Details of the Target
General Information of Target
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| HT115 | SNV: p.K121T; p.R296I | . | |||
| IGROV1 | SNV: p.L164P | . | |||
| P31FUJ | SNV: p.I146T | DBIA Probe Info | |||
| SNU1 | SNV: p.E253A; p.L289F | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
TH211 Probe Info |
 |
Y182(20.00); Y35(13.99) | LDD0257 | [1] | |
|
TH214 Probe Info |
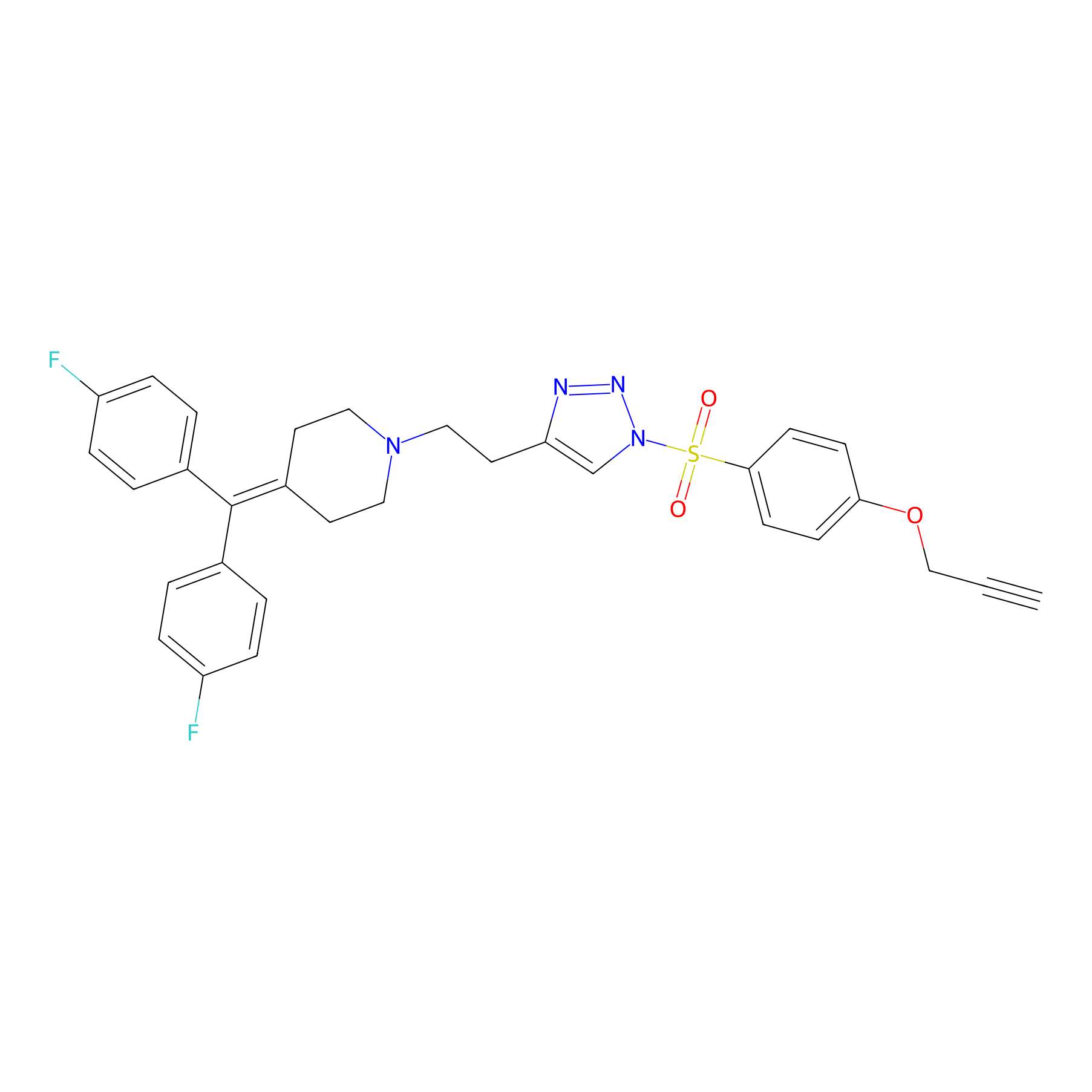 |
Y182(20.00) | LDD0258 | [1] | |
|
TH216 Probe Info |
 |
Y182(20.00) | LDD0259 | [1] | |
|
STPyne Probe Info |
 |
K165(2.06); K295(1.75) | LDD0277 | [2] | |
|
m-APA Probe Info |
 |
13.62 | LDD0403 | [3] | |
|
Dasatinib-CA-4PAP Probe Info |
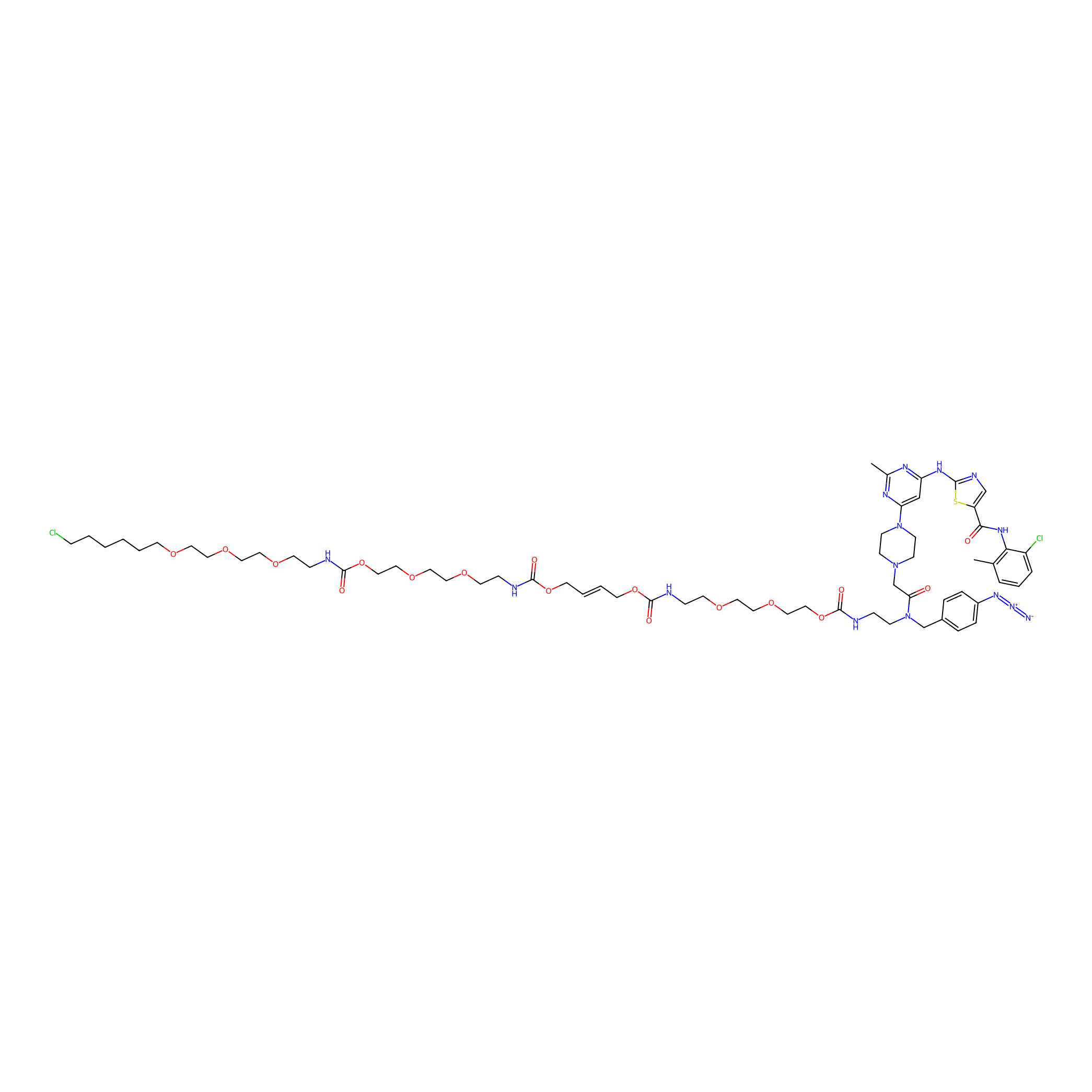 |
17.07 | LDD0270 | [4] | |
|
BTD Probe Info |
 |
C162(0.61) | LDD2095 | [5] | |
|
AHL-Pu-1 Probe Info |
 |
C162(2.46) | LDD0168 | [6] | |
|
HHS-465 Probe Info |
 |
Y182(10.00) | LDD2237 | [7] | |
|
DBIA Probe Info |
 |
C39(1.54) | LDD0531 | [8] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [9] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C162(0.00); C39(0.00) | LDD0038 | [10] | |
|
IA-alkyne Probe Info |
 |
C162(0.00); C39(0.00) | LDD0036 | [10] | |
|
Lodoacetamide azide Probe Info |
 |
C162(0.00); C39(0.00) | LDD0037 | [10] | |
|
JW-RF-010 Probe Info |
 |
C39(0.00); C162(0.00) | LDD0026 | [11] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [11] | |
|
Compound 10 Probe Info |
 |
N.A. | LDD2216 | [12] | |
|
IPM Probe Info |
 |
N.A. | LDD2156 | [13] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [14] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [15] | |
|
AOyne Probe Info |
 |
11.30 | LDD0443 | [16] | |
|
NAIA_5 Probe Info |
 |
C162(0.00); C119(0.00); C39(0.00) | LDD2223 | [17] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C158 Probe Info |
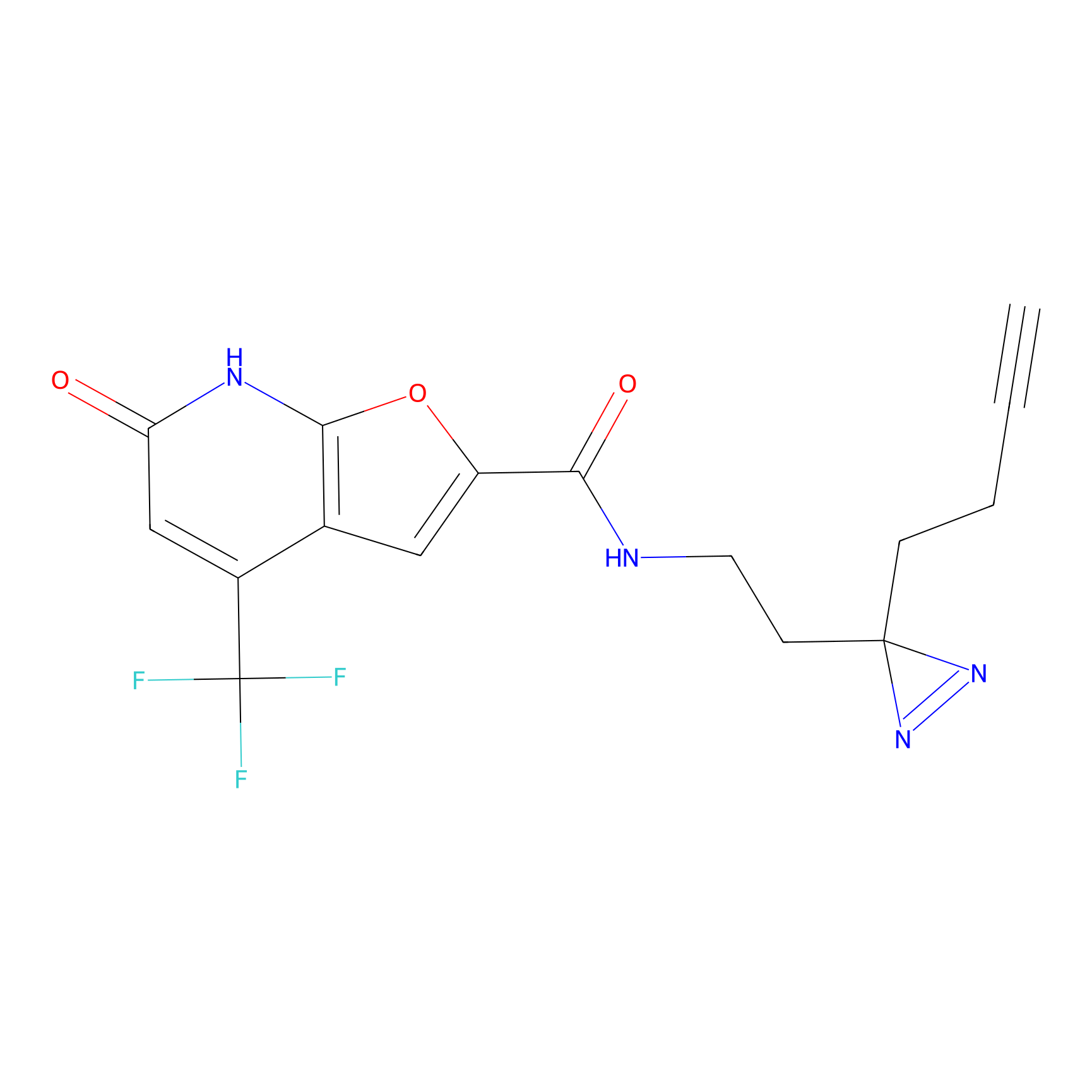 |
9.51 | LDD1838 | [18] | |
|
C275 Probe Info |
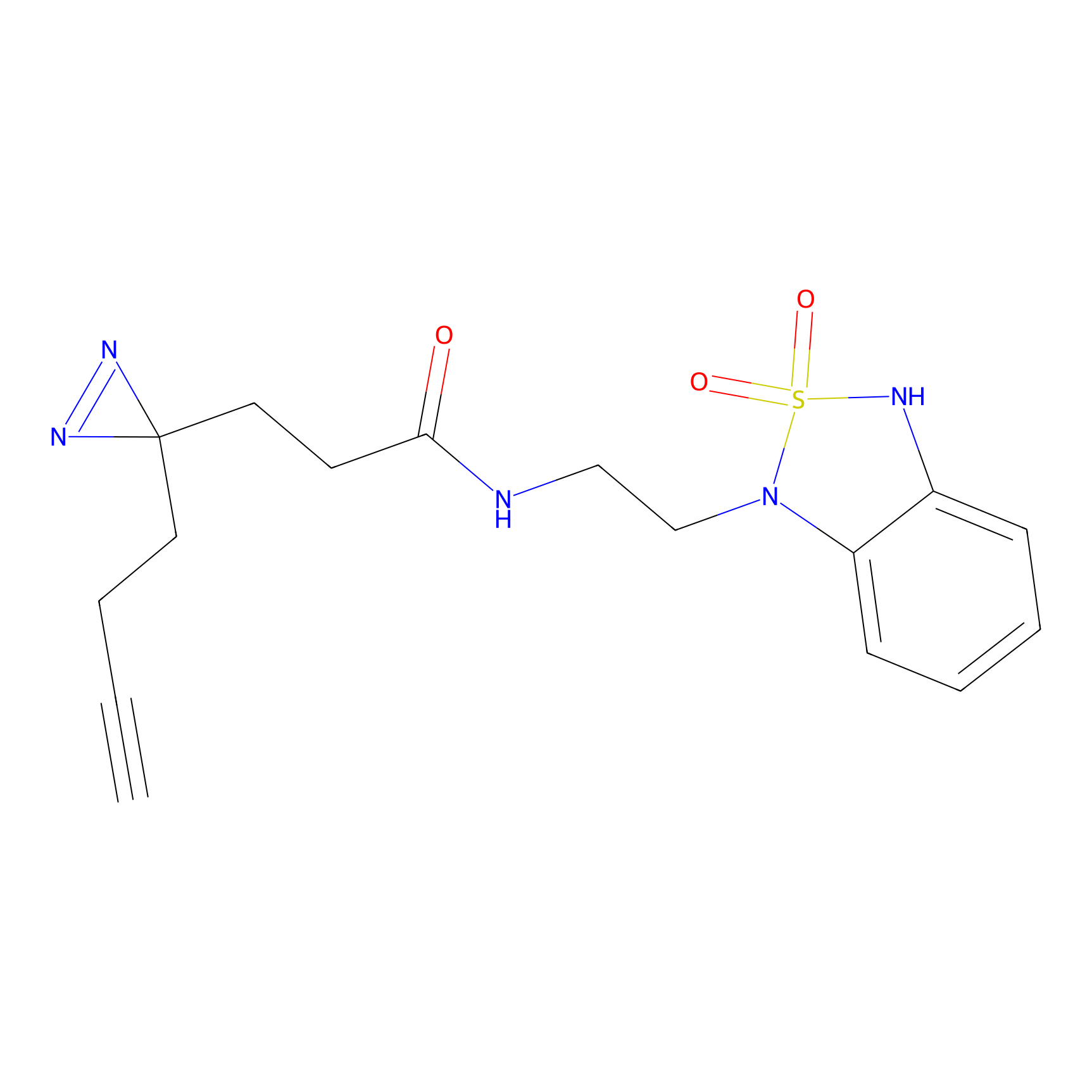 |
4.92 | LDD1945 | [18] | |
|
C282 Probe Info |
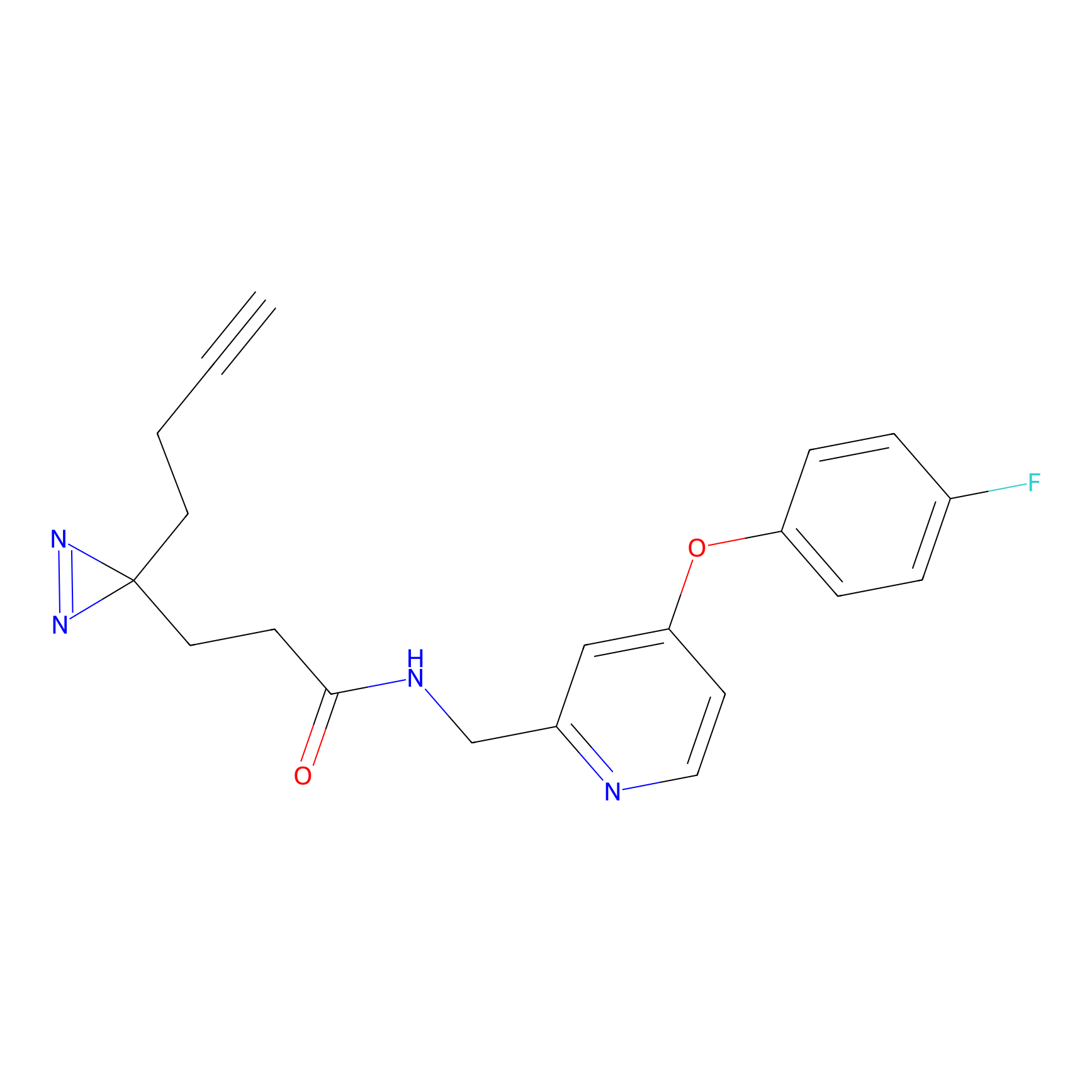 |
23.10 | LDD1952 | [18] | |
|
Dasatinib-CA-3PAP Probe Info |
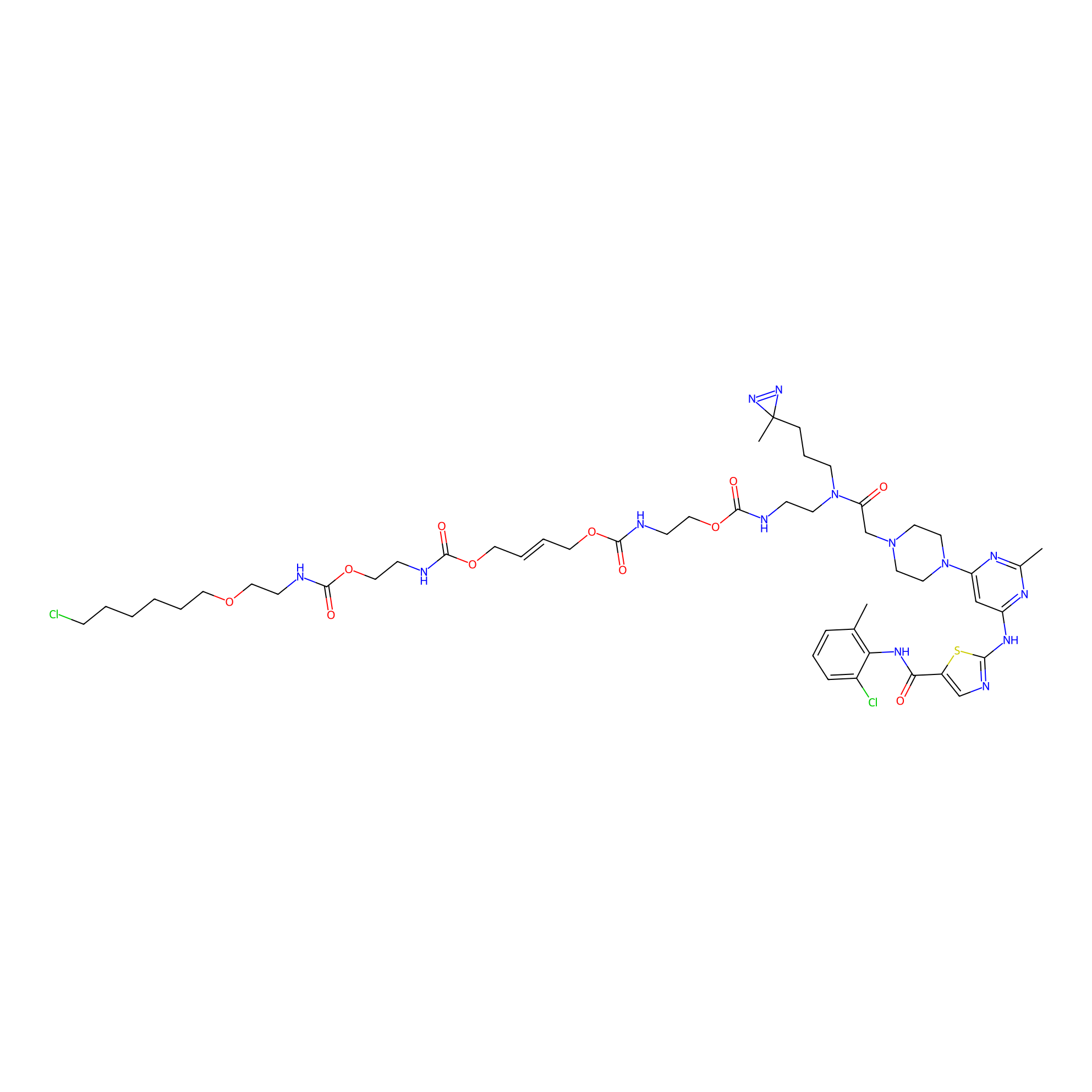 |
N.A. | LDD0365 | [4] | |
|
Dasatinib-CA-8PAP Probe Info |
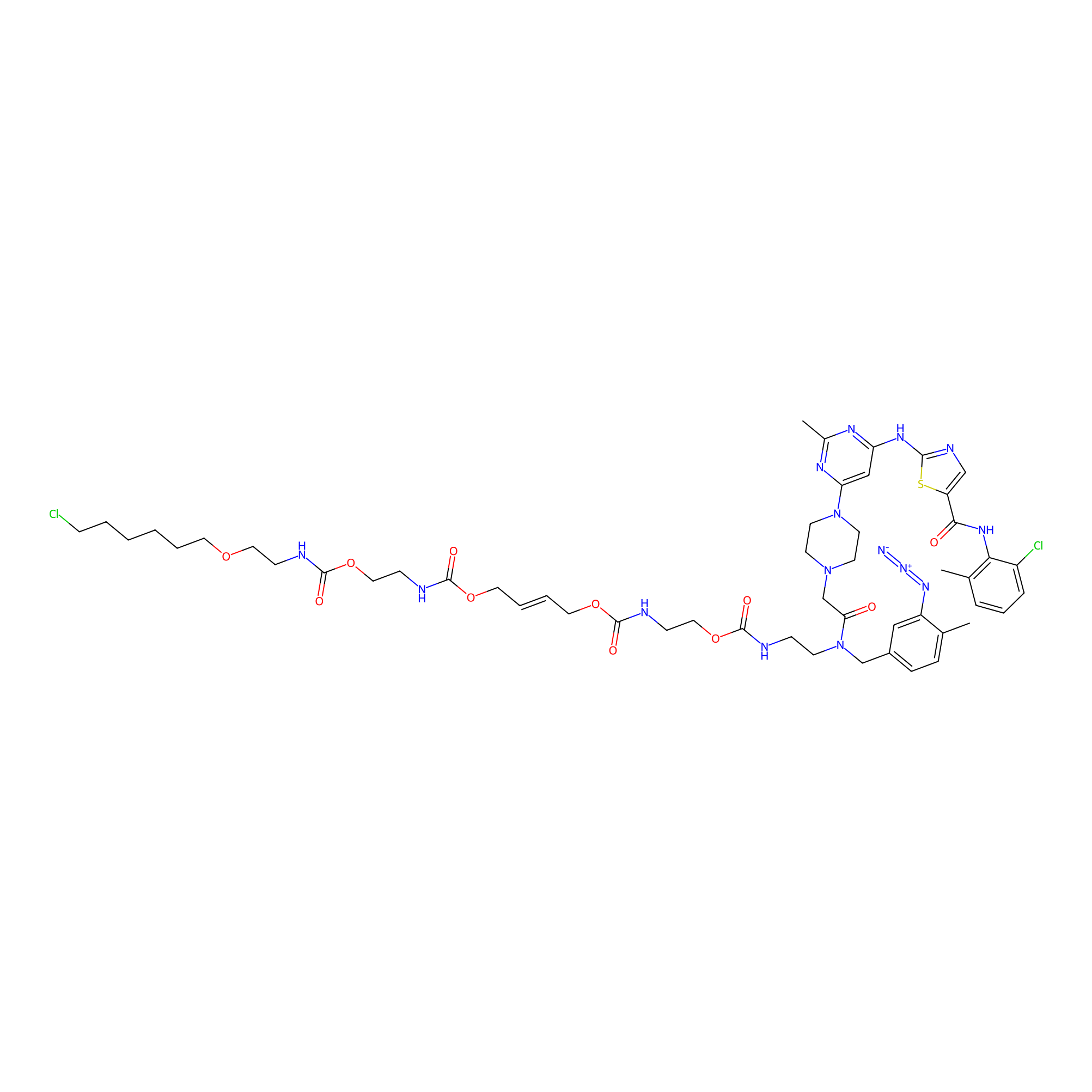 |
5.02; 0.00 | LDD0364 | [4] | |
|
DA-2 Probe Info |
 |
N.A. | LDD0070 | [19] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0502 | 1-(Cyanoacetyl)piperidine | MDA-MB-231 | C162(0.61) | LDD2095 | [5] |
| LDCM0537 | 2-Cyano-N,N-dimethylacetamide | MDA-MB-231 | C162(1.05) | LDD2130 | [5] |
| LDCM0539 | 3-(4-Isopropylpiperazin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C162(0.69) | LDD2132 | [5] |
| LDCM0025 | 4SU-RNA | HEK-293T | C162(2.46) | LDD0168 | [6] |
| LDCM0214 | AC1 | HCT 116 | C39(1.54) | LDD0531 | [8] |
| LDCM0215 | AC10 | PaTu 8988t | C39(0.30) | LDD1094 | [8] |
| LDCM0216 | AC100 | HCT 116 | C39(0.38) | LDD0533 | [8] |
| LDCM0217 | AC101 | HCT 116 | C39(0.43) | LDD0534 | [8] |
