Details of the Target
General Information of Target
| Target ID | LDTP12318 | |||||
|---|---|---|---|---|---|---|
| Target Name | Calcium-independent phospholipase A2-gamma (PNPLA8) | |||||
| Gene Name | PNPLA8 | |||||
| Gene ID | 50640 | |||||
| Synonyms |
IPLA22; IPLA2G; Calcium-independent phospholipase A2-gamma; EC 3.1.1.-; EC 3.1.1.5; Intracellular membrane-associated calcium-independent phospholipase A2 gamma; iPLA2-gamma; PNPLA-gamma; Patatin-like phospholipase domain-containing protein 8; iPLA2-2
|
|||||
| 3D Structure | ||||||
| Sequence |
MAALAPLPPLPAQFKSIQHHLRTAQEHDKRDPVVAYYCRLYAMQTGMKIDSKTPECRKFL
SKLMDQLEALKKQLGDNEAITQEIVGCAHLENYALKMFLYADNEDRAGRFHKNMIKSFYT ASLLIDVITVFGELTDENVKHRKYARWKATYIHNCLKNGETPQAGPVGIEEDNDIEENED AGAASLPTQPTQPSSSSTYDPSNMPSGNYTGIQIPPGAHAPANTPAEVPHSTGVASNTIQ PTPQTIPAIDPALFNTISQGDVRLTPEDFARAQKYCKYAGSALQYEDVSTAVQNLQKALK LLTTGRE |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Subcellular location |
Endoplasmic reticulum membrane
|
|||||
| Function |
Calcium-independent and membrane-bound phospholipase, that catalyzes the esterolytic cleavage of fatty acids from glycerophospholipids to yield free fatty acids and lysophospholipids, hence regulating membrane physical properties and the release of lipid second messengers and growth factors. Hydrolyzes phosphatidylethanolamine, phosphatidylcholine and probably phosphatidylinositol with a possible preference for the former. Has also a broad substrate specificity in terms of fatty acid moieties, hydrolyzing saturated and mono-unsaturated fatty acids at nearly equal rates from either the sn-1 or sn-2 position in diacyl phosphatidylcholine. However, has a weak activity toward polyunsaturated fatty acids at the sn-2 position, and thereby favors the production of 2-arachidonoyl lysophosphatidylcholine, a key branch point metabolite in eicosanoid signaling. On the other hand, can produce arachidonic acid from the sn-1 position of diacyl phospholipid and from the sn-2 position of arachidonate-containing plasmalogen substrates. Therefore, plays an important role in the mobilization of arachidonic acid in response to cellular stimuli and the generation of lipid second messengers. Can also hydrolyze lysophosphatidylcholine. In the mitochondrial compartment, catalyzes the hydrolysis and release of oxidized aliphatic chains from cardiolipin and integrates mitochondrial bioenergetics and signaling. It is essential for maintaining efficient bioenergetic mitochondrial function through tailoring mitochondrial membrane lipid metabolism and composition.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| HEC1B | SNV: p.L313S | . | |||
