Details of the Target
General Information of Target
| Target ID | LDTP00751 | |||||
|---|---|---|---|---|---|---|
| Target Name | Receptor-interacting serine/threonine-protein kinase 2 (RIPK2) | |||||
| Gene Name | RIPK2 | |||||
| Gene ID | 8767 | |||||
| Synonyms |
CARDIAK; RICK; RIP2; Receptor-interacting serine/threonine-protein kinase 2; EC 2.7.11.1; CARD-containing interleukin-1 beta-converting enzyme-associated kinase; CARD-containing IL-1 beta ICE-kinase; RIP-like-interacting CLARP kinase; Receptor-interacting protein 2; RIP-2; Tyrosine-protein kinase RIPK2; EC 2.7.10.2
|
|||||
| 3D Structure | ||||||
| Sequence |
MNGEAICSALPTIPYHKLADLRYLSRGASGTVSSARHADWRVQVAVKHLHIHTPLLDSER
KDVLREAEILHKARFSYILPILGICNEPEFLGIVTEYMPNGSLNELLHRKTEYPDVAWPL RFRILHEIALGVNYLHNMTPPLLHHDLKTQNILLDNEFHVKIADFGLSKWRMMSLSQSRS SKSAPEGGTIIYMPPENYEPGQKSRASIKHDIYSYAVITWEVLSRKQPFEDVTNPLQIMY SVSQGHRPVINEESLPYDIPHRARMISLIESGWAQNPDERPSFLKCLIELEPVLRTFEEI TFLEAVIQLKKTKLQSVSSAIHLCDKKKMELSLNIPVNHGPQEESCGSSQLHENSGSPET SRSLPAPQDNDFLSRKAQDCYFMKLHHCPGNHSWDSTISGSQRAAFCDHKTTPCSSAIIN PLSTAGNSERLQPGIAQQWIQSKREDIVNQMTEACLNQSLDALLSRDLIMKEDYELVSTK PTRTSKVRQLLDTTDIQGEEFAKVIVQKLKDNKQMGLQPYPEILVVSRSPSLNLLQNKSM |
|||||
| Target Type |
Clinical trial
|
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Protein kinase superfamily, TKL Ser/Thr protein kinase family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Serine/threonine/tyrosine-protein kinase that plays an essential role in modulation of innate and adaptive immune responses . Acts as a key effector of NOD1 and NOD2 signaling pathways: upon activation by bacterial peptidoglycans, NOD1 and NOD2 oligomerize and recruit RIPK2 via CARD-CARD domains, leading to the formation of RIPK2 filaments. Once recruited, RIPK2 autophosphorylates and undergoes 'Lys-63'-linked polyubiquitination by E3 ubiquitin ligases XIAP, BIRC2 and BIRC3, as well as 'Met-1'-linked (linear) polyubiquitination by the LUBAC complex, becoming a scaffolding protein for downstream effectors . 'Met-1'-linked polyubiquitin chains attached to RIPK2 recruit IKBKG/NEMO, which undergoes 'Lys-63'-linked polyubiquitination in a RIPK2-dependent process. 'Lys-63'-linked polyubiquitin chains attached to RIPK2 serve as docking sites for TAB2 and TAB3 and mediate the recruitment of MAP3K7/TAK1 to IKBKG/NEMO, inducing subsequent activation of IKBKB/IKKB. In turn, NF-kappa-B is released from NF-kappa-B inhibitors and translocates into the nucleus where it activates the transcription of hundreds of genes involved in immune response, growth control, or protection against apoptosis. The protein kinase activity is dispensable for the NOD1 and NOD2 signaling pathways. Contributes to the tyrosine phosphorylation of the guanine exchange factor ARHGEF2 through Src tyrosine kinase leading to NF-kappa-B activation by NOD2. Also involved in adaptive immunity: plays a role during engagement of the T-cell receptor (TCR) in promoting BCL10 phosphorylation and subsequent NF-kappa-B activation. Plays a role in the inactivation of RHOA in response to NGFR signaling.
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| A172 | SNV: p.I418L | DBIA Probe Info | |||
| DU145 | SNV: p.I259M | DBIA Probe Info | |||
| HCT15 | Deletion: p.L373YfsTer11 | . | |||
| HT1197 | Insertion: p.Q497PfsTer16 | . | |||
| LNCaP clone FGC | SNV: p.H144Q; p.A216S | . | |||
| SW480 | SNV: p.P228A | . | |||
| SW620 | SNV: p.P228A | . | |||