| LDCM0218 | AC102 | HCT 116 | C39(0.56) | LDD0535 | [8] |
| LDCM0219 | AC103 | HCT 116 | C39(0.34) | LDD0536 | [8] |
| LDCM0220 | AC104 | HCT 116 | C39(0.68) | LDD0537 | [8] |
| LDCM0221 | AC105 | HCT 116 | C39(0.41) | LDD0538 | [8] |
| LDCM0222 | AC106 | HCT 116 | C39(0.35) | LDD0539 | [8] |
| LDCM0223 | AC107 | HCT 116 | C39(0.41) | LDD0540 | [8] |
| LDCM0224 | AC108 | HCT 116 | C39(0.35) | LDD0541 | [8] |
| LDCM0225 | AC109 | HCT 116 | C39(0.84) | LDD0542 | [8] |
| LDCM0226 | AC11 | PaTu 8988t | C39(0.32) | LDD1105 | [8] |
| LDCM0227 | AC110 | HCT 116 | C39(0.34) | LDD0544 | [8] |
| LDCM0228 | AC111 | HCT 116 | C39(0.33) | LDD0545 | [8] |
| LDCM0229 | AC112 | HCT 116 | C39(0.32) | LDD0546 | [8] |
| LDCM0230 | AC113 | HCT 116 | C162(0.74); C39(0.94) | LDD0547 | [8] |
| LDCM0231 | AC114 | HCT 116 | C162(0.80); C39(0.84) | LDD0548 | [8] |
| LDCM0232 | AC115 | HCT 116 | C162(0.71); C39(0.65) | LDD0549 | [8] |
| LDCM0233 | AC116 | HCT 116 | C162(0.93); C39(0.95) | LDD0550 | [8] |
| LDCM0234 | AC117 | HCT 116 | C162(0.85); C39(0.94) | LDD0551 | [8] |
| LDCM0235 | AC118 | HCT 116 | C162(0.76); C39(1.08) | LDD0552 | [8] |
| LDCM0236 | AC119 | HCT 116 | C162(0.71); C39(1.08) | LDD0553 | [8] |
| LDCM0237 | AC12 | PaTu 8988t | C39(0.32) | LDD1116 | [8] |
| LDCM0238 | AC120 | HCT 116 | C162(0.79); C39(0.87) | LDD0555 | [8] |
| LDCM0239 | AC121 | HCT 116 | C162(0.72); C39(1.50) | LDD0556 | [8] |
| LDCM0240 | AC122 | HCT 116 | C162(0.86); C39(1.05) | LDD0557 | [8] |
| LDCM0241 | AC123 | HCT 116 | C162(0.70); C39(1.31) | LDD0558 | [8] |
| LDCM0242 | AC124 | HCT 116 | C162(0.45); C39(1.39) | LDD0559 | [8] |
| LDCM0243 | AC125 | HCT 116 | C162(0.87); C39(1.48) | LDD0560 | [8] |
| LDCM0244 | AC126 | HCT 116 | C162(0.56); C39(1.41) | LDD0561 | [8] |
| LDCM0245 | AC127 | HCT 116 | C162(0.57); C39(1.45) | LDD0562 | [8] |
| LDCM0246 | AC128 | HCT 116 | C39(0.28) | LDD0563 | [8] |
| LDCM0247 | AC129 | HCT 116 | C39(1.20) | LDD0564 | [8] |
| LDCM0249 | AC130 | HCT 116 | C39(0.50) | LDD0566 | [8] |
| LDCM0250 | AC131 | HCT 116 | C39(1.36) | LDD0567 | [8] |
| LDCM0251 | AC132 | HCT 116 | C39(1.32) | LDD0568 | [8] |
| LDCM0252 | AC133 | HCT 116 | C39(0.56) | LDD0569 | [8] |
| LDCM0253 | AC134 | HCT 116 | C39(0.59) | LDD0570 | [8] |
| LDCM0254 | AC135 | HCT 116 | C39(0.51) | LDD0571 | [8] |
| LDCM0255 | AC136 | HCT 116 | C39(0.78) | LDD0572 | [8] |
| LDCM0256 | AC137 | HCT 116 | C39(1.15) | LDD0573 | [8] |
| LDCM0257 | AC138 | HCT 116 | C39(0.67) | LDD0574 | [8] |
| LDCM0258 | AC139 | HCT 116 | C39(0.84) | LDD0575 | [8] |
| LDCM0259 | AC14 | PaTu 8988t | C39(0.75) | LDD1138 | [8] |
| LDCM0260 | AC140 | HCT 116 | C39(0.59) | LDD0577 | [8] |
| LDCM0261 | AC141 | HCT 116 | C39(0.87) | LDD0578 | [8] |
| LDCM0262 | AC142 | HCT 116 | C39(1.01) | LDD0579 | [8] |
| LDCM0263 | AC143 | HCT 116 | C39(0.93) | LDD0580 | [8] |
| LDCM0264 | AC144 | HCT 116 | C39(0.62) | LDD0581 | [8] |
| LDCM0265 | AC145 | HCT 116 | C39(1.19) | LDD0582 | [8] |
