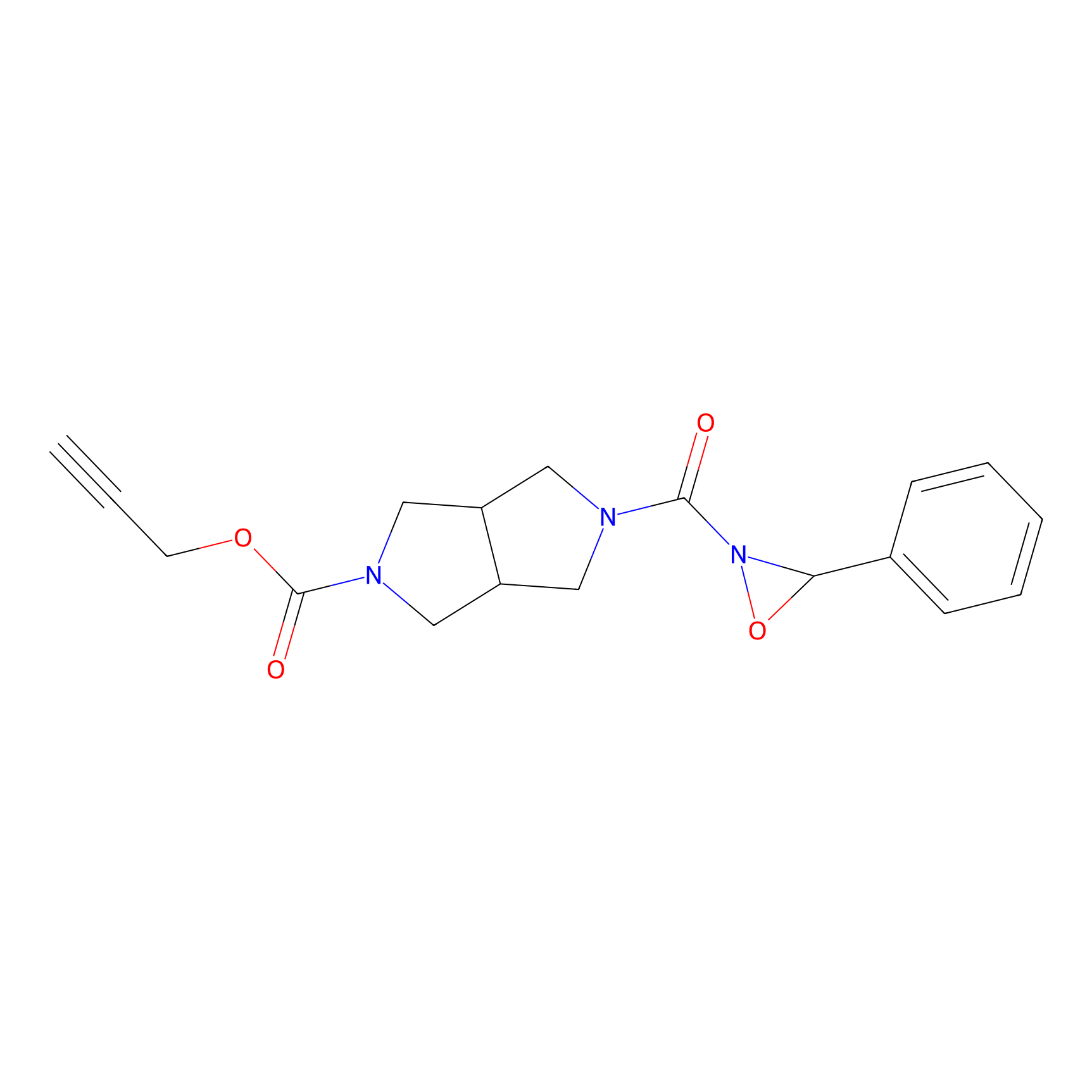 |
N.A. | LDD0193 | [4] | |
|
AZ-9 Probe Info |
 |
E141(10.00); D167(10.00) | LDD2208 | [5] | |
|
ONAyne Probe Info |
 |
K38(0.00); K43(0.00) | LDD0273 | [6] | |
|
HPAP Probe Info |
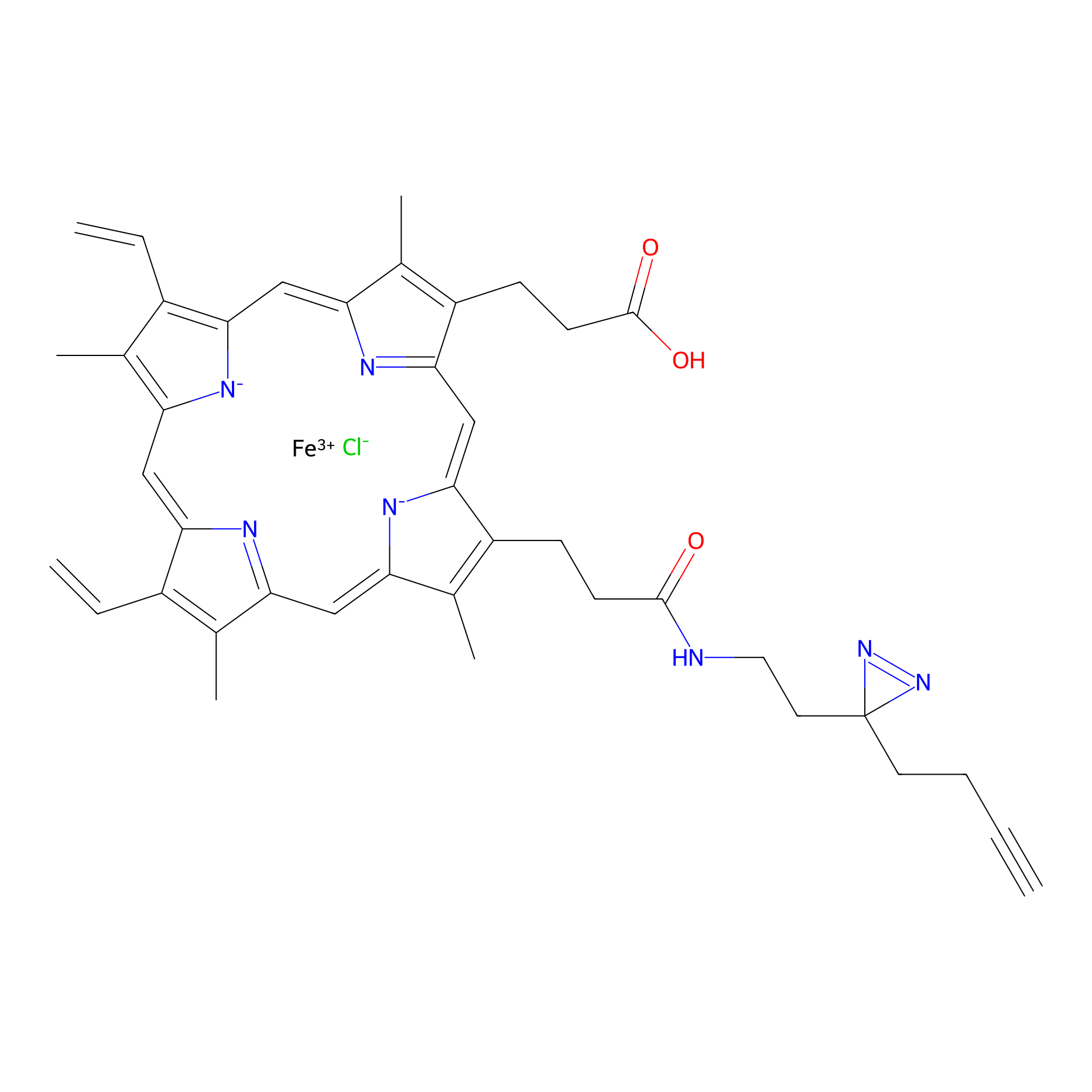 |
3.11 | LDD0064 | [7] | |
|
DBIA Probe Info |
 |
C52(1.04) | LDD1511 | [8] | |
|
AMP probe Probe Info |
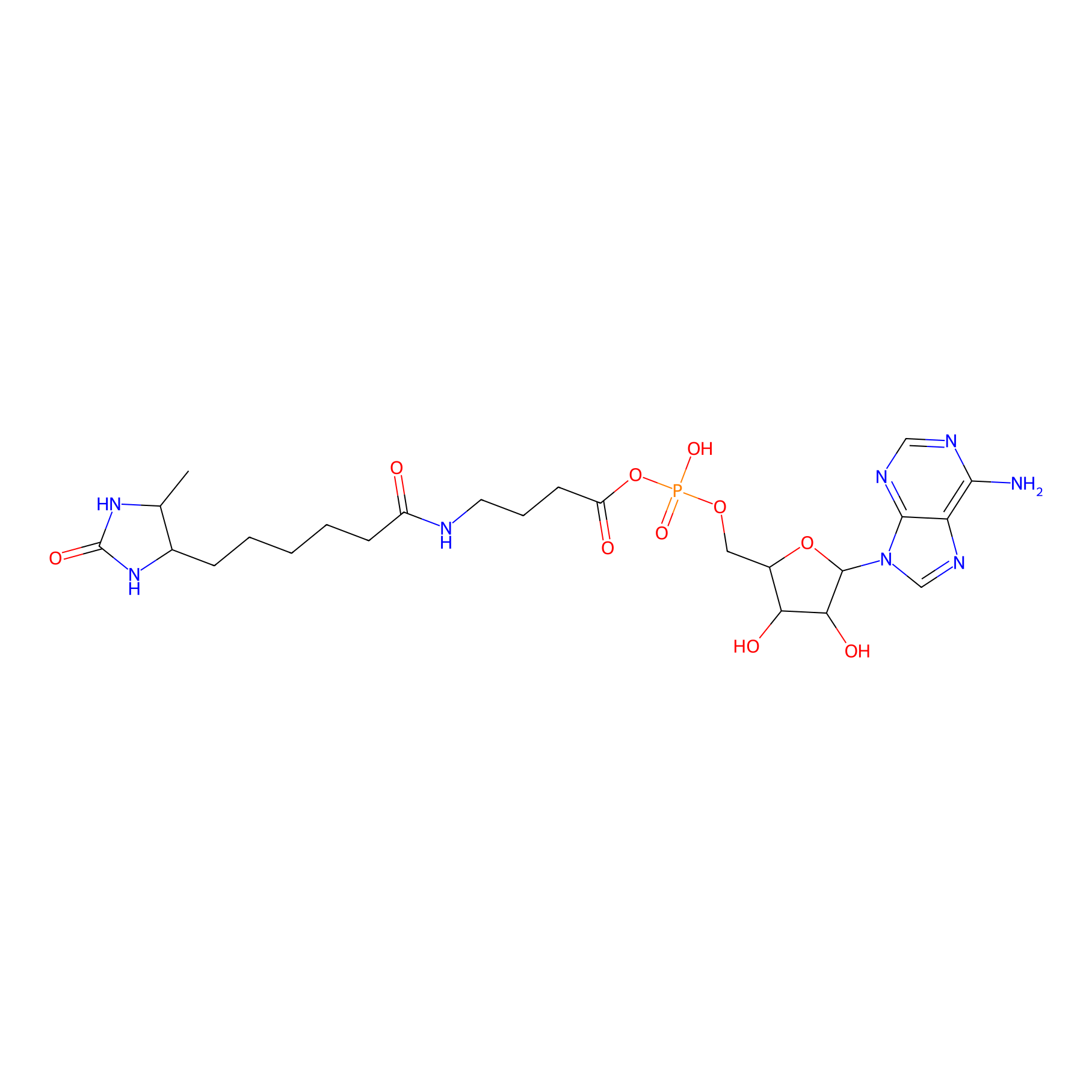 |
N.A. | LDD0200 | [9] | |
|
ATP probe Probe Info |
 |
K38(0.00); K169(0.00); K43(0.00) | LDD0199 | [9] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [10] | |
|
SF Probe Info |
 |
K43(0.00); Y110(0.00) | LDD0028 | [11] | |
|
STPyne Probe Info |
 |
K178(0.00); K169(0.00); K38(0.00) | LDD0009 | [10] | |
|
W1 Probe Info |
 |
N.A. | LDD0236 | [12] | |
|
MPP-AC Probe Info |