| SW756 | SNV: p.D115N | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
C-Sul Probe Info |
 |
3.58 | LDD0066 | [2] | |
|
JZ128-DTB Probe Info |
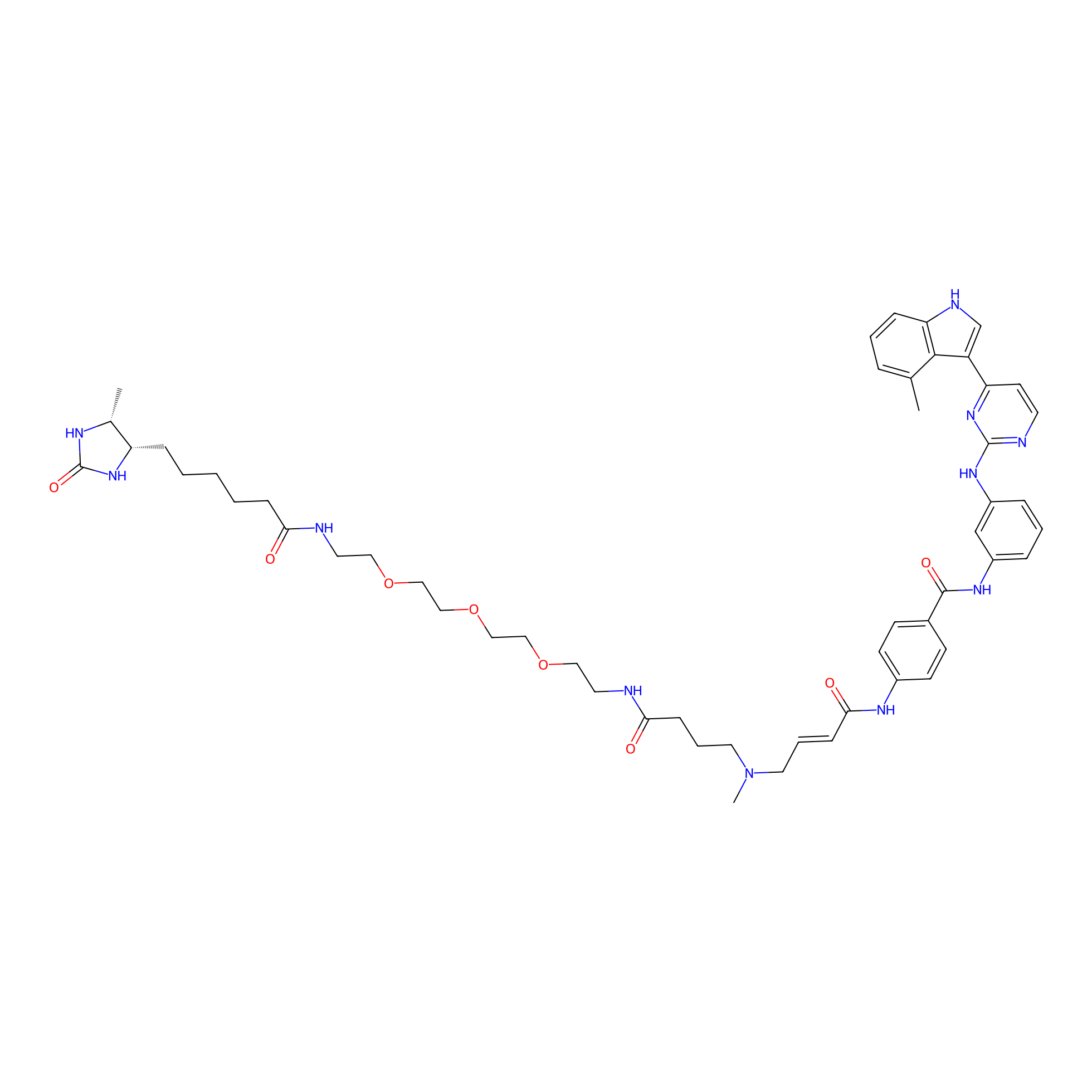 |
C324(0.00); C414(0.00); C346(0.00) | LDD0462 | [3] | |
|
DA-P3 Probe Info |
 |
14.82 | LDD0184 | [4] | |
|
AHL-Pu-1 Probe Info |
 |
C346(2.78) | LDD0170 | [5] | |
|
DBIA Probe Info |
 |
C380(5.52) | LDD0080 | [6] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0162 | [7] | |
|
NAIA_4 Probe Info |
 |
C324(0.00); C346(0.00) | LDD2226 | [8] | |
|
IPM Probe Info |
 |
N.A. | LDD0147 | [9] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [9] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [10] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [11] | |
|
NAIA_5 Probe Info |
 |
C455(0.00); C7(0.00) | LDD2223 | [8] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FFF probe3 Probe Info |
 |
20.00 | LDD0465 | [12] | |
|
FFF probe6 Probe Info |
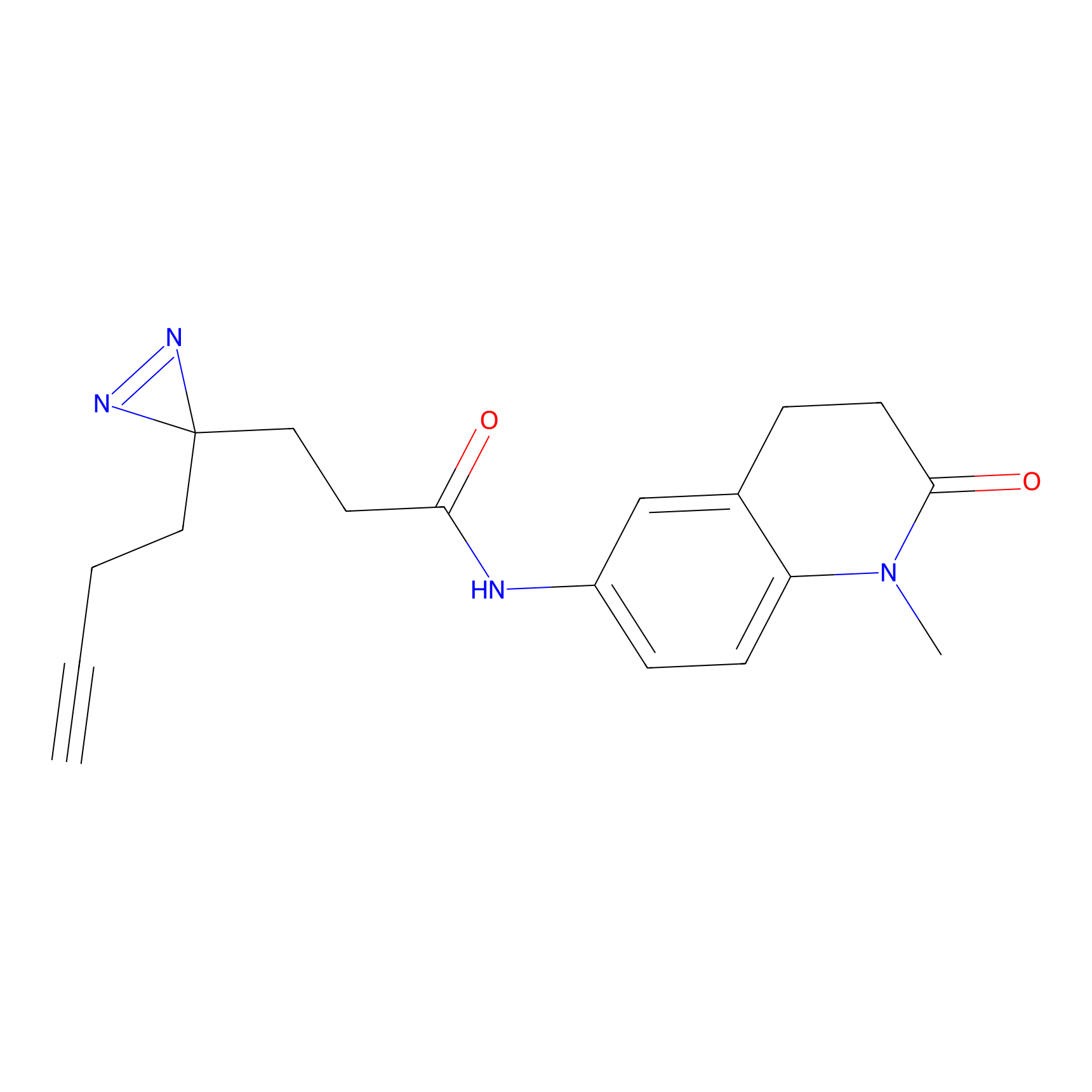 |
20.00 | LDD0468 | [12] | |
|
Dasatinib-CA-3PAP Probe Info |
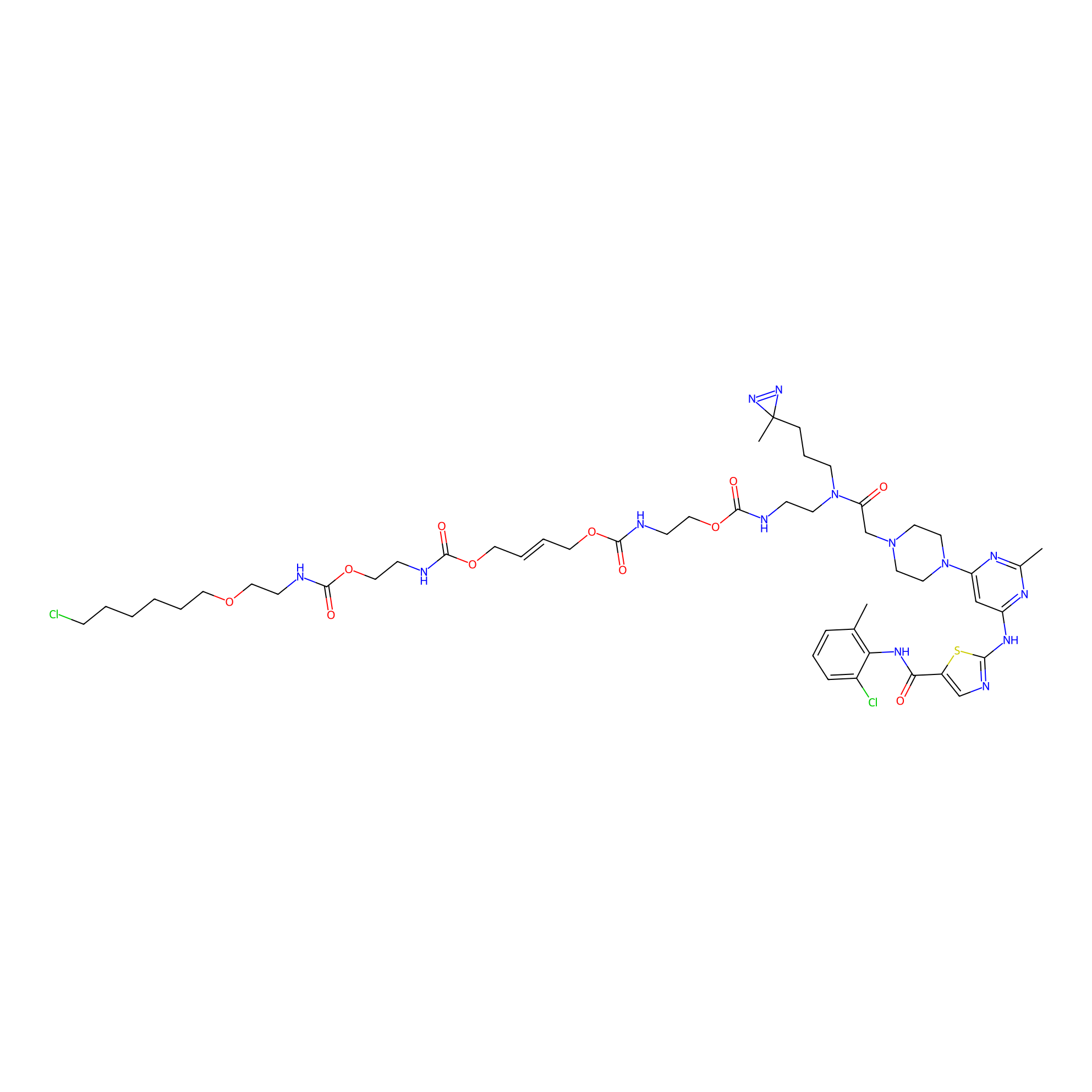 |
N.A. | LDD0365 | [13] | |
|
Dasatinib-CA-8PAP Probe Info |
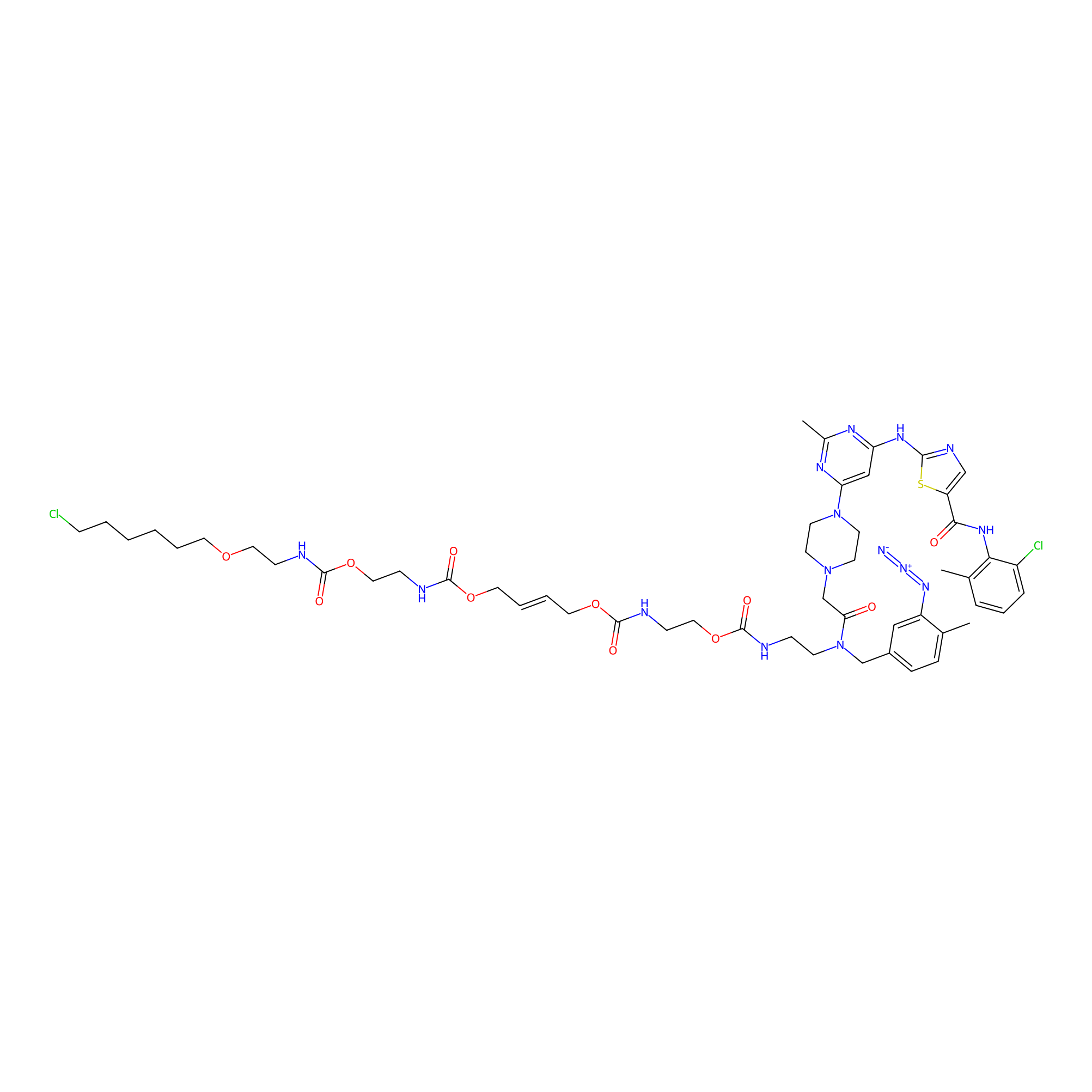 |
N.A. | LDD0364 | [13] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | DM93 | C346(2.78) | LDD0170 | [5] |
| LDCM0214 | AC1 | HCT 116 | C380(1.04); C286(1.25); C407(0.94); C414(1.01) | LDD0531 | [6] |
| LDCM0215 | AC10 | HCT 116 | C380(0.78); C286(0.85); C407(0.52); C414(0.94) | LDD0532 | [6] |
| LDCM0216 | AC100 | HCT 116 | C286(0.48) | LDD0533 | [6] |
| LDCM0217 | AC101 | HCT 116 | C286(0.63) | LDD0534 | [6] |
| LDCM0218 | AC102 | HCT 116 | C286(0.43) | LDD0535 | [6] |
| LDCM0219 | AC103 | HCT 116 | C286(0.43) | LDD0536 | [6] |
| LDCM0220 | AC104 | HCT 116 | C286(0.49) | LDD0537 | [6] |
| LDCM0221 | AC105 | HCT 116 | C286(0.47) | LDD0538 | [6] |
| LDCM0222 | AC106 | HCT 116 | C286(0.48) | LDD0539 | [6] |
| LDCM0223 | AC107 | HCT 116 | C286(0.48) | LDD0540 | [6] |
| LDCM0224 | AC108 | HCT 116 | C286(0.56) | LDD0541 | [6] |
| LDCM0225 | AC109 | HCT 116 | C286(0.60) | LDD0542 | [6] |
| LDCM0226 | AC11 | HCT 116 | C380(0.90); C286(0.79); C407(0.61); C414(1.04) | LDD0543 | [6] |
| LDCM0227 | AC110 | HCT 116 | C286(0.55) | LDD0544 | [6] |
| LDCM0228 | AC111 | HCT 116 | C286(0.58) | LDD0545 | [6] |
| LDCM0229 | AC112 | HCT 116 | C286(0.47) | LDD0546 | [6] |
| LDCM0230 | AC113 | HCT 116 | C380(0.84); C286(1.00); C407(0.85); C414(1.13) | LDD0547 | [6] |
| LDCM0231 | AC114 | HCT 116 | C380(0.98); C286(0.87); C407(0.71); C414(0.97) | LDD0548 | [6] |
| LDCM0232 | AC115 | HCT 116 | C380(0.96); C286(0.86); C407(0.98); C414(1.14) | LDD0549 | [6] |
| LDCM0233 | AC116 | HCT 116 | C380(1.11); C286(0.88); C407(0.73); C414(1.15) | LDD0550 | [6] |
| LDCM0234 | AC117 | HCT 116 | C380(0.86); C286(0.90); C407(0.67); C414(1.32) | LDD0551 | [6] |
| LDCM0235 | AC118 | HCT 116 | C380(0.88); C286(0.99); C407(0.66); C414(1.23) | LDD0552 | [6] |
| LDCM0236 | AC119 | HCT 116 | C380(1.03); C286(0.85); C407(0.69); C414(1.21) | LDD0553 | [6] |
| LDCM0237 | AC12 | HCT 116 | C380(0.84); C286(0.82); C407(0.56); C414(0.89) | LDD0554 | [6] |
| LDCM0238 | AC120 | HCT 116 | C380(1.10); C286(0.83); C407(0.73); C414(1.13) | LDD0555 | [6] |
| LDCM0239 | AC121 | HCT 116 | C380(0.77); C286(1.01); C407(0.74); C414(0.97) | LDD0556 | [6] |
| LDCM0240 | AC122 | HCT 116 | C380(0.95); C286(1.04); C407(0.89); C414(1.46) | LDD0557 | [6] |