| LDCM0266 | AC146 | HCT 116 | C39(0.57) | LDD0583 | [8] |
| LDCM0267 | AC147 | HCT 116 | C39(0.49) | LDD0584 | [8] |
| LDCM0268 | AC148 | HCT 116 | C39(0.19) | LDD0585 | [8] |
| LDCM0269 | AC149 | HCT 116 | C39(0.38) | LDD0586 | [8] |
| LDCM0270 | AC15 | PaTu 8988t | C39(0.52) | LDD1149 | [8] |
| LDCM0271 | AC150 | HCT 116 | C39(1.01) | LDD0588 | [8] |
| LDCM0272 | AC151 | HCT 116 | C39(1.06) | LDD0589 | [8] |
| LDCM0273 | AC152 | HCT 116 | C39(0.37) | LDD0590 | [8] |
| LDCM0274 | AC153 | HCT 116 | C39(0.19) | LDD0591 | [8] |
| LDCM0621 | AC154 | HCT 116 | C39(0.63) | LDD2158 | [8] |
| LDCM0622 | AC155 | HCT 116 | C39(0.76) | LDD2159 | [8] |
| LDCM0623 | AC156 | HCT 116 | C39(1.43) | LDD2160 | [8] |
| LDCM0624 | AC157 | HCT 116 | C39(1.19) | LDD2161 | [8] |
| LDCM0276 | AC17 | HCT 116 | C39(1.05); C162(1.18) | LDD0593 | [8] |
| LDCM0277 | AC18 | HCT 116 | C39(0.39); C162(1.19) | LDD0594 | [8] |
| LDCM0278 | AC19 | HCT 116 | C39(0.83); C162(1.28) | LDD0595 | [8] |
| LDCM0279 | AC2 | HCT 116 | C39(1.09) | LDD0596 | [8] |
| LDCM0280 | AC20 | HCT 116 | C39(1.02); C162(1.04) | LDD0597 | [8] |
| LDCM0281 | AC21 | HCT 116 | C39(0.83); C162(1.06) | LDD0598 | [8] |
| LDCM0282 | AC22 | HCT 116 | C162(1.09); C39(1.29) | LDD0599 | [8] |
| LDCM0283 | AC23 | HCT 116 | C162(0.87); C39(1.01) | LDD0600 | [8] |
| LDCM0284 | AC24 | HCT 116 | C162(1.04); C39(1.27) | LDD0601 | [8] |
| LDCM0285 | AC25 | HCT 116 | C162(1.17); C39(1.31) | LDD0602 | [8] |
| LDCM0286 | AC26 | HCT 116 | C39(0.73); C162(1.35) | LDD0603 | [8] |
| LDCM0287 | AC27 | HCT 116 | C39(0.76); C162(1.28) | LDD0604 | [8] |
| LDCM0288 | AC28 | HCT 116 | C39(0.72); C162(1.50) | LDD0605 | [8] |
| LDCM0289 | AC29 | HCT 116 | C39(0.49); C162(1.01) | LDD0606 | [8] |
| LDCM0290 | AC3 | HCT 116 | C39(1.21) | LDD0607 | [8] |
| LDCM0291 | AC30 | HCT 116 | C39(0.50); C162(1.07) | LDD0608 | [8] |
| LDCM0292 | AC31 | HCT 116 | C39(0.60); C162(1.37) | LDD0609 | [8] |
| LDCM0293 | AC32 | HCT 116 | C39(0.47); C162(1.19) | LDD0610 | [8] |
| LDCM0294 | AC33 | HCT 116 | C162(0.84); C39(0.90) | LDD0611 | [8] |
| LDCM0295 | AC34 | HCT 116 | C162(0.48); C39(0.79) | LDD0612 | [8] |
| LDCM0296 | AC35 | HCT 116 | C162(1.12); C39(1.35) | LDD0613 | [8] |
| LDCM0297 | AC36 | HCT 116 | C39(0.91); C162(1.00) | LDD0614 | [8] |
| LDCM0298 | AC37 | HCT 116 | C162(0.98); C39(1.55) | LDD0615 | [8] |
| LDCM0299 | AC38 | HCT 116 | C162(0.96); C39(0.98) | LDD0616 | [8] |
| LDCM0300 | AC39 | HCT 116 | C162(0.66); C39(0.86) | LDD0617 | [8] |
| LDCM0301 | AC4 | HCT 116 | C39(1.37) | LDD0618 | [8] |
| LDCM0302 | AC40 | HCT 116 | C162(0.76); C39(0.96) | LDD0619 | [8] |
| LDCM0303 | AC41 | HCT 116 | C39(1.05); C162(1.13) | LDD0620 | [8] |
| LDCM0304 | AC42 | HCT 116 | C39(1.03); C162(1.11) | LDD0621 | [8] |
| LDCM0305 | AC43 | HCT 116 | C39(0.89); C162(1.58) | LDD0622 | [8] |
| LDCM0306 | AC44 | HCT 116 | C162(1.12); C39(1.86) | LDD0623 | [8] |
| LDCM0307 | AC45 | HCT 116 | C162(1.13); C39(1.42) | LDD0624 | [8] |