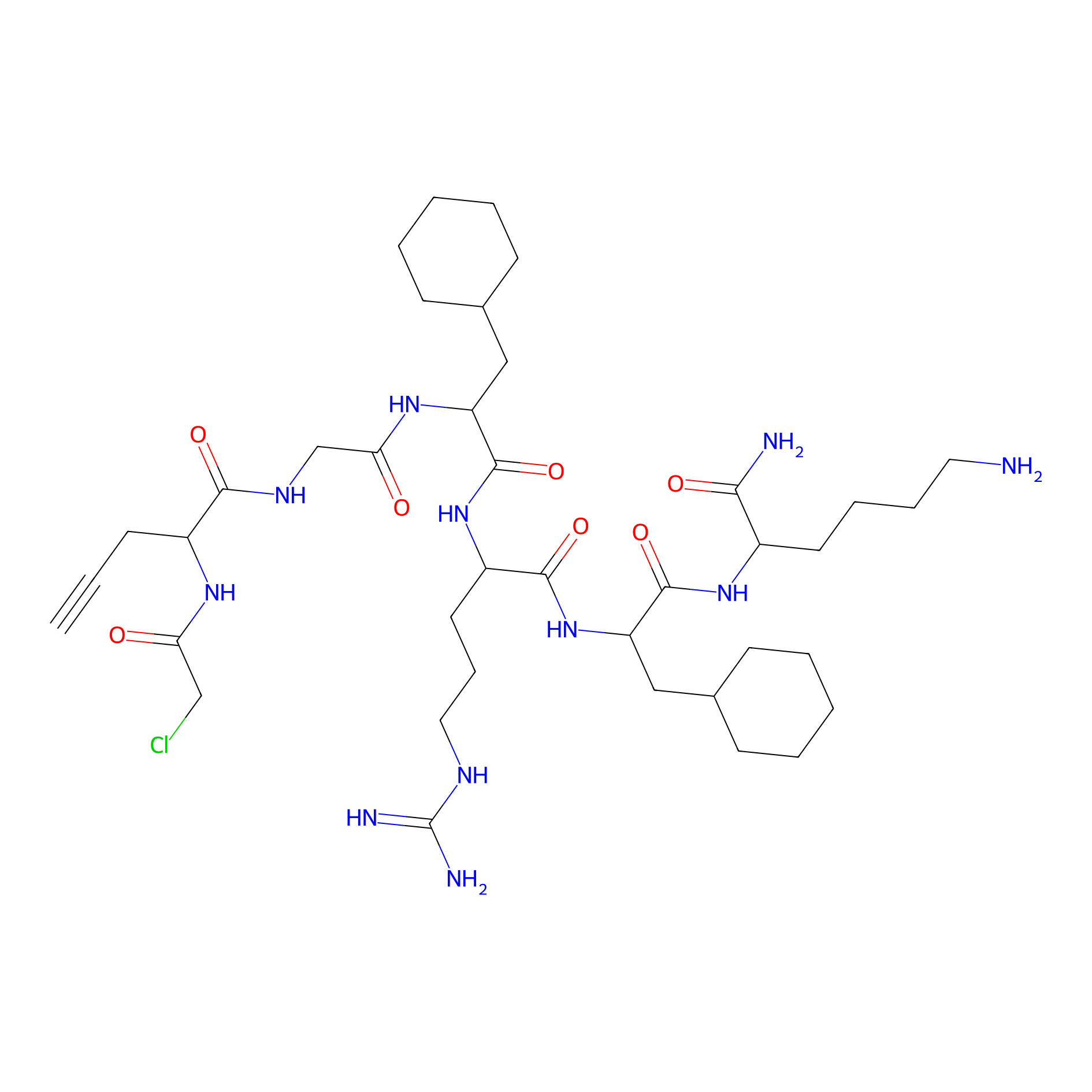 |
N.A. | LDD0428 | [13] | |
|
NAIA_5 Probe Info |
 |
C52(0.00); C75(0.00); C70(0.00) | LDD2223 | [14] | |
|
TER-AC Probe Info |
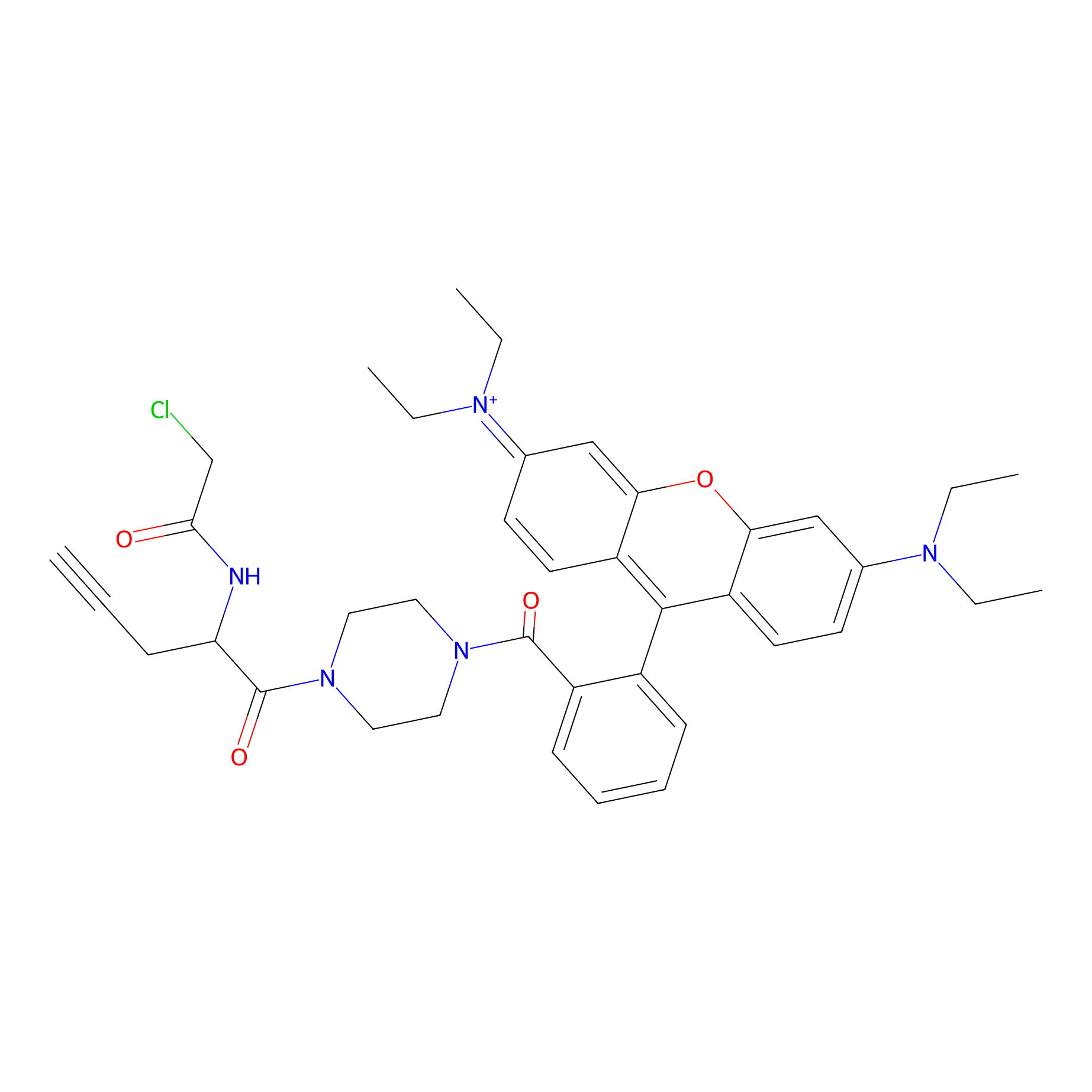 |
N.A. | LDD0426 | [13] | |
|
TPP-AC Probe Info |
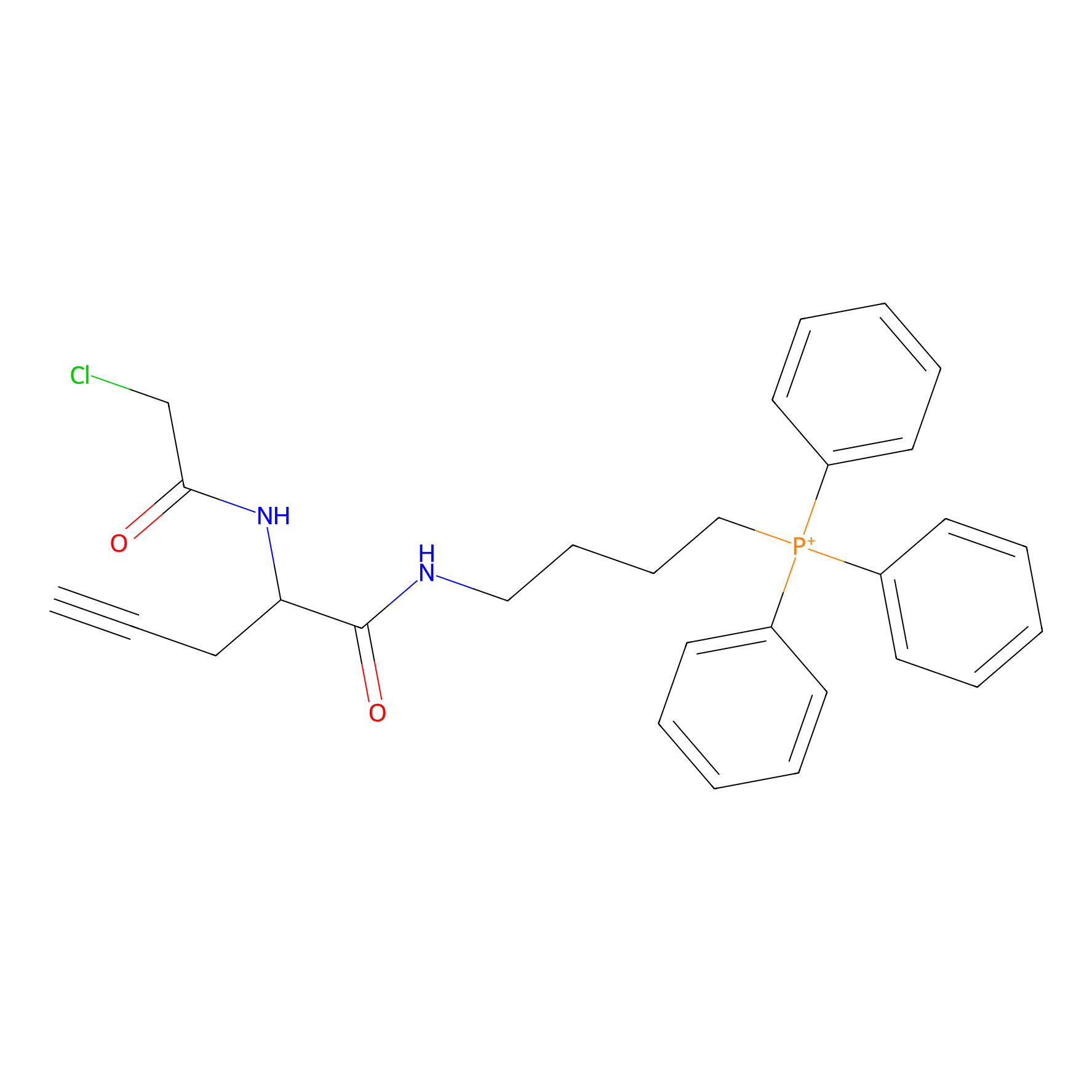 |
N.A. | LDD0427 | [13] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C094 Probe Info |
 |
23.92 | LDD1785 | [15] | |
|
C134 Probe Info |
 |
20.11 | LDD1816 | [15] | |
|
C187 Probe Info |
 |
20.68 | LDD1865 | [15] | |
|
C191 Probe Info |
 |
16.00 | LDD1868 | [15] | |
|
C193 Probe Info |
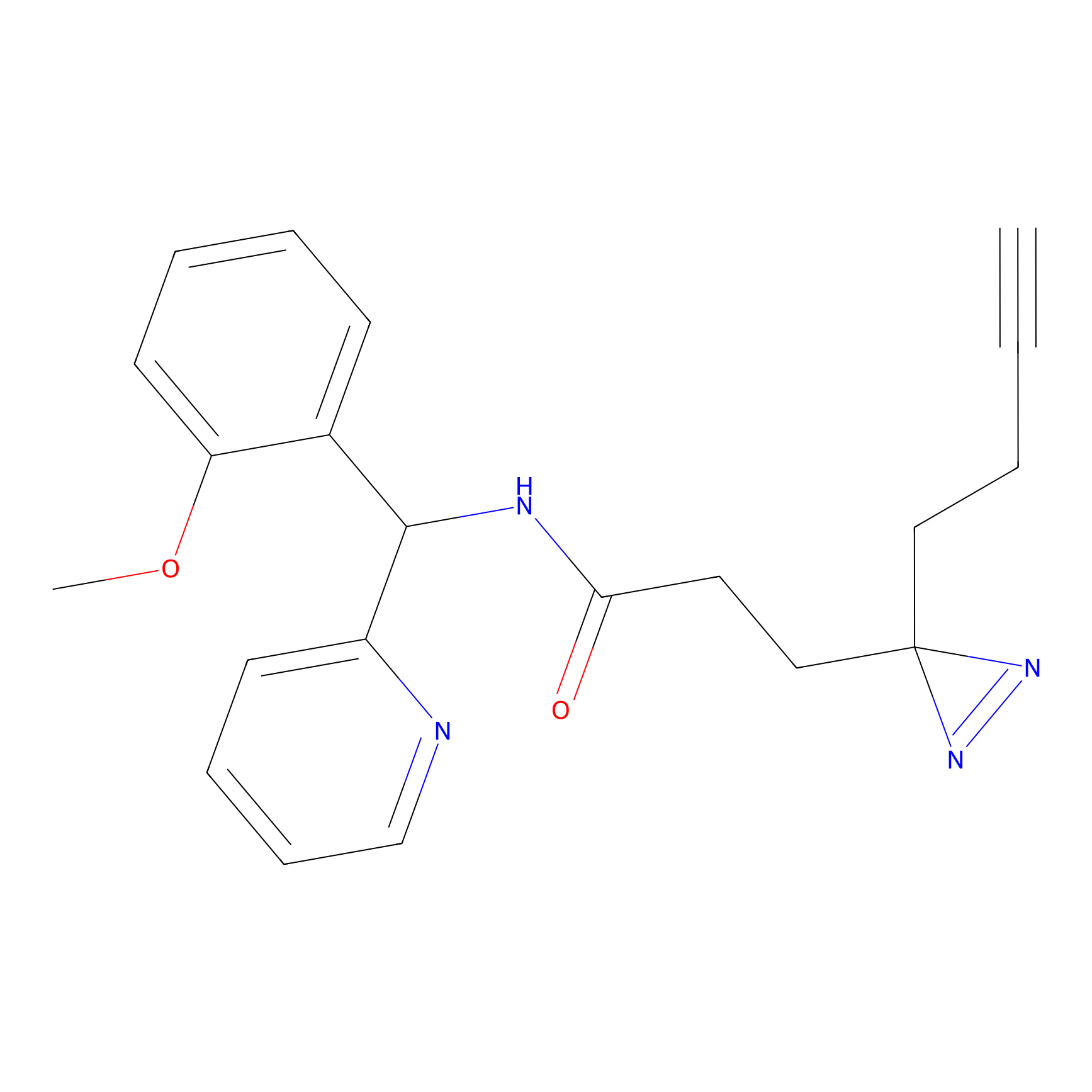 |
7.31 | LDD1869 | [15] | |
|
C289 Probe Info |
 |
27.86 | LDD1959 | [15] | |
|
C350 Probe Info |
 |
24.08 | LDD2011 | [15] | |
|
C388 Probe Info |
 |
35.75 | LDD2047 | [15] | |