| LDCM0241 | AC123 | HCT 116 | C380(0.95); C286(0.96); C407(0.73); C414(1.05) | LDD0558 | [6] |
| LDCM0242 | AC124 | HCT 116 | C380(1.00); C286(0.97); C407(0.77); C414(0.97) | LDD0559 | [6] |
| LDCM0243 | AC125 | HCT 116 | C380(1.07); C286(1.02); C407(0.82); C414(1.06) | LDD0560 | [6] |
| LDCM0244 | AC126 | HCT 116 | C380(1.18); C286(0.88); C407(0.72); C414(1.14) | LDD0561 | [6] |
| LDCM0245 | AC127 | HCT 116 | C380(1.19); C286(0.90); C407(0.74); C414(1.19) | LDD0562 | [6] |
| LDCM0246 | AC128 | HCT 116 | C380(0.65); C286(1.39) | LDD0563 | [6] |
| LDCM0247 | AC129 | HCT 116 | C380(0.58); C286(1.59) | LDD0564 | [6] |
| LDCM0249 | AC130 | HCT 116 | C380(0.65); C286(1.59) | LDD0566 | [6] |
| LDCM0250 | AC131 | HCT 116 | C380(0.74); C286(1.29) | LDD0567 | [6] |
| LDCM0251 | AC132 | HCT 116 | C380(0.62); C286(1.50) | LDD0568 | [6] |
| LDCM0252 | AC133 | HCT 116 | C380(0.57); C286(1.36) | LDD0569 | [6] |
| LDCM0253 | AC134 | HCT 116 | C380(0.64); C286(1.17) | LDD0570 | [6] |
| LDCM0254 | AC135 | HCT 116 | C380(0.70); C286(1.25) | LDD0571 | [6] |
| LDCM0255 | AC136 | HCT 116 | C380(0.67); C286(1.24) | LDD0572 | [6] |
| LDCM0256 | AC137 | HCT 116 | C380(0.50); C286(1.12) | LDD0573 | [6] |
| LDCM0257 | AC138 | HCT 116 | C380(0.55); C286(0.81) | LDD0574 | [6] |
| LDCM0258 | AC139 | HCT 116 | C380(0.62); C286(1.20) | LDD0575 | [6] |
| LDCM0259 | AC14 | HCT 116 | C380(0.66); C286(0.87); C407(0.53); C414(1.32) | LDD0576 | [6] |
| LDCM0260 | AC140 | HCT 116 | C380(0.59); C286(1.18) | LDD0577 | [6] |
| LDCM0261 | AC141 | HCT 116 | C380(0.73); C286(1.00) | LDD0578 | [6] |
| LDCM0262 | AC142 | HCT 116 | C380(0.60); C286(1.18) | LDD0579 | [6] |
| LDCM0263 | AC143 | HCT 116 | C380(1.14); C286(0.80); C407(1.16) | LDD0580 | [6] |
| LDCM0264 | AC144 | HCT 116 | C286(0.71); C407(1.38); C380(1.38) | LDD0581 | [6] |
| LDCM0265 | AC145 | HCT 116 | C286(0.85); C380(1.08); C407(1.16) | LDD0582 | [6] |
| LDCM0266 | AC146 | HCT 116 | C286(0.84); C380(1.17); C407(1.27) | LDD0583 | [6] |
| LDCM0267 | AC147 | HCT 116 | C286(0.80); C380(1.21); C407(1.59) | LDD0584 | [6] |
| LDCM0268 | AC148 | HCT 116 | C286(1.17); C380(1.43); C407(1.89) | LDD0585 | [6] |
| LDCM0269 | AC149 | HCT 116 | C286(0.90); C380(1.33); C407(1.45) | LDD0586 | [6] |
| LDCM0270 | AC15 | HCT 116 | C407(0.48); C286(0.78); C380(0.92); C414(1.05) | LDD0587 | [6] |
| LDCM0271 | AC150 | HCT 116 | C286(0.88); C380(1.04); C407(1.18) | LDD0588 | [6] |
| LDCM0272 | AC151 | HCT 116 | C286(0.91); C380(1.12); C407(1.15) | LDD0589 | [6] |
| LDCM0273 | AC152 | HCT 116 | C286(0.79); C380(1.22); C407(1.35) | LDD0590 | [6] |
| LDCM0274 | AC153 | HCT 116 | C286(0.93); C407(1.95); C380(1.99) | LDD0591 | [6] |
| LDCM0621 | AC154 | HCT 116 | C380(1.36); C286(0.81); C407(1.41) | LDD2158 | [6] |
| LDCM0622 | AC155 | HCT 116 | C380(1.27); C286(0.80); C407(1.53) | LDD2159 | [6] |
| LDCM0623 | AC156 | HCT 116 | C380(1.36); C286(1.04); C407(1.36) | LDD2160 | [6] |
| LDCM0624 | AC157 | HCT 116 | C380(1.09); C286(0.97); C407(1.18) | LDD2161 | [6] |
| LDCM0276 | AC17 | HCT 116 | C380(0.34); C414(1.07); C286(1.08) | LDD0593 | [6] |
| LDCM0277 | AC18 | HCT 116 | C380(0.44); C286(0.82); C414(1.04) | LDD0594 | [6] |
| LDCM0278 | AC19 | HCT 116 | C380(0.35); C286(1.04); C414(1.06) | LDD0595 | [6] |
| LDCM0279 | AC2 | HCT 116 | C414(0.95); C407(0.97); C380(1.06); C286(1.12) | LDD0596 | [6] |
| LDCM0280 | AC20 | HCT 116 | C380(0.42); C286(0.95); C414(1.18) | LDD0597 | [6] |