| LDCM0308 | AC46 | HCT 116 | C39(0.96) | LDD0625 | [8] |
| LDCM0309 | AC47 | HCT 116 | C39(1.04) | LDD0626 | [8] |
| LDCM0310 | AC48 | HCT 116 | C39(0.90) | LDD0627 | [8] |
| LDCM0311 | AC49 | HCT 116 | C39(0.88) | LDD0628 | [8] |
| LDCM0312 | AC5 | HCT 116 | C39(1.55) | LDD0629 | [8] |
| LDCM0313 | AC50 | HCT 116 | C39(0.84) | LDD0630 | [8] |
| LDCM0314 | AC51 | HCT 116 | C39(0.93) | LDD0631 | [8] |
| LDCM0315 | AC52 | HCT 116 | C39(0.90) | LDD0632 | [8] |
| LDCM0316 | AC53 | HCT 116 | C39(1.23) | LDD0633 | [8] |
| LDCM0317 | AC54 | HCT 116 | C39(0.94) | LDD0634 | [8] |
| LDCM0318 | AC55 | HCT 116 | C39(1.42) | LDD0635 | [8] |
| LDCM0319 | AC56 | HCT 116 | C39(0.85) | LDD0636 | [8] |
| LDCM0320 | AC57 | HCT 116 | C39(0.24); C162(1.01) | LDD0637 | [8] |
| LDCM0321 | AC58 | HCT 116 | C39(0.18); C162(1.04) | LDD0638 | [8] |
| LDCM0322 | AC59 | HCT 116 | C39(0.19); C162(1.07) | LDD0639 | [8] |
| LDCM0323 | AC6 | PaTu 8988t | C39(0.50) | LDD1202 | [8] |
| LDCM0324 | AC60 | HCT 116 | C39(0.18); C162(1.35) | LDD0641 | [8] |
| LDCM0325 | AC61 | HCT 116 | C39(0.17); C162(1.01) | LDD0642 | [8] |
| LDCM0326 | AC62 | HCT 116 | C39(0.14); C162(0.93) | LDD0643 | [8] |
| LDCM0327 | AC63 | HCT 116 | C39(0.20); C162(1.03) | LDD0644 | [8] |
| LDCM0328 | AC64 | HCT 116 | C39(0.14); C162(1.61) | LDD0645 | [8] |
| LDCM0329 | AC65 | HCT 116 | C39(0.23); C162(1.39) | LDD0646 | [8] |
| LDCM0330 | AC66 | HCT 116 | C39(0.22); C162(1.39) | LDD0647 | [8] |
| LDCM0331 | AC67 | HCT 116 | C39(0.08); C162(0.94) | LDD0648 | [8] |
| LDCM0332 | AC68 | HCT 116 | C39(0.65) | LDD0649 | [8] |
| LDCM0333 | AC69 | HCT 116 | C39(0.55) | LDD0650 | [8] |
| LDCM0334 | AC7 | PaTu 8988t | C39(1.10) | LDD1213 | [8] |
| LDCM0335 | AC70 | HCT 116 | C39(0.40) | LDD0652 | [8] |
| LDCM0336 | AC71 | HCT 116 | C39(0.82) | LDD0653 | [8] |
| LDCM0337 | AC72 | HCT 116 | C39(0.60) | LDD0654 | [8] |
| LDCM0338 | AC73 | HCT 116 | C39(0.35) | LDD0655 | [8] |
| LDCM0339 | AC74 | HCT 116 | C39(0.45) | LDD0656 | [8] |
| LDCM0340 | AC75 | HCT 116 | C39(0.41) | LDD0657 | [8] |
| LDCM0341 | AC76 | HCT 116 | C39(0.59) | LDD0658 | [8] |
| LDCM0342 | AC77 | HCT 116 | C39(0.52) | LDD0659 | [8] |
| LDCM0343 | AC78 | HCT 116 | C39(0.65) | LDD0660 | [8] |
| LDCM0344 | AC79 | HCT 116 | C39(0.66) | LDD0661 | [8] |
| LDCM0345 | AC8 | PaTu 8988t | C39(0.76) | LDD1224 | [8] |
| LDCM0346 | AC80 | HCT 116 | C39(0.70) | LDD0663 | [8] |
| LDCM0347 | AC81 | HCT 116 | C39(0.79) | LDD0664 | [8] |
| LDCM0348 | AC82 | HCT 116 | C39(0.44) | LDD0665 | [8] |
| LDCM0349 | AC83 | HCT 116 | C39(0.40) | LDD0666 | [8] |
| LDCM0350 | AC84 | HCT 116 | C39(0.44) | LDD0667 | [8] |
| LDCM0351 | AC85 | HCT 116 | C39(0.65) | LDD0668 | [8] |
| LDCM0352 | AC86 | HCT 116 | C39(0.83) | LDD0669 | [8] |
| LDCM0353 | AC87 | HCT 116 | C39(0.98) | LDD0670 | [8] |
| LDCM0354 | AC88 | HCT 116 | C39(0.96) | LDD0671 | [8] |
| LDCM0355 | AC89 | HCT 116 | C39(0.65) | LDD0672 | [8] |