| JHH6 | SNV: p.V20I | . | |||
| MFE319 | Deletion: p.M221CfsTer21 | . | |||
| RKO | Deletion: p.M221CfsTer21 | . | |||
| RL | SNV: p.N682D | . | |||
| SUPT1 | SNV: p.F393S | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K769(0.74) | LDD0277 | [1] | |
|
m-APA Probe Info |
 |
12.02 | LDD0403 | [2] | |
|
DBIA Probe Info |
 |
C714(1.71) | LDD3312 | [3] | |
|
BTD Probe Info |
 |
C559(1.00) | LDD2099 | [4] | |
|
AHL-Pu-1 Probe Info |
 |
C714(2.07) | LDD0168 | [5] | |
|
IA-alkyne Probe Info |
 |
C645(0.00); C597(0.00) | LDD0165 | [6] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [7] | |
|
IPM Probe Info |
 |
N.A. | LDD2156 | [8] | |
|
PF-06672131 Probe Info |
 |
N.A. | LDD0152 | [9] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [10] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [11] | |
|
NAIA_5 Probe Info |
 |
C643(0.00); C559(0.00) | LDD2223 | [12] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C004 Probe Info |
 |
5.66 | LDD1714 | [13] | |
|
C106 Probe Info |
 |
16.11 | LDD1793 | [13] | |
|
C228 Probe Info |
 |
19.43 | LDD1901 | [13] | |
|
C243 Probe Info |
 |
13.83 | LDD1916 | [13] | |
|
C246 Probe Info |
 |
13.00 | LDD1919 | [13] | |
|
C270 Probe Info |
 |
7.36 | LDD1940 | [13] | |
|
C288 Probe Info |
 |
8.63 | LDD1958 | [13] | |
|
C293 Probe Info |
 |
32.00 | LDD1963 | [13] | |
|
C296 Probe Info |
 |
15.14 | LDD1966 | [13] | |
|
C343 Probe Info |
 |
14.93 | LDD2005 | [13] | |
|
C382 Probe Info |
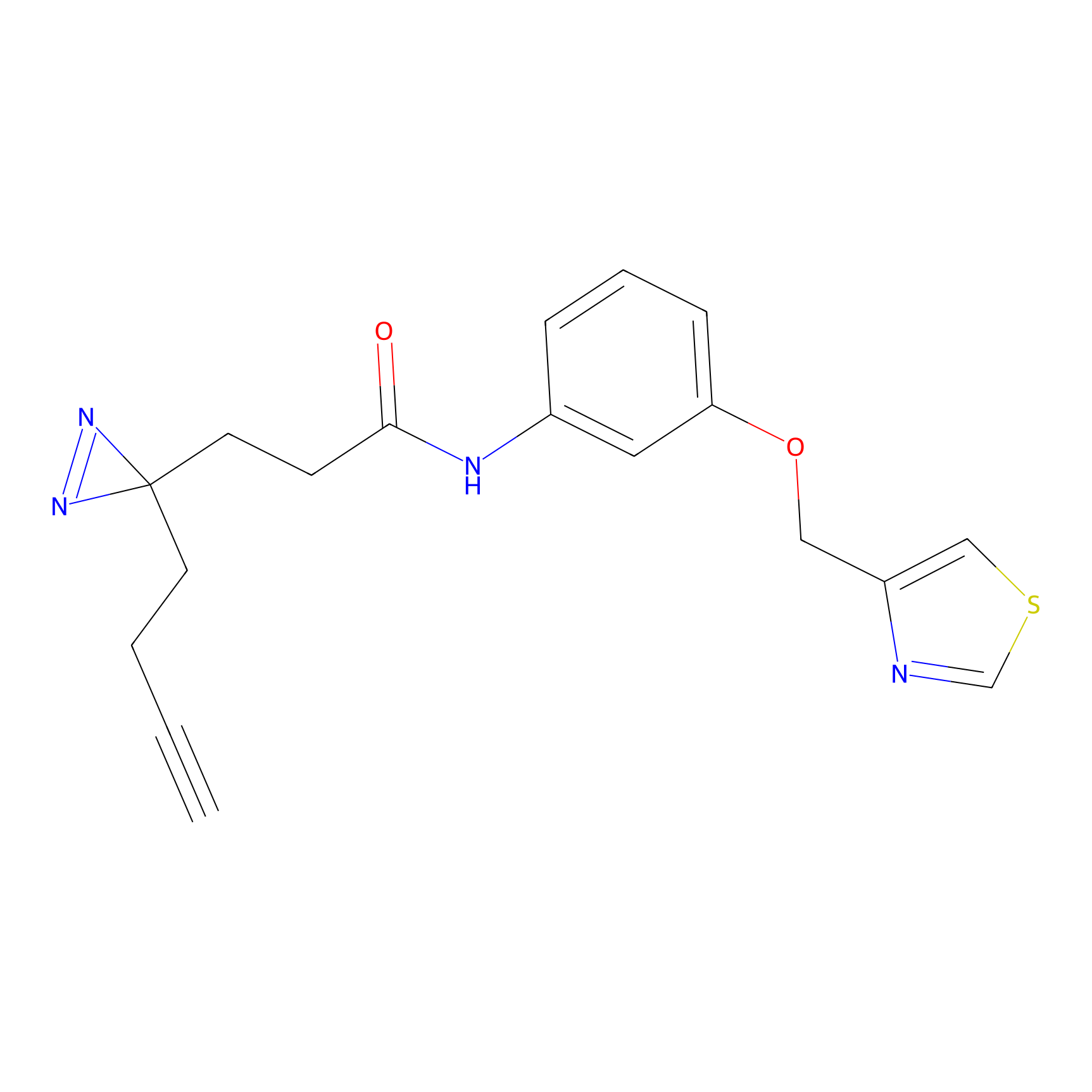 |
8.94 | LDD2041 | [13] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | HEK-293T | C714(2.07) | LDD0168 | [5] |
| LDCM0214 | AC1 | HEK-293T | C714(1.07); C100(1.17) | LDD1507 | [14] |
| LDCM0215 | AC10 | HEK-293T | C714(1.00); C100(0.91) | LDD1508 | [14] |
| LDCM0226 | AC11 | HEK-293T | C714(1.00); C100(0.95) | LDD1509 | [14] |
| LDCM0237 | AC12 | HEK-293T | C714(1.00); C100(1.03) | LDD1510 | [14] |
| LDCM0259 | AC14 | HEK-293T | C714(1.07); C645(0.94) | LDD1512 | [14] |
| LDCM0270 | AC15 | HEK-293T | C714(0.97) | LDD1513 | [14] |
| LDCM0276 | AC17 | HEK-293T | C714(1.12); C100(1.19) | LDD1515 | [14] |
| LDCM0277 | AC18 | HEK-293T | C714(1.07); C100(0.93) | LDD1516 | [14] |
| LDCM0278 | AC19 | HEK-293T | C714(0.87); C100(0.85) | LDD1517 | [14] |