| LDCM0281 | AC21 | HCT 116 | C380(0.40); C414(0.92); C286(0.99) | LDD0598 | [6] |
| LDCM0282 | AC22 | HCT 116 | C380(0.39); C414(0.99); C286(1.03) | LDD0599 | [6] |
| LDCM0283 | AC23 | HCT 116 | C380(0.40); C286(0.84); C414(0.86) | LDD0600 | [6] |
| LDCM0284 | AC24 | HCT 116 | C380(0.32); C286(1.04); C414(1.24) | LDD0601 | [6] |
| LDCM0285 | AC25 | HCT 116 | C286(0.99); C414(1.03); C407(1.16) | LDD0602 | [6] |
| LDCM0286 | AC26 | HCT 116 | C286(0.99); C407(1.08); C414(1.35) | LDD0603 | [6] |
| LDCM0287 | AC27 | HCT 116 | C286(0.92); C407(1.08); C414(1.27) | LDD0604 | [6] |
| LDCM0288 | AC28 | HCT 116 | C407(0.86); C286(0.92); C414(1.20) | LDD0605 | [6] |
| LDCM0289 | AC29 | HCT 116 | C407(0.96); C286(0.99); C414(1.31) | LDD0606 | [6] |
| LDCM0290 | AC3 | HCT 116 | C414(0.88); C407(0.94); C380(1.11); C286(1.12) | LDD0607 | [6] |
| LDCM0291 | AC30 | HCT 116 | C407(0.78); C286(0.87); C414(1.26) | LDD0608 | [6] |
| LDCM0292 | AC31 | HCT 116 | C407(0.72); C286(0.94); C414(1.15) | LDD0609 | [6] |
| LDCM0293 | AC32 | HCT 116 | C407(0.68); C286(0.91); C414(1.26) | LDD0610 | [6] |
| LDCM0294 | AC33 | HCT 116 | C407(0.83); C286(0.86); C414(1.14) | LDD0611 | [6] |
| LDCM0295 | AC34 | HCT 116 | C407(0.83); C286(0.84); C414(1.30) | LDD0612 | [6] |
| LDCM0296 | AC35 | HCT 116 | C407(1.10); C286(1.12); C414(1.33) | LDD0613 | [6] |
| LDCM0297 | AC36 | HCT 116 | C286(0.96); C414(1.29); C407(1.42) | LDD0614 | [6] |
| LDCM0298 | AC37 | HCT 116 | C286(0.88); C414(1.28); C407(1.46) | LDD0615 | [6] |
| LDCM0299 | AC38 | HCT 116 | C286(0.90); C414(1.16); C407(1.17) | LDD0616 | [6] |
| LDCM0300 | AC39 | HCT 116 | C407(1.00); C286(1.10); C414(1.14) | LDD0617 | [6] |
| LDCM0301 | AC4 | HCT 116 | C414(0.93); C407(0.95); C380(1.12); C286(1.25) | LDD0618 | [6] |
| LDCM0302 | AC40 | HCT 116 | C286(0.88); C407(1.22); C414(1.39) | LDD0619 | [6] |
| LDCM0303 | AC41 | HCT 116 | C407(0.83); C286(0.86); C414(1.46) | LDD0620 | [6] |
| LDCM0304 | AC42 | HCT 116 | C407(0.67); C286(0.90); C414(1.14) | LDD0621 | [6] |
| LDCM0305 | AC43 | HCT 116 | C407(0.76); C286(1.01); C414(1.30) | LDD0622 | [6] |
| LDCM0306 | AC44 | HCT 116 | C407(0.80); C286(0.98); C414(1.48) | LDD0623 | [6] |
| LDCM0307 | AC45 | HCT 116 | C407(0.86); C286(0.92); C414(1.41) | LDD0624 | [6] |
| LDCM0308 | AC46 | HCT 116 | C414(1.03); C286(1.03) | LDD0625 | [6] |
| LDCM0309 | AC47 | HCT 116 | C414(0.96); C286(1.05) | LDD0626 | [6] |
| LDCM0310 | AC48 | HCT 116 | C286(1.10); C414(1.15) | LDD0627 | [6] |
| LDCM0311 | AC49 | HCT 116 | C414(1.01); C286(1.08) | LDD0628 | [6] |
| LDCM0312 | AC5 | HCT 116 | C414(0.86); C407(0.98); C380(1.01); C286(1.24) | LDD0629 | [6] |
| LDCM0313 | AC50 | HCT 116 | C286(1.02); C414(1.23) | LDD0630 | [6] |
| LDCM0314 | AC51 | HCT 116 | C414(0.98); C286(1.14) | LDD0631 | [6] |
| LDCM0315 | AC52 | HCT 116 | C286(1.13); C414(1.14) | LDD0632 | [6] |
| LDCM0316 | AC53 | HCT 116 | C414(1.08); C286(1.20) | LDD0633 | [6] |
| LDCM0317 | AC54 | HCT 116 | C414(0.96); C286(0.99) | LDD0634 | [6] |
| LDCM0318 | AC55 | HCT 116 | C286(1.12); C414(1.18) | LDD0635 | [6] |
| LDCM0319 | AC56 | HCT 116 | C414(1.05); C286(1.19) | LDD0636 | [6] |
| LDCM0320 | AC57 | HCT 116 | C286(1.28) | LDD0637 | [6] |
| LDCM0321 | AC58 | HCT 116 | C286(1.02) | LDD0638 | [6] |
| LDCM0322 | AC59 | HCT 116 | C286(1.26) | LDD0639 | [6] |
| LDCM0323 | AC6 | HCT 116 | C380(0.75); C286(0.78); C407(0.90); C414(0.98) | LDD0640 | [6] |