| LDCM0357 | AC90 | HCT 116 | C39(2.73) | LDD0674 | [8] |
| LDCM0358 | AC91 | HCT 116 | C39(0.44) | LDD0675 | [8] |
| LDCM0359 | AC92 | HCT 116 | C39(0.43) | LDD0676 | [8] |
| LDCM0360 | AC93 | HCT 116 | C39(0.93) | LDD0677 | [8] |
| LDCM0361 | AC94 | HCT 116 | C39(0.80) | LDD0678 | [8] |
| LDCM0362 | AC95 | HCT 116 | C39(0.62) | LDD0679 | [8] |
| LDCM0363 | AC96 | HCT 116 | C39(0.64) | LDD0680 | [8] |
| LDCM0364 | AC97 | HCT 116 | C39(0.46) | LDD0681 | [8] |
| LDCM0365 | AC98 | HCT 116 | C39(0.32) | LDD0682 | [8] |
| LDCM0366 | AC99 | HCT 116 | C39(0.43) | LDD0683 | [8] |
| LDCM0248 | AKOS034007472 | PaTu 8988t | C39(0.33) | LDD1127 | [8] |
| LDCM0356 | AKOS034007680 | PaTu 8988t | C39(0.28) | LDD1235 | [8] |
| LDCM0275 | AKOS034007705 | PaTu 8988t | C39(0.29) | LDD1154 | [8] |
| LDCM0156 | Aniline | NCI-H1299 | 13.62 | LDD0403 | [3] |
| LDCM0108 | Chloroacetamide | HeLa | C162(0.00); C39(0.00) | LDD0222 | [15] |
| LDCM0367 | CL1 | HEK-293T | C162(1.11) | LDD1571 | [20] |
| LDCM0368 | CL10 | HEK-293T | C162(0.78); C39(1.12) | LDD1572 | [20] |
| LDCM0369 | CL100 | HCT 116 | C39(1.59) | LDD0686 | [8] |
| LDCM0370 | CL101 | PaTu 8988t | C39(0.65) | LDD1249 | [8] |
| LDCM0371 | CL102 | PaTu 8988t | C39(0.61) | LDD1250 | [8] |
| LDCM0372 | CL103 | PaTu 8988t | C39(0.24) | LDD1251 | [8] |
| LDCM0373 | CL104 | PaTu 8988t | C39(0.56) | LDD1252 | [8] |
| LDCM0374 | CL105 | HCT 116 | C39(0.63); C162(1.10) | LDD0691 | [8] |
| LDCM0375 | CL106 | HCT 116 | C39(0.19); C162(0.92) | LDD0692 | [8] |
| LDCM0376 | CL107 | HCT 116 | C39(0.35); C162(1.08) | LDD0693 | [8] |
| LDCM0377 | CL108 | HCT 116 | C39(0.29); C162(1.13) | LDD0694 | [8] |
| LDCM0378 | CL109 | HCT 116 | C39(0.73); C162(0.92) | LDD0695 | [8] |
| LDCM0379 | CL11 | HEK-293T | C162(0.88) | LDD1583 | [20] |
| LDCM0380 | CL110 | HCT 116 | C39(0.44); C162(0.92) | LDD0697 | [8] |
| LDCM0381 | CL111 | HCT 116 | C39(0.80); C162(1.00) | LDD0698 | [8] |
| LDCM0382 | CL112 | HCT 116 | C162(0.93); C39(0.94) | LDD0699 | [8] |
| LDCM0383 | CL113 | HCT 116 | C39(0.48); C162(0.71) | LDD0700 | [8] |
| LDCM0384 | CL114 | HCT 116 | C39(0.51); C162(0.58) | LDD0701 | [8] |
| LDCM0385 | CL115 | HCT 116 | C39(0.68); C162(0.77) | LDD0702 | [8] |
| LDCM0386 | CL116 | HCT 116 | C162(0.75); C39(0.89) | LDD0703 | [8] |
| LDCM0387 | CL117 | HCT 116 | C162(0.86); C39(0.86) | LDD0704 | [8] |
| LDCM0388 | CL118 | HCT 116 | C39(0.83); C162(1.31) | LDD0705 | [8] |
| LDCM0389 | CL119 | HCT 116 | C39(0.91); C162(1.49) | LDD0706 | [8] |
| LDCM0390 | CL12 | HEK-293T | C162(1.01) | LDD1594 | [20] |
| LDCM0391 | CL120 | HCT 116 | C39(0.84); C162(1.53) | LDD0708 | [8] |
| LDCM0392 | CL121 | HCT 116 | C39(1.09) | LDD0709 | [8] |
| LDCM0393 | CL122 | HCT 116 | C39(1.01) | LDD0710 | [8] |
| LDCM0394 | CL123 | HCT 116 | C39(0.98) | LDD0711 | [8] |
| LDCM0395 | CL124 | HCT 116 | C39(1.03) | LDD0712 | [8] |
| LDCM0396 | CL125 | HCT 116 | C39(0.82); C162(1.31) | LDD0713 | [8] |
| LDCM0397 | CL126 | HCT 116 | C162(1.16); C39(1.50) | LDD0714 | [8] |