|
FFF probe11 Probe Info |
 |
16.78 | LDD0472 | [16] | |
|
FFF probe13 Probe Info |
 |
9.22 | LDD0476 | [16] | |
|
FFF probe3 Probe Info |
 |
11.54 | LDD0465 | [16] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0138 | [17] | |
|
DA-2 Probe Info |
 |
N.A. | LDD0071 | [18] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0068 | [19] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0259 | AC14 | HEK-293T | C52(0.95) | LDD1512 | [8] |
| LDCM0281 | AC21 | HEK-293T | C52(0.86) | LDD1520 | [8] |
| LDCM0282 | AC22 | HEK-293T | C52(0.80) | LDD1521 | [8] |
| LDCM0289 | AC29 | HEK-293T | C52(0.90) | LDD1528 | [8] |
| LDCM0291 | AC30 | HEK-293T | C52(0.67) | LDD1530 | [8] |
| LDCM0298 | AC37 | HEK-293T | C52(1.11) | LDD1537 | [8] |
| LDCM0299 | AC38 | HEK-293T | C52(0.76) | LDD1538 | [8] |
| LDCM0307 | AC45 | HEK-293T | C52(1.07) | LDD1546 | [8] |
| LDCM0308 | AC46 | HEK-293T | C52(0.78) | LDD1547 | [8] |
| LDCM0312 | AC5 | HEK-293T | C52(0.85) | LDD1551 | [8] |
| LDCM0316 | AC53 | HEK-293T | C52(0.91) | LDD1555 | [8] |
| LDCM0317 | AC54 | HEK-293T | C52(0.74) | LDD1556 | [8] |
| LDCM0323 | AC6 | HEK-293T | C52(0.71) | LDD1562 | [8] |
| LDCM0325 | AC61 | HEK-293T | C52(1.08) | LDD1564 | [8] |
| LDCM0326 | AC62 | HEK-293T | C52(0.87) | LDD1565 | [8] |
| LDCM0248 | AKOS034007472 | HEK-293T | C52(1.04) | LDD1511 | [8] |
| LDCM0367 | CL1 | HEK-293T | C52(0.88) | LDD1571 | [8] |
| LDCM0368 | CL10 | HEK-293T | C52(0.97) | LDD1572 | [8] |
| LDCM0370 | CL101 | HEK-293T | C52(0.98) | LDD1574 | [8] |
| LDCM0372 | CL103 | HEK-293T | C52(0.92) | LDD1576 | [8] |