| LDCM0324 | AC60 | HCT 116 | C286(1.19) | LDD0641 | [6] |
| LDCM0325 | AC61 | HCT 116 | C286(1.20) | LDD0642 | [6] |
| LDCM0326 | AC62 | HCT 116 | C286(1.06) | LDD0643 | [6] |
| LDCM0327 | AC63 | HCT 116 | C286(0.96) | LDD0644 | [6] |
| LDCM0328 | AC64 | HCT 116 | C286(1.00) | LDD0645 | [6] |
| LDCM0329 | AC65 | HCT 116 | C286(1.19) | LDD0646 | [6] |
| LDCM0330 | AC66 | HCT 116 | C286(1.50) | LDD0647 | [6] |
| LDCM0331 | AC67 | HCT 116 | C286(1.18) | LDD0648 | [6] |
| LDCM0332 | AC68 | HCT 116 | C407(0.76); C414(0.92); C286(0.98); C380(1.27) | LDD0649 | [6] |
| LDCM0333 | AC69 | HCT 116 | C407(0.73); C286(0.90); C414(0.94); C380(1.52) | LDD0650 | [6] |
| LDCM0334 | AC7 | HCT 116 | C286(0.82); C380(0.83); C414(0.94); C407(1.12) | LDD0651 | [6] |
| LDCM0335 | AC70 | HCT 116 | C407(0.73); C286(0.96); C414(0.96); C380(1.63) | LDD0652 | [6] |
| LDCM0336 | AC71 | HCT 116 | C407(0.57); C414(0.75); C286(0.88); C380(1.16) | LDD0653 | [6] |
| LDCM0337 | AC72 | HCT 116 | C407(0.56); C414(0.93); C286(0.99); C380(1.65) | LDD0654 | [6] |
| LDCM0338 | AC73 | HCT 116 | C407(0.69); C414(0.97); C286(1.08); C380(1.76) | LDD0655 | [6] |
| LDCM0339 | AC74 | HCT 116 | C407(0.50); C414(0.83); C286(1.03); C380(1.76) | LDD0656 | [6] |
| LDCM0340 | AC75 | HCT 116 | C407(0.62); C286(1.10); C414(1.20); C380(2.15) | LDD0657 | [6] |
| LDCM0341 | AC76 | HCT 116 | C407(0.57); C286(1.01); C414(1.20); C380(1.54) | LDD0658 | [6] |
| LDCM0342 | AC77 | HCT 116 | C407(0.59); C286(1.05); C414(1.17); C380(1.30) | LDD0659 | [6] |
| LDCM0343 | AC78 | HCT 116 | C407(0.59); C286(0.78); C414(0.86); C380(1.27) | LDD0660 | [6] |
| LDCM0344 | AC79 | HCT 116 | C407(0.53); C414(0.87); C286(0.92); C380(1.38) | LDD0661 | [6] |
| LDCM0345 | AC8 | HCT 116 | C286(0.73); C407(0.77); C380(0.78); C414(0.96) | LDD0662 | [6] |
| LDCM0346 | AC80 | HCT 116 | C407(0.58); C286(0.86); C414(0.86); C380(1.30) | LDD0663 | [6] |
| LDCM0347 | AC81 | HCT 116 | C407(0.47); C286(0.90); C414(0.91); C380(1.52) | LDD0664 | [6] |
| LDCM0348 | AC82 | HCT 116 | C407(0.52); C414(0.87); C286(1.24); C380(2.31) | LDD0665 | [6] |
| LDCM0349 | AC83 | HCT 116 | C286(0.74); C414(1.10); C407(1.14); C380(1.35) | LDD0666 | [6] |
| LDCM0350 | AC84 | HCT 116 | C286(0.78); C414(0.97); C407(1.05); C380(1.27) | LDD0667 | [6] |
| LDCM0351 | AC85 | HCT 116 | C286(0.72); C414(0.94); C407(1.11); C380(1.31) | LDD0668 | [6] |
| LDCM0352 | AC86 | HCT 116 | C286(0.93); C414(0.99); C407(1.09); C380(1.11) | LDD0669 | [6] |
| LDCM0353 | AC87 | HCT 116 | C286(0.87); C407(0.96); C380(1.00); C414(1.00) | LDD0670 | [6] |
| LDCM0354 | AC88 | HCT 116 | C286(0.87); C414(0.94); C407(1.05); C380(1.30) | LDD0671 | [6] |
| LDCM0355 | AC89 | HCT 116 | C286(0.80); C407(0.81); C414(1.04); C380(1.38) | LDD0672 | [6] |
| LDCM0357 | AC90 | HCT 116 | C407(0.93); C380(1.04); C414(1.20); C286(1.26) | LDD0674 | [6] |
| LDCM0358 | AC91 | HCT 116 | C407(0.75); C286(0.89); C414(1.27); C380(1.35) | LDD0675 | [6] |
| LDCM0359 | AC92 | HCT 116 | C286(0.79); C407(0.81); C414(1.15); C380(1.38) | LDD0676 | [6] |
| LDCM0360 | AC93 | HCT 116 | C407(0.88); C286(0.88); C414(1.06); C380(1.22) | LDD0677 | [6] |
| LDCM0361 | AC94 | HCT 116 | C407(0.65); C286(0.78); C414(1.08); C380(1.24) | LDD0678 | [6] |
| LDCM0362 | AC95 | HCT 116 | C286(0.80); C407(0.98); C414(1.00); C380(1.19) | LDD0679 | [6] |
| LDCM0363 | AC96 | HCT 116 | C407(0.62); C286(0.72); C380(1.24); C414(1.25) | LDD0680 | [6] |