| LDCM0398 | CL127 | HCT 116 | C39(0.46); C162(1.36) | LDD0715 | [8] |
| LDCM0399 | CL128 | HCT 116 | C39(0.24); C162(0.98) | LDD0716 | [8] |
| LDCM0400 | CL13 | HEK-293T | C162(1.06) | LDD1604 | [20] |
| LDCM0401 | CL14 | HEK-293T | C162(1.17) | LDD1605 | [20] |
| LDCM0402 | CL15 | HEK-293T | C162(1.12); C39(1.10) | LDD1606 | [20] |
| LDCM0403 | CL16 | HCT 116 | C39(0.51) | LDD0720 | [8] |
| LDCM0404 | CL17 | HCT 116 | C39(0.37) | LDD0721 | [8] |
| LDCM0405 | CL18 | HCT 116 | C39(0.17) | LDD0722 | [8] |
| LDCM0406 | CL19 | HCT 116 | C39(0.28) | LDD0723 | [8] |
| LDCM0407 | CL2 | HEK-293T | C162(1.08) | LDD1611 | [20] |
| LDCM0408 | CL20 | HCT 116 | C39(0.24) | LDD0725 | [8] |
| LDCM0409 | CL21 | HCT 116 | C39(0.15) | LDD0726 | [8] |
| LDCM0410 | CL22 | HCT 116 | C39(0.11) | LDD0727 | [8] |
| LDCM0411 | CL23 | HCT 116 | C39(0.40) | LDD0728 | [8] |
| LDCM0412 | CL24 | HCT 116 | C39(0.13) | LDD0729 | [8] |
| LDCM0413 | CL25 | HCT 116 | C39(0.18) | LDD0730 | [8] |
| LDCM0414 | CL26 | HCT 116 | C39(0.34) | LDD0731 | [8] |
| LDCM0415 | CL27 | HCT 116 | C39(0.44) | LDD0732 | [8] |
| LDCM0416 | CL28 | HCT 116 | C39(0.26) | LDD0733 | [8] |
| LDCM0417 | CL29 | HCT 116 | C39(0.23) | LDD0734 | [8] |
| LDCM0418 | CL3 | HEK-293T | C162(1.11); C39(1.06) | LDD1622 | [20] |
| LDCM0419 | CL30 | HCT 116 | C39(0.36) | LDD0736 | [8] |
| LDCM0420 | CL31 | HCT 116 | C39(0.64) | LDD0737 | [8] |
| LDCM0421 | CL32 | HCT 116 | C39(0.34) | LDD0738 | [8] |
| LDCM0422 | CL33 | HCT 116 | C39(0.40) | LDD0739 | [8] |
| LDCM0423 | CL34 | HCT 116 | C39(0.23) | LDD0740 | [8] |
| LDCM0424 | CL35 | HCT 116 | C39(0.25) | LDD0741 | [8] |
| LDCM0425 | CL36 | HCT 116 | C39(0.32) | LDD0742 | [8] |
| LDCM0426 | CL37 | HCT 116 | C39(0.22) | LDD0743 | [8] |
| LDCM0428 | CL39 | HCT 116 | C39(0.25) | LDD0745 | [8] |
| LDCM0429 | CL4 | HEK-293T | C162(0.90); C39(1.23) | LDD1633 | [20] |
| LDCM0430 | CL40 | HCT 116 | C39(0.37) | LDD0747 | [8] |
| LDCM0431 | CL41 | HCT 116 | C39(0.41) | LDD0748 | [8] |
| LDCM0432 | CL42 | HCT 116 | C39(0.15) | LDD0749 | [8] |
| LDCM0433 | CL43 | HCT 116 | C39(0.19) | LDD0750 | [8] |
| LDCM0434 | CL44 | HCT 116 | C39(0.27) | LDD0751 | [8] |
| LDCM0435 | CL45 | HCT 116 | C39(0.19) | LDD0752 | [8] |
| LDCM0436 | CL46 | HCT 116 | C162(1.46) | LDD0753 | [8] |
| LDCM0437 | CL47 | HCT 116 | C162(1.55) | LDD0754 | [8] |
| LDCM0438 | CL48 | HCT 116 | C162(1.39) | LDD0755 | [8] |
| LDCM0439 | CL49 | HCT 116 | C162(1.49) | LDD0756 | [8] |
| LDCM0440 | CL5 | HEK-293T | C162(0.99) | LDD1644 | [20] |
| LDCM0441 | CL50 | HCT 116 | C162(1.18) | LDD0758 | [8] |
| LDCM0442 | CL51 | HCT 116 | C162(1.32) | LDD0759 | [8] |
| LDCM0443 | CL52 | HCT 116 | C162(1.32) | LDD0760 | [8] |
| LDCM0444 | CL53 | HCT 116 | C162(0.74) | LDD0761 | [8] |
| LDCM0445 | CL54 | HCT 116 | C162(1.12) | LDD0762 | [8] |
| LDCM0446 | CL55 | HCT 116 | C162(0.93) | LDD0763 | [8] |
| LDCM0447 | CL56 | HCT 116 | C162(1.04) | LDD0764 | [8] |