| LDCM0279 | AC2 | HEK-293T | C714(1.00); C100(0.97) | LDD1518 | [14] |
| LDCM0280 | AC20 | HEK-293T | C714(1.02); C100(1.00) | LDD1519 | [14] |
| LDCM0281 | AC21 | HEK-293T | C714(1.06); C100(0.86) | LDD1520 | [14] |
| LDCM0282 | AC22 | HEK-293T | C714(1.01); C645(0.97) | LDD1521 | [14] |
| LDCM0283 | AC23 | HEK-293T | C714(1.00) | LDD1522 | [14] |
| LDCM0284 | AC24 | HEK-293T | C714(1.01); C100(0.95) | LDD1523 | [14] |
| LDCM0285 | AC25 | HEK-293T | C714(1.02); C100(1.10) | LDD1524 | [14] |
| LDCM0286 | AC26 | HEK-293T | C714(1.09); C100(0.90) | LDD1525 | [14] |
| LDCM0287 | AC27 | HEK-293T | C714(1.02); C100(0.90) | LDD1526 | [14] |
| LDCM0288 | AC28 | HEK-293T | C714(0.95); C100(0.95) | LDD1527 | [14] |
| LDCM0289 | AC29 | HEK-293T | C714(1.13); C100(1.05) | LDD1528 | [14] |
| LDCM0290 | AC3 | HEK-293T | C714(1.21); C100(0.88) | LDD1529 | [14] |
| LDCM0291 | AC30 | HEK-293T | C714(0.99); C645(0.98) | LDD1530 | [14] |
| LDCM0292 | AC31 | HEK-293T | C714(1.01) | LDD1531 | [14] |
| LDCM0293 | AC32 | HEK-293T | C714(0.88); C100(0.72) | LDD1532 | [14] |
| LDCM0294 | AC33 | HEK-293T | C714(1.14); C100(0.86) | LDD1533 | [14] |
| LDCM0295 | AC34 | HEK-293T | C714(1.09); C100(0.87) | LDD1534 | [14] |
| LDCM0296 | AC35 | HEK-293T | C714(0.92); C100(1.17) | LDD1535 | [14] |
| LDCM0297 | AC36 | HEK-293T | C714(1.02); C100(0.89) | LDD1536 | [14] |
| LDCM0298 | AC37 | HEK-293T | C714(1.03); C100(0.83) | LDD1537 | [14] |
| LDCM0299 | AC38 | HEK-293T | C714(1.06); C645(0.76) | LDD1538 | [14] |
| LDCM0300 | AC39 | HEK-293T | C714(1.01) | LDD1539 | [14] |
| LDCM0301 | AC4 | HEK-293T | C714(0.95); C100(0.96) | LDD1540 | [14] |
| LDCM0302 | AC40 | HEK-293T | C714(0.92); C100(1.20) | LDD1541 | [14] |
| LDCM0303 | AC41 | HEK-293T | C714(1.13); C100(1.58) | LDD1542 | [14] |
| LDCM0304 | AC42 | HEK-293T | C714(1.11); C100(1.09) | LDD1543 | [14] |
| LDCM0305 | AC43 | HEK-293T | C714(0.93); C100(0.96) | LDD1544 | [14] |
| LDCM0306 | AC44 | HEK-293T | C714(0.98); C100(0.99) | LDD1545 | [14] |
| LDCM0307 | AC45 | HEK-293T | C714(1.06); C100(1.23) | LDD1546 | [14] |
| LDCM0308 | AC46 | HEK-293T | C714(1.07); C645(1.19) | LDD1547 | [14] |
| LDCM0309 | AC47 | HEK-293T | C714(1.02) | LDD1548 | [14] |
| LDCM0310 | AC48 | HEK-293T | C714(0.90); C100(0.97) | LDD1549 | [14] |
| LDCM0311 | AC49 | HEK-293T | C714(1.08); C100(1.02) | LDD1550 | [14] |
| LDCM0312 | AC5 | HEK-293T | C714(1.03); C100(1.22) | LDD1551 | [14] |