| LDCM0364 | AC97 | HCT 116 | C286(0.66); C407(0.99); C414(1.23); C380(1.61) | LDD0681 | [6] |
| LDCM0365 | AC98 | HCT 116 | C286(0.45) | LDD0682 | [6] |
| LDCM0366 | AC99 | HCT 116 | C286(0.66) | LDD0683 | [6] |
| LDCM0248 | AKOS034007472 | HCT 116 | C380(0.80); C286(0.91); C407(0.52); C414(1.29) | LDD0565 | [6] |
| LDCM0356 | AKOS034007680 | HCT 116 | C407(0.80); C286(0.83); C380(0.93); C414(0.96) | LDD0673 | [6] |
| LDCM0275 | AKOS034007705 | HCT 116 | C407(0.54); C286(0.77); C414(1.12); C380(1.36) | LDD0592 | [6] |
| LDCM0156 | Aniline | NCI-H1299 | 11.98 | LDD0403 | [1] |
| LDCM0632 | CL-Sc | Hep-G2 | C286(20.00); C7(1.65); C324(1.48) | LDD2227 | [8] |
| LDCM0367 | CL1 | HCT 116 | C286(1.16); C380(1.38) | LDD0684 | [6] |
| LDCM0368 | CL10 | HCT 116 | C286(1.03); C380(1.38) | LDD0685 | [6] |
| LDCM0369 | CL100 | HCT 116 | C407(0.97); C414(0.99); C286(1.12); C380(1.52) | LDD0686 | [6] |
| LDCM0370 | CL101 | HCT 116 | C407(0.66); C286(0.81); C414(1.04); C380(1.08) | LDD0687 | [6] |
| LDCM0371 | CL102 | HCT 116 | C407(0.54); C286(0.86); C380(0.93); C414(1.19) | LDD0688 | [6] |
| LDCM0372 | CL103 | HCT 116 | C407(0.52); C286(0.93); C380(1.00); C414(1.13) | LDD0689 | [6] |
| LDCM0373 | CL104 | HCT 116 | C407(0.57); C286(0.79); C414(0.94); C380(1.01) | LDD0690 | [6] |
| LDCM0374 | CL105 | HCT 116 | C380(0.40); C414(0.85); C286(1.00) | LDD0691 | [6] |
| LDCM0375 | CL106 | HCT 116 | C380(0.35); C414(0.61); C286(0.86) | LDD0692 | [6] |
| LDCM0376 | CL107 | HCT 116 | C380(0.40); C414(0.79); C286(0.88) | LDD0693 | [6] |
| LDCM0377 | CL108 | HCT 116 | C380(0.33); C414(0.75); C286(0.93) | LDD0694 | [6] |
| LDCM0378 | CL109 | HCT 116 | C380(0.42); C286(0.91); C414(0.99) | LDD0695 | [6] |
| LDCM0379 | CL11 | HCT 116 | C286(0.97); C380(1.74) | LDD0696 | [6] |
| LDCM0380 | CL110 | HCT 116 | C380(0.30); C414(0.83); C286(0.93) | LDD0697 | [6] |
| LDCM0381 | CL111 | HCT 116 | C380(0.38); C414(0.92); C286(0.99) | LDD0698 | [6] |
| LDCM0382 | CL112 | HCT 116 | C407(0.85); C286(1.06); C414(1.09) | LDD0699 | [6] |
| LDCM0383 | CL113 | HCT 116 | C407(0.74); C286(0.89); C414(1.15) | LDD0700 | [6] |
| LDCM0384 | CL114 | HCT 116 | C407(0.87); C286(0.90); C414(1.32) | LDD0701 | [6] |
| LDCM0385 | CL115 | HCT 116 | C407(0.76); C286(0.94); C414(1.22) | LDD0702 | [6] |
| LDCM0386 | CL116 | HCT 116 | C407(0.85); C286(0.97); C414(1.18) | LDD0703 | [6] |
| LDCM0387 | CL117 | HCT 116 | C407(0.88); C286(1.00); C414(1.34) | LDD0704 | [6] |
| LDCM0388 | CL118 | HCT 116 | C407(0.60); C286(0.88); C414(1.28) | LDD0705 | [6] |
| LDCM0389 | CL119 | HCT 116 | C407(0.68); C286(0.87); C414(1.19) | LDD0706 | [6] |
| LDCM0390 | CL12 | HCT 116 | C286(1.07); C380(1.34) | LDD0707 | [6] |
| LDCM0391 | CL120 | HCT 116 | C407(0.70); C286(0.80); C414(1.23) | LDD0708 | [6] |
| LDCM0392 | CL121 | HCT 116 | C286(1.09); C414(1.13) | LDD0709 | [6] |
| LDCM0393 | CL122 | HCT 116 | C414(0.99); C286(1.00) | LDD0710 | [6] |
| LDCM0394 | CL123 | HCT 116 | C414(0.98); C286(1.01) | LDD0711 | [6] |
| LDCM0395 | CL124 | HCT 116 | C414(1.17); C286(1.21) | LDD0712 | [6] |
| LDCM0396 | CL125 | HCT 116 | C286(1.21) | LDD0713 | [6] |
| LDCM0397 | CL126 | HCT 116 | C286(1.02) | LDD0714 | [6] |
| LDCM0398 | CL127 | HCT 116 | C286(1.18) | LDD0715 | [6] |
| LDCM0399 | CL128 | HCT 116 | C286(1.31) | LDD0716 | [6] |
| LDCM0400 | CL13 | HCT 116 | C286(0.97); C380(1.27) | LDD0717 | [6] |