| LDCM0374 | CL105 | HEK-293T | C52(0.85) | LDD1578 | [8] |
| LDCM0376 | CL107 | HEK-293T | C52(0.97) | LDD1580 | [8] |
| LDCM0378 | CL109 | HEK-293T | C52(0.90) | LDD1582 | [8] |
| LDCM0381 | CL111 | HEK-293T | C52(0.98) | LDD1585 | [8] |
| LDCM0383 | CL113 | HEK-293T | C52(0.86) | LDD1587 | [8] |
| LDCM0385 | CL115 | HEK-293T | C52(0.94) | LDD1589 | [8] |
| LDCM0387 | CL117 | HEK-293T | C52(0.81) | LDD1591 | [8] |
| LDCM0389 | CL119 | HEK-293T | C52(1.12) | LDD1593 | [8] |
| LDCM0392 | CL121 | HEK-293T | C52(0.85) | LDD1596 | [8] |
| LDCM0394 | CL123 | HEK-293T | C52(0.82) | LDD1598 | [8] |
| LDCM0396 | CL125 | HEK-293T | C52(1.27) | LDD1600 | [8] |
| LDCM0398 | CL127 | HEK-293T | C52(1.01) | LDD1602 | [8] |
| LDCM0400 | CL13 | HEK-293T | C52(0.88) | LDD1604 | [8] |