| LDCM0313 | AC50 | HEK-293T | C714(1.11); C100(0.90) | LDD1552 | [14] |
| LDCM0314 | AC51 | HEK-293T | C714(1.03); C100(0.89) | LDD1553 | [14] |
| LDCM0315 | AC52 | HEK-293T | C714(1.00); C100(0.82) | LDD1554 | [14] |
| LDCM0316 | AC53 | HEK-293T | C714(1.06); C100(0.93) | LDD1555 | [14] |
| LDCM0317 | AC54 | HEK-293T | C714(1.10); C645(0.91) | LDD1556 | [14] |
| LDCM0318 | AC55 | HEK-293T | C714(1.00) | LDD1557 | [14] |
| LDCM0319 | AC56 | HEK-293T | C714(0.95); C100(1.04) | LDD1558 | [14] |
| LDCM0320 | AC57 | HEK-293T | C714(1.13); C100(0.96) | LDD1559 | [14] |
| LDCM0321 | AC58 | HEK-293T | C714(1.04); C100(0.85) | LDD1560 | [14] |
| LDCM0322 | AC59 | HEK-293T | C714(1.04); C100(0.95) | LDD1561 | [14] |
| LDCM0323 | AC6 | HEK-293T | C714(1.09); C645(0.84) | LDD1562 | [14] |
| LDCM0324 | AC60 | HEK-293T | C714(1.09); C100(0.99) | LDD1563 | [14] |
| LDCM0325 | AC61 | HEK-293T | C714(1.09); C100(0.89) | LDD1564 | [14] |
| LDCM0326 | AC62 | HEK-293T | C714(1.08); C645(0.99) | LDD1565 | [14] |
| LDCM0327 | AC63 | HEK-293T | C714(0.95) | LDD1566 | [14] |
| LDCM0328 | AC64 | HEK-293T | C714(0.94); C100(0.85) | LDD1567 | [14] |
| LDCM0334 | AC7 | HEK-293T | C714(0.94) | LDD1568 | [14] |
| LDCM0345 | AC8 | HEK-293T | C714(0.96); C100(1.06) | LDD1569 | [14] |
| LDCM0248 | AKOS034007472 | HEK-293T | C714(1.01); C100(1.17) | LDD1511 | [14] |
| LDCM0356 | AKOS034007680 | HEK-293T | C714(1.15); C100(1.04) | LDD1570 | [14] |
| LDCM0275 | AKOS034007705 | HEK-293T | C714(0.98); C100(0.89) | LDD1514 | [14] |
| LDCM0156 | Aniline | NCI-H1299 | 12.02 | LDD0403 | [2] |
| LDCM0200 | BPK-25 | T cell | C714(4.38) | LDD0529 | [15] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [10] |
| LDCM0367 | CL1 | HEK-293T | C714(1.12) | LDD1571 | [14] |
| LDCM0368 | CL10 | HEK-293T | C714(1.34); C645(0.92) | LDD1572 | [14] |
| LDCM0369 | CL100 | HEK-293T | C714(1.04) | LDD1573 | [14] |
| LDCM0370 | CL101 | HEK-293T | C714(0.96) | LDD1574 | [14] |
| LDCM0371 | CL102 | HEK-293T | C714(1.03); C100(1.62) | LDD1575 | [14] |
| LDCM0372 | CL103 | HEK-293T | C714(0.94); C100(1.10) | LDD1576 | [14] |
| LDCM0373 | CL104 | HEK-293T | C714(1.17) | LDD1577 | [14] |
| LDCM0374 | CL105 | HEK-293T | C714(1.02) | LDD1578 | [14] |
| LDCM0375 | CL106 | HEK-293T | C714(0.95); C100(1.12) | LDD1579 | [14] |
| LDCM0376 | CL107 | HEK-293T | C714(0.94); C100(0.91) | LDD1580 | [14] |
| LDCM0377 | CL108 | HEK-293T | C714(0.96) | LDD1581 | [14] |
| LDCM0378 | CL109 | HEK-293T | C714(0.91) | LDD1582 | [14] |