| LDCM0401 | CL14 | HCT 116 | C286(1.01); C380(1.11) | LDD0718 | [6] |
| LDCM0402 | CL15 | HCT 116 | C286(0.92); C380(1.72) | LDD0719 | [6] |
| LDCM0403 | CL16 | HCT 116 | C407(0.90); C286(0.94); C380(1.50) | LDD0720 | [6] |
| LDCM0404 | CL17 | HCT 116 | C286(0.86); C407(0.87); C414(1.35); C380(1.36) | LDD0721 | [6] |
| LDCM0405 | CL18 | HCT 116 | C286(0.68); C407(1.01); C380(1.08); C414(1.23) | LDD0722 | [6] |
| LDCM0406 | CL19 | HCT 116 | C286(0.78); C380(0.96); C407(1.07); C414(1.13) | LDD0723 | [6] |
| LDCM0407 | CL2 | HCT 116 | C286(1.08); C380(1.36) | LDD0724 | [6] |
| LDCM0408 | CL20 | HCT 116 | C286(0.93); C380(1.11); C407(1.14); C414(1.36) | LDD0725 | [6] |
| LDCM0409 | CL21 | HCT 116 | C286(0.85); C407(1.15); C414(1.15); C380(1.21) | LDD0726 | [6] |
| LDCM0410 | CL22 | HCT 116 | C414(1.00); C286(1.03); C380(1.29); C407(1.79) | LDD0727 | [6] |
| LDCM0411 | CL23 | HCT 116 | C286(0.87); C414(1.18); C380(1.28); C407(1.35) | LDD0728 | [6] |
| LDCM0412 | CL24 | HCT 116 | C380(1.12); C286(0.85); C407(1.34); C414(1.07) | LDD0729 | [6] |
| LDCM0413 | CL25 | HCT 116 | C380(1.06); C286(0.89); C407(1.21); C414(1.23) | LDD0730 | [6] |
| LDCM0414 | CL26 | HCT 116 | C380(1.49); C286(0.84); C407(0.90); C414(1.11) | LDD0731 | [6] |
| LDCM0415 | CL27 | HCT 116 | C380(1.10); C286(0.97); C407(1.15); C414(1.01) | LDD0732 | [6] |
| LDCM0416 | CL28 | HCT 116 | C380(1.24); C286(0.78); C407(1.06); C414(1.13) | LDD0733 | [6] |
| LDCM0417 | CL29 | HCT 116 | C380(1.12); C286(0.81); C407(1.13); C414(1.18) | LDD0734 | [6] |
| LDCM0418 | CL3 | HCT 116 | C380(1.18); C286(0.93) | LDD0735 | [6] |
| LDCM0419 | CL30 | HCT 116 | C380(1.12); C286(0.84); C407(1.09); C414(0.91) | LDD0736 | [6] |
| LDCM0420 | CL31 | HCT 116 | C380(0.90); C286(0.96); C414(1.13) | LDD0737 | [6] |
| LDCM0421 | CL32 | HCT 116 | C380(1.48); C286(0.90) | LDD0738 | [6] |
| LDCM0422 | CL33 | HCT 116 | C380(2.18); C286(0.66) | LDD0739 | [6] |
| LDCM0423 | CL34 | HCT 116 | C380(1.31); C286(0.92) | LDD0740 | [6] |
| LDCM0424 | CL35 | HCT 116 | C380(0.92); C286(0.90) | LDD0741 | [6] |
| LDCM0425 | CL36 | HCT 116 | C380(1.16); C286(0.95) | LDD0742 | [6] |
| LDCM0426 | CL37 | HCT 116 | C380(0.99); C286(0.91) | LDD0743 | [6] |
| LDCM0428 | CL39 | HCT 116 | C380(2.10); C286(0.99) | LDD0745 | [6] |
| LDCM0429 | CL4 | HCT 116 | C380(1.53); C286(0.94) | LDD0746 | [6] |
| LDCM0430 | CL40 | HCT 116 | C380(1.01); C286(0.86) | LDD0747 | [6] |
| LDCM0431 | CL41 | HCT 116 | C380(1.31); C286(0.94) | LDD0748 | [6] |
| LDCM0432 | CL42 | HCT 116 | C380(1.20); C286(1.03) | LDD0749 | [6] |
| LDCM0433 | CL43 | HCT 116 | C380(1.04); C286(0.95) | LDD0750 | [6] |
| LDCM0434 | CL44 | HCT 116 | C380(0.93); C286(0.89) | LDD0751 | [6] |
| LDCM0435 | CL45 | HCT 116 | C380(0.91); C286(0.99) | LDD0752 | [6] |
| LDCM0436 | CL46 | HCT 116 | C380(1.31); C286(0.91); C414(1.27) | LDD0753 | [6] |
| LDCM0437 | CL47 | HCT 116 | C380(0.80); C286(0.85); C414(1.10) | LDD0754 | [6] |
| LDCM0438 | CL48 | HCT 116 | C380(1.04); C286(0.98); C414(1.17) | LDD0755 | [6] |
| LDCM0439 | CL49 | HCT 116 | C380(0.92); C286(1.23); C414(1.05) | LDD0756 | [6] |
| LDCM0440 | CL5 | HCT 116 | C380(1.04); C286(1.08) | LDD0757 | [6] |
| LDCM0441 | CL50 | HCT 116 | C380(0.96); C286(1.12); C414(1.21) | LDD0758 | [6] |
| LDCM0442 | CL51 | HCT 116 | C380(0.97); C286(0.98); C414(1.15) | LDD0759 | [6] |
| LDCM0443 | CL52 | HCT 116 | C380(1.17); C286(1.02); C414(1.25) | LDD0760 | [6] |