| LDCM0402 | CL15 | HEK-293T | C52(0.93) | LDD1606 | [8] |
| LDCM0409 | CL21 | HEK-293T | C52(0.94) | LDD1613 | [8] |
| LDCM0410 | CL22 | HEK-293T | C52(0.90) | LDD1614 | [8] |
| LDCM0413 | CL25 | HEK-293T | C52(1.08) | LDD1617 | [8] |
| LDCM0415 | CL27 | HEK-293T | C52(0.96) | LDD1619 | [8] |
| LDCM0418 | CL3 | HEK-293T | C52(1.08) | LDD1622 | [8] |
| LDCM0422 | CL33 | HEK-293T | C52(0.85) | LDD1626 | [8] |
| LDCM0423 | CL34 | HEK-293T | C52(0.91) | LDD1627 | [8] |
| LDCM0426 | CL37 | HEK-293T | C52(0.95) | LDD1630 | [8] |
| LDCM0428 | CL39 | HEK-293T | C52(0.84) | LDD1632 | [8] |
| LDCM0435 | CL45 | HEK-293T | C52(0.68) | LDD1639 | [8] |
| LDCM0436 | CL46 | HEK-293T | C52(0.80) | LDD1640 | [8] |
| LDCM0439 | CL49 | HEK-293T | C52(0.87) | LDD1643 | [8] |
| LDCM0448 | CL57 | HEK-293T | C52(0.86) | LDD1651 | [8] |