| LDCM0379 | CL11 | HEK-293T | C714(1.15) | LDD1583 | [14] |
| LDCM0380 | CL110 | HEK-293T | C714(0.99); C100(1.64) | LDD1584 | [14] |
| LDCM0381 | CL111 | HEK-293T | C714(0.91); C100(1.40) | LDD1585 | [14] |
| LDCM0382 | CL112 | HEK-293T | C714(1.14) | LDD1586 | [14] |
| LDCM0383 | CL113 | HEK-293T | C714(0.99) | LDD1587 | [14] |
| LDCM0384 | CL114 | HEK-293T | C714(0.97); C100(3.09) | LDD1588 | [14] |
| LDCM0385 | CL115 | HEK-293T | C714(1.04); C100(1.12) | LDD1589 | [14] |
| LDCM0386 | CL116 | HEK-293T | C714(0.99) | LDD1590 | [14] |
| LDCM0387 | CL117 | HEK-293T | C714(1.03) | LDD1591 | [14] |
| LDCM0388 | CL118 | HEK-293T | C714(0.94); C100(1.08) | LDD1592 | [14] |
| LDCM0389 | CL119 | HEK-293T | C714(0.99); C100(0.98) | LDD1593 | [14] |
| LDCM0390 | CL12 | HEK-293T | C714(0.88); C100(1.02) | LDD1594 | [14] |
| LDCM0391 | CL120 | HEK-293T | C714(1.04) | LDD1595 | [14] |
| LDCM0392 | CL121 | HEK-293T | C714(1.01) | LDD1596 | [14] |
| LDCM0393 | CL122 | HEK-293T | C714(1.09); C100(0.98) | LDD1597 | [14] |
| LDCM0394 | CL123 | HEK-293T | C714(0.93); C100(1.32) | LDD1598 | [14] |
| LDCM0395 | CL124 | HEK-293T | C714(1.10) | LDD1599 | [14] |
| LDCM0396 | CL125 | HEK-293T | C714(1.15) | LDD1600 | [14] |
| LDCM0397 | CL126 | HEK-293T | C714(0.87); C100(1.20) | LDD1601 | [14] |
| LDCM0398 | CL127 | HEK-293T | C714(1.06); C100(1.16) | LDD1602 | [14] |
| LDCM0399 | CL128 | HEK-293T | C714(0.97) | LDD1603 | [14] |
| LDCM0400 | CL13 | HEK-293T | C714(1.20) | LDD1604 | [14] |
| LDCM0401 | CL14 | HEK-293T | C714(0.99); C100(0.95) | LDD1605 | [14] |
| LDCM0402 | CL15 | HEK-293T | C714(0.97); C100(2.65) | LDD1606 | [14] |
| LDCM0403 | CL16 | HEK-293T | C714(1.13) | LDD1607 | [14] |
| LDCM0404 | CL17 | HEK-293T | C714(1.13); C100(2.04) | LDD1608 | [14] |
| LDCM0405 | CL18 | HEK-293T | C714(1.18); C100(1.00) | LDD1609 | [14] |
| LDCM0406 | CL19 | HEK-293T | C714(0.97); C100(0.95) | LDD1610 | [14] |
| LDCM0407 | CL2 | HEK-293T | C714(1.05); C100(0.86) | LDD1611 | [14] |
| LDCM0408 | CL20 | HEK-293T | C714(1.20); C100(0.88) | LDD1612 | [14] |
| LDCM0409 | CL21 | HEK-293T | C714(1.20); C100(1.70) | LDD1613 | [14] |
| LDCM0410 | CL22 | HEK-293T | C714(1.15); C645(0.99) | LDD1614 | [14] |
| LDCM0411 | CL23 | HEK-293T | C714(1.22) | LDD1615 | [14] |
| LDCM0412 | CL24 | HEK-293T | C714(0.94); C100(1.00) | LDD1616 | [14] |
| LDCM0413 | CL25 | HEK-293T | C714(0.93) | LDD1617 | [14] |