| LDCM0448 | CL57 | HCT 116 | C162(0.95) | LDD0765 | [8] |
| LDCM0449 | CL58 | HCT 116 | C162(1.39) | LDD0766 | [8] |
| LDCM0450 | CL59 | HCT 116 | C162(1.09) | LDD0767 | [8] |
| LDCM0451 | CL6 | HEK-293T | C162(1.13) | LDD1654 | [20] |
| LDCM0452 | CL60 | HCT 116 | C162(1.52) | LDD0769 | [8] |
| LDCM0453 | CL61 | HCT 116 | C162(1.07) | LDD0770 | [8] |
| LDCM0454 | CL62 | HCT 116 | C162(1.37) | LDD0771 | [8] |
| LDCM0455 | CL63 | HCT 116 | C162(0.87) | LDD0772 | [8] |
| LDCM0456 | CL64 | HCT 116 | C162(1.10) | LDD0773 | [8] |
| LDCM0457 | CL65 | HCT 116 | C162(0.98) | LDD0774 | [8] |
| LDCM0458 | CL66 | HCT 116 | C162(1.18) | LDD0775 | [8] |
| LDCM0459 | CL67 | HCT 116 | C162(0.87) | LDD0776 | [8] |
| LDCM0460 | CL68 | HCT 116 | C162(0.71) | LDD0777 | [8] |
| LDCM0461 | CL69 | HCT 116 | C162(0.83) | LDD0778 | [8] |
| LDCM0462 | CL7 | HEK-293T | C162(1.21) | LDD1665 | [20] |
| LDCM0463 | CL70 | HCT 116 | C162(0.77) | LDD0780 | [8] |
| LDCM0464 | CL71 | HCT 116 | C162(0.78) | LDD0781 | [8] |
| LDCM0465 | CL72 | HCT 116 | C162(0.85) | LDD0782 | [8] |
| LDCM0466 | CL73 | HCT 116 | C162(0.90) | LDD0783 | [8] |
| LDCM0467 | CL74 | HCT 116 | C162(0.80) | LDD0784 | [8] |
| LDCM0469 | CL76 | HCT 116 | C162(0.71); C39(0.39) | LDD0786 | [8] |
| LDCM0470 | CL77 | HCT 116 | C162(0.67); C39(0.66) | LDD0787 | [8] |
| LDCM0471 | CL78 | HCT 116 | C162(0.74); C39(0.64) | LDD0788 | [8] |
| LDCM0472 | CL79 | HCT 116 | C162(0.84); C39(0.27) | LDD0789 | [8] |
| LDCM0473 | CL8 | HEK-293T | C162(1.31); C39(0.99) | LDD1676 | [20] |
| LDCM0474 | CL80 | HCT 116 | C162(0.84); C39(0.40) | LDD0791 | [8] |
| LDCM0475 | CL81 | HCT 116 | C162(0.98); C39(0.45) | LDD0792 | [8] |
| LDCM0476 | CL82 | HCT 116 | C162(1.04); C39(0.14) | LDD0793 | [8] |
| LDCM0477 | CL83 | HCT 116 | C162(0.74); C39(0.18) | LDD0794 | [8] |
| LDCM0478 | CL84 | HCT 116 | C162(0.78); C39(0.13) | LDD0795 | [8] |
| LDCM0479 | CL85 | HCT 116 | C162(0.65); C39(0.66) | LDD0796 | [8] |
| LDCM0480 | CL86 | HCT 116 | C162(0.67); C39(0.99) | LDD0797 | [8] |
| LDCM0481 | CL87 | HCT 116 | C162(0.75); C39(0.49) | LDD0798 | [8] |
| LDCM0482 | CL88 | HCT 116 | C162(0.64); C39(0.20) | LDD0799 | [8] |
| LDCM0483 | CL89 | HCT 116 | C162(0.76); C39(0.13) | LDD0800 | [8] |
| LDCM0484 | CL9 | HEK-293T | C162(0.90) | LDD1687 | [20] |
| LDCM0485 | CL90 | HCT 116 | C162(0.64); C39(0.85) | LDD0802 | [8] |
| LDCM0486 | CL91 | HCT 116 | C39(0.80) | LDD0803 | [8] |
| LDCM0487 | CL92 | HCT 116 | C39(0.89) | LDD0804 | [8] |
| LDCM0488 | CL93 | HCT 116 | C39(1.32) | LDD0805 | [8] |
| LDCM0489 | CL94 | HCT 116 | C39(1.52) | LDD0806 | [8] |
| LDCM0490 | CL95 | HCT 116 | C39(0.90) | LDD0807 | [8] |
| LDCM0491 | CL96 | HCT 116 | C39(0.88) | LDD0808 | [8] |
| LDCM0492 | CL97 | HCT 116 | C39(0.89) | LDD0809 | [8] |
| LDCM0493 | CL98 | HCT 116 | C39(1.27) | LDD0810 | [8] |
| LDCM0494 | CL99 | HCT 116 | C39(1.39) | LDD0811 | [8] |
| LDCM0097 | Dasatinib | K562 | 17.07 | LDD0270 | [4] |