| LDCM0449 | CL58 | HEK-293T | C52(0.84) | LDD1652 | [8] |
| LDCM0453 | CL61 | HEK-293T | C52(1.04) | LDD1656 | [8] |
| LDCM0455 | CL63 | HEK-293T | C52(1.13) | LDD1658 | [8] |
| LDCM0461 | CL69 | HEK-293T | C52(1.19) | LDD1664 | [8] |
| LDCM0463 | CL70 | HEK-293T | C52(0.84) | LDD1666 | [8] |
| LDCM0466 | CL73 | HEK-293T | C52(0.95) | LDD1669 | [8] |
| LDCM0475 | CL81 | HEK-293T | C52(1.48) | LDD1678 | [8] |
| LDCM0476 | CL82 | HEK-293T | C52(0.98) | LDD1679 | [8] |
| LDCM0479 | CL85 | HEK-293T | C52(1.92) | LDD1682 | [8] |
| LDCM0481 | CL87 | HEK-293T | C52(0.98) | LDD1684 | [8] |
| LDCM0484 | CL9 | HEK-293T | C52(1.18) | LDD1687 | [8] |
| LDCM0488 | CL93 | HEK-293T | C52(1.02) | LDD1691 | [8] |
| LDCM0489 | CL94 | HEK-293T | C52(0.85) | LDD1692 | [8] |
| LDCM0492 | CL97 | HEK-293T | C52(0.80) | LDD1695 | [8] |
| LDCM0494 | CL99 | HEK-293T | C52(0.89) | LDD1697 | [8] |
| LDCM0495 | E2913 | HEK-293T | C52(0.87) | LDD1698 | [8] |
| LDCM0468 | Fragment33 | HEK-293T | C52(1.06) | LDD1671 | [8] |
| LDCM0014 | Panhematin | hPBMC | 3.11 | LDD0064 | [7] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Cyclic AMP-responsive element-binding protein 3-like protein 1 (CREB3L1) | BZIP family | Q96BA8 | |||
GPCR
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Probable G-protein coupled receptor 152 (GPR152) | G-protein coupled receptor 1 family | Q8TDT2 | |||
Immunoglobulin
Cytokine and receptor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Interferon gamma receptor 2 (IFNGR2) | Type II cytokine receptor family | P38484 | |||
Other
References