| LDCM0414 | CL26 | HEK-293T | C714(0.91); C100(0.92) | LDD1618 | [14] |
| LDCM0415 | CL27 | HEK-293T | C714(0.99); C100(1.22) | LDD1619 | [14] |
| LDCM0416 | CL28 | HEK-293T | C714(0.96) | LDD1620 | [14] |
| LDCM0417 | CL29 | HEK-293T | C714(1.06); C100(0.92) | LDD1621 | [14] |
| LDCM0418 | CL3 | HEK-293T | C714(1.04); C100(1.35) | LDD1622 | [14] |
| LDCM0419 | CL30 | HEK-293T | C714(1.10); C100(1.00) | LDD1623 | [14] |
| LDCM0420 | CL31 | HEK-293T | C714(1.01); C100(1.02) | LDD1624 | [14] |
| LDCM0421 | CL32 | HEK-293T | C714(1.08); C100(0.86) | LDD1625 | [14] |
| LDCM0422 | CL33 | HEK-293T | C714(0.58); C100(1.59) | LDD1626 | [14] |
| LDCM0423 | CL34 | HEK-293T | C714(1.15); C645(0.78) | LDD1627 | [14] |
| LDCM0424 | CL35 | HEK-293T | C714(1.10) | LDD1628 | [14] |
| LDCM0425 | CL36 | HEK-293T | C714(0.93); C100(1.14) | LDD1629 | [14] |
| LDCM0426 | CL37 | HEK-293T | C714(1.02) | LDD1630 | [14] |
| LDCM0428 | CL39 | HEK-293T | C714(1.06); C100(1.56) | LDD1632 | [14] |
| LDCM0429 | CL4 | HEK-293T | C714(1.12) | LDD1633 | [14] |
| LDCM0430 | CL40 | HEK-293T | C714(0.98) | LDD1634 | [14] |
| LDCM0431 | CL41 | HEK-293T | C714(1.06); C100(1.60) | LDD1635 | [14] |
| LDCM0432 | CL42 | HEK-293T | C714(1.13); C100(0.86) | LDD1636 | [14] |
| LDCM0433 | CL43 | HEK-293T | C714(0.91); C100(1.15) | LDD1637 | [14] |
| LDCM0434 | CL44 | HEK-293T | C714(1.09); C100(1.04) | LDD1638 | [14] |
| LDCM0435 | CL45 | HEK-293T | C714(1.09); C100(1.45) | LDD1639 | [14] |
| LDCM0436 | CL46 | HEK-293T | C714(1.14); C645(0.87) | LDD1640 | [14] |
| LDCM0437 | CL47 | HEK-293T | C714(1.04) | LDD1641 | [14] |
| LDCM0438 | CL48 | HEK-293T | C714(0.79); C100(1.03) | LDD1642 | [14] |
| LDCM0439 | CL49 | HEK-293T | C714(1.01) | LDD1643 | [14] |
| LDCM0440 | CL5 | HEK-293T | C714(1.20); C100(1.03) | LDD1644 | [14] |
| LDCM0441 | CL50 | HEK-293T | C714(0.96); C100(1.28) | LDD1645 | [14] |
| LDCM0443 | CL52 | HEK-293T | C714(0.95) | LDD1646 | [14] |
| LDCM0444 | CL53 | HEK-293T | C714(1.20); C100(2.28) | LDD1647 | [14] |
| LDCM0445 | CL54 | HEK-293T | C714(0.93); C100(2.15) | LDD1648 | [14] |
| LDCM0446 | CL55 | HEK-293T | C714(0.60); C100(0.96) | LDD1649 | [14] |
| LDCM0447 | CL56 | HEK-293T | C714(1.15); C100(0.96) | LDD1650 | [14] |
| LDCM0448 | CL57 | HEK-293T | C714(1.12); C100(1.56) | LDD1651 | [14] |
| LDCM0449 | CL58 | HEK-293T | C714(1.22); C645(0.97) | LDD1652 | [14] |
| LDCM0450 | CL59 | HEK-293T | C714(1.18) | LDD1653 | [14] |
| LDCM0451 | CL6 | HEK-293T | C714(1.14); C100(1.57) | LDD1654 | [14] |