| LDCM0495 | E2913 | HEK-293T | C162(1.01); C39(1.00) | LDD1698 | [20] |
| LDCM0625 | F8 | Ramos | C162(1.09) | LDD2187 | [21] |
| LDCM0572 | Fragment10 | Ramos | C162(0.89) | LDD2189 | [21] |
| LDCM0573 | Fragment11 | Ramos | C162(0.29) | LDD2190 | [21] |
| LDCM0574 | Fragment12 | Ramos | C162(1.27) | LDD2191 | [21] |
| LDCM0575 | Fragment13 | Ramos | C162(1.19) | LDD2192 | [21] |
| LDCM0576 | Fragment14 | Ramos | C162(1.17) | LDD2193 | [21] |
| LDCM0579 | Fragment20 | Ramos | C162(1.33) | LDD2194 | [21] |
| LDCM0580 | Fragment21 | Ramos | C162(0.81) | LDD2195 | [21] |
| LDCM0582 | Fragment23 | Ramos | C162(0.55) | LDD2196 | [21] |
| LDCM0578 | Fragment27 | Ramos | C162(0.73) | LDD2197 | [21] |
| LDCM0586 | Fragment28 | Ramos | C162(1.67) | LDD2198 | [21] |
| LDCM0588 | Fragment30 | Ramos | C162(1.03) | LDD2199 | [21] |
| LDCM0589 | Fragment31 | Ramos | C162(0.68) | LDD2200 | [21] |
| LDCM0590 | Fragment32 | Ramos | C162(1.03) | LDD2201 | [21] |
| LDCM0468 | Fragment33 | HCT 116 | C162(0.99) | LDD0785 | [8] |
| LDCM0596 | Fragment38 | Ramos | C162(1.62) | LDD2203 | [21] |
| LDCM0566 | Fragment4 | Ramos | C162(0.99) | LDD2184 | [21] |
| LDCM0427 | Fragment51 | HCT 116 | C39(0.15) | LDD0744 | [8] |
| LDCM0610 | Fragment52 | Ramos | C162(1.82) | LDD2204 | [21] |
| LDCM0614 | Fragment56 | Ramos | C162(1.23) | LDD2205 | [21] |
| LDCM0569 | Fragment7 | Ramos | C162(1.11) | LDD2186 | [21] |
| LDCM0571 | Fragment9 | Ramos | C162(1.39) | LDD2188 | [21] |
| LDCM0022 | KB02 | HEK-293T | C162(0.94) | LDD1492 | [20] |
| LDCM0023 | KB03 | HEK-293T | C162(1.00) | LDD1497 | [20] |
| LDCM0024 | KB05 | COLO792 | C39(1.73) | LDD3310 | [22] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C162(1.53) | LDD2107 | [5] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C162(1.29) | LDD2127 | [5] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C162(0.86) | LDD2143 | [5] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C162(2.24) | LDD2144 | [5] |
| LDCM0553 | Nucleophilic fragment 6b | MDA-MB-231 | C162(1.90) | LDD2147 | [5] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C119(2.17) | LDD2207 | [23] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Nuclear pore complex protein Nup153 (NUP153) | NUP153 family | P49790 | |||
Other
The Drug(s) Related To This Target
Approved
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Dasatinib | Small molecular drug | DB01254 | |||
| Fostamatinib | Small molecular drug | DB12010 | |||
| Minocycline | Small molecular drug | DB01017 | |||
Phase 4
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Ozagrel | Small molecular drug | D0C7AA | |||
Phase 3
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Losmapimod | Small molecular drug | D0C8DD | |||
Phase 2
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Dilmapimod | Small molecular drug | D0K0JQ | |||
| Vx-702 | Small molecular drug | D0D9AH | |||
| Vx-745 | Small molecular drug | D02NNV | |||
Investigative
Patented
Discontinued
References