| LDCM0452 | CL60 | HEK-293T | C714(0.95); C100(1.06) | LDD1655 | [14] |
| LDCM0453 | CL61 | HEK-293T | C714(1.06) | LDD1656 | [14] |
| LDCM0454 | CL62 | HEK-293T | C714(0.97); C100(0.82) | LDD1657 | [14] |
| LDCM0455 | CL63 | HEK-293T | C714(1.02); C100(0.96) | LDD1658 | [14] |
| LDCM0456 | CL64 | HEK-293T | C714(0.95) | LDD1659 | [14] |
| LDCM0457 | CL65 | HEK-293T | C714(1.07); C100(1.15) | LDD1660 | [14] |
| LDCM0458 | CL66 | HEK-293T | C714(1.16); C100(1.20) | LDD1661 | [14] |
| LDCM0459 | CL67 | HEK-293T | C714(1.03); C100(1.00) | LDD1662 | [14] |
| LDCM0460 | CL68 | HEK-293T | C714(1.04); C100(1.02) | LDD1663 | [14] |
| LDCM0461 | CL69 | HEK-293T | C714(1.09); C100(1.13) | LDD1664 | [14] |
| LDCM0462 | CL7 | HEK-293T | C714(0.99); C100(0.99) | LDD1665 | [14] |
| LDCM0463 | CL70 | HEK-293T | C714(1.11); C645(1.19) | LDD1666 | [14] |
| LDCM0464 | CL71 | HEK-293T | C714(1.20) | LDD1667 | [14] |
| LDCM0465 | CL72 | HEK-293T | C714(0.91); C100(1.21) | LDD1668 | [14] |
| LDCM0466 | CL73 | HEK-293T | C714(0.95) | LDD1669 | [14] |
| LDCM0467 | CL74 | HEK-293T | C714(0.96); C100(0.96) | LDD1670 | [14] |
| LDCM0469 | CL76 | HEK-293T | C714(1.01) | LDD1672 | [14] |
| LDCM0470 | CL77 | HEK-293T | C714(1.14); C100(2.06) | LDD1673 | [14] |
| LDCM0471 | CL78 | HEK-293T | C714(1.07); C100(0.95) | LDD1674 | [14] |
| LDCM0472 | CL79 | HEK-293T | C714(1.00); C100(1.12) | LDD1675 | [14] |
| LDCM0473 | CL8 | HEK-293T | C714(1.14); C100(2.60) | LDD1676 | [14] |
| LDCM0474 | CL80 | HEK-293T | C714(1.03); C100(0.99) | LDD1677 | [14] |
| LDCM0475 | CL81 | HEK-293T | C714(1.11); C100(1.05) | LDD1678 | [14] |
| LDCM0476 | CL82 | HEK-293T | C714(1.10); C645(0.82) | LDD1679 | [14] |
| LDCM0477 | CL83 | HEK-293T | C714(1.00) | LDD1680 | [14] |
| LDCM0478 | CL84 | HEK-293T | C714(0.96); C100(1.16) | LDD1681 | [14] |
| LDCM0479 | CL85 | HEK-293T | C714(0.99) | LDD1682 | [14] |
| LDCM0480 | CL86 | HEK-293T | C714(0.97); C100(0.97) | LDD1683 | [14] |
| LDCM0481 | CL87 | HEK-293T | C714(1.07); C100(1.18) | LDD1684 | [14] |
| LDCM0482 | CL88 | HEK-293T | C714(1.09) | LDD1685 | [14] |
| LDCM0483 | CL89 | HEK-293T | C714(1.11); C100(0.98) | LDD1686 | [14] |
| LDCM0484 | CL9 | HEK-293T | C714(1.09); C100(1.03) | LDD1687 | [14] |
| LDCM0485 | CL90 | HEK-293T | C714(1.04); C100(1.92) | LDD1688 | [14] |
| LDCM0486 | CL91 | HEK-293T | C714(0.92); C100(1.05) | LDD1689 | [14] |
| LDCM0487 | CL92 | HEK-293T | C714(1.03); C100(1.13) | LDD1690 | [14] |
| LDCM0488 | CL93 | HEK-293T | C714(1.07); C100(1.01) | LDD1691 | [14] |
| LDCM0489 | CL94 | HEK-293T | C714(1.09); C645(0.81) | LDD1692 | [14] |
| LDCM0490 | CL95 | HEK-293T | C714(1.11) | LDD1693 | [14] |
| LDCM0491 | CL96 | HEK-293T | C714(0.84); C100(1.56) | LDD1694 | [14] |
| LDCM0492 | CL97 | HEK-293T | C714(1.02) | LDD1695 | [14] |
| LDCM0493 | CL98 | HEK-293T | C714(0.96); C100(1.22) | LDD1696 | [14] |
| LDCM0494 | CL99 | HEK-293T | C714(0.94); C100(1.47) | LDD1697 | [14] |
| LDCM0495 | E2913 | HEK-293T | C714(1.05); C100(1.51) | LDD1698 | [14] |
| LDCM0203 | EV-96 | T cell | C714(5.24) | LDD0523 | [15] |
| LDCM0625 | F8 | Ramos | C714(0.78) | LDD2187 | [16] |
| LDCM0572 | Fragment10 | Ramos | C714(0.83) | LDD2189 | [16] |
| LDCM0574 | Fragment12 | Ramos | C714(0.88) | LDD2191 | [16] |
| LDCM0575 | Fragment13 | Ramos | C714(0.80) | LDD2192 | [16] |
| LDCM0576 | Fragment14 | Ramos | C714(1.05) | LDD2193 | [16] |
| LDCM0580 | Fragment21 | Ramos | C714(0.56) | LDD2195 | [16] |
| LDCM0582 | Fragment23 | Ramos | C714(0.63) | LDD2196 | [16] |
| LDCM0578 | Fragment27 | Ramos | C714(0.76) | LDD2197 | [16] |
| LDCM0586 | Fragment28 | Ramos | C714(0.77) | LDD2198 | [16] |
| LDCM0588 | Fragment30 | Ramos | C714(0.64) | LDD2199 | [16] |
| LDCM0589 | Fragment31 | Ramos | C714(0.56) | LDD2200 | [16] |
| LDCM0590 | Fragment32 | Ramos | C714(1.91) | LDD2201 | [16] |
| LDCM0468 | Fragment33 | HEK-293T | C714(0.96); C100(0.99) | LDD1671 | [14] |
| LDCM0596 | Fragment38 | Ramos | C714(0.32) | LDD2203 | [16] |
| LDCM0566 | Fragment4 | Ramos | C714(1.09) | LDD2184 | [16] |
| LDCM0427 | Fragment51 | HEK-293T | C714(1.05); C100(1.04) | LDD1631 | [14] |
| LDCM0610 | Fragment52 | Ramos | C714(0.62) | LDD2204 | [16] |
| LDCM0614 | Fragment56 | Ramos | C714(0.69) | LDD2205 | [16] |
| LDCM0569 | Fragment7 | Ramos | C714(3.14) | LDD2186 | [16] |
| LDCM0022 | KB02 | HEK-293T | C714(0.98) | LDD1492 | [14] |
| LDCM0023 | KB03 | HEK-293T | C714(1.07) | LDD1497 | [14] |
| LDCM0024 | KB05 | HMCB | C714(1.71) | LDD3312 | [3] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C559(1.00) | LDD2099 | [4] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C559(0.68) | LDD2109 | [4] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C559(0.76) | LDD2127 | [4] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C559(0.82) | LDD2136 | [4] |
| LDCM0131 | RA190 | MM1.R | C714(1.65) | LDD0304 | [17] |
References