| LDCM0444 | CL53 | HCT 116 | C380(1.02); C286(0.82); C414(1.04) | LDD0761 | [6] |
| LDCM0445 | CL54 | HCT 116 | C380(1.25); C286(1.01); C414(1.12) | LDD0762 | [6] |
| LDCM0446 | CL55 | HCT 116 | C380(1.29); C286(1.06); C414(1.14) | LDD0763 | [6] |
| LDCM0447 | CL56 | HCT 116 | C380(0.93); C286(0.95); C414(0.96) | LDD0764 | [6] |
| LDCM0448 | CL57 | HCT 116 | C380(1.29); C286(1.01); C414(1.01) | LDD0765 | [6] |
| LDCM0449 | CL58 | HCT 116 | C380(1.51); C286(1.13); C414(1.03) | LDD0766 | [6] |
| LDCM0450 | CL59 | HCT 116 | C380(1.16); C286(1.12); C414(0.99) | LDD0767 | [6] |
| LDCM0451 | CL6 | HCT 116 | C380(1.40); C286(0.84) | LDD0768 | [6] |
| LDCM0452 | CL60 | HCT 116 | C380(1.14); C286(1.08); C414(1.12) | LDD0769 | [6] |
| LDCM0453 | CL61 | HCT 116 | C380(0.96); C286(0.86); C407(0.90); C414(0.59) | LDD0770 | [6] |
| LDCM0454 | CL62 | HCT 116 | C380(0.94); C286(0.79); C407(0.91); C414(0.91) | LDD0771 | [6] |
| LDCM0455 | CL63 | HCT 116 | C380(0.89); C286(0.82); C407(0.99); C414(0.83) | LDD0772 | [6] |
| LDCM0456 | CL64 | HCT 116 | C380(0.72); C286(0.87); C407(0.93); C414(0.89) | LDD0773 | [6] |
| LDCM0457 | CL65 | HCT 116 | C380(0.84); C286(0.96); C407(0.76); C414(0.81) | LDD0774 | [6] |
| LDCM0458 | CL66 | HCT 116 | C380(0.83); C286(0.92); C407(0.98); C414(0.75) | LDD0775 | [6] |
| LDCM0459 | CL67 | HCT 116 | C380(0.86); C286(0.92); C407(0.81); C414(0.77) | LDD0776 | [6] |
| LDCM0460 | CL68 | HCT 116 | C380(0.67); C286(0.87); C407(0.81); C414(0.76) | LDD0777 | [6] |
| LDCM0461 | CL69 | HCT 116 | C380(0.94); C286(0.94); C407(0.75); C414(0.77) | LDD0778 | [6] |
| LDCM0462 | CL7 | HCT 116 | C380(1.05); C286(1.00) | LDD0779 | [6] |
| LDCM0463 | CL70 | HCT 116 | C380(0.85); C286(0.85); C407(0.71); C414(0.70) | LDD0780 | [6] |
| LDCM0464 | CL71 | HCT 116 | C380(0.88); C286(0.88); C407(0.80); C414(0.87) | LDD0781 | [6] |
| LDCM0465 | CL72 | HCT 116 | C380(0.84); C286(0.89); C407(0.74); C414(0.92) | LDD0782 | [6] |
| LDCM0466 | CL73 | HCT 116 | C380(0.81); C286(0.84); C407(0.75); C414(0.80) | LDD0783 | [6] |
| LDCM0467 | CL74 | HCT 116 | C380(0.97); C286(0.88); C407(0.77); C414(0.84) | LDD0784 | [6] |
| LDCM0469 | CL76 | HCT 116 | C407(0.76) | LDD0786 | [6] |
| LDCM0470 | CL77 | HCT 116 | C407(0.78) | LDD0787 | [6] |
| LDCM0471 | CL78 | HCT 116 | C407(0.69) | LDD0788 | [6] |
| LDCM0472 | CL79 | HCT 116 | C407(0.72) | LDD0789 | [6] |
| LDCM0473 | CL8 | HCT 116 | C380(5.47); C286(0.97) | LDD0790 | [6] |
| LDCM0474 | CL80 | HCT 116 | C407(0.55) | LDD0791 | [6] |
| LDCM0475 | CL81 | HCT 116 | C407(0.57) | LDD0792 | [6] |
| LDCM0476 | CL82 | HCT 116 | C407(0.46) | LDD0793 | [6] |
| LDCM0477 | CL83 | HCT 116 | C407(0.49) | LDD0794 | [6] |
| LDCM0478 | CL84 | HCT 116 | C407(0.54) | LDD0795 | [6] |
| LDCM0479 | CL85 | HCT 116 | C407(0.54) | LDD0796 | [6] |
| LDCM0480 | CL86 | HCT 116 | C407(0.51) | LDD0797 | [6] |
| LDCM0481 | CL87 | HCT 116 | C407(0.46) | LDD0798 | [6] |
| LDCM0482 | CL88 | HCT 116 | C407(0.51) | LDD0799 | [6] |
| LDCM0483 | CL89 | HCT 116 | C407(0.55) | LDD0800 | [6] |
| LDCM0484 | CL9 | HCT 116 | C380(1.76); C286(1.00) | LDD0801 | [6] |
| LDCM0485 | CL90 | HCT 116 | C407(0.53) | LDD0802 | [6] |
| LDCM0486 | CL91 | HCT 116 | C380(1.12); C286(1.00); C407(0.98); C414(0.83) | LDD0803 | [6] |
| LDCM0487 | CL92 | HCT 116 | C380(0.88); C286(1.11); C407(0.90); C414(0.88) | LDD0804 | [6] |
| LDCM0488 | CL93 | HCT 116 | C380(1.12); C286(1.26); C407(0.77); C414(0.88) | LDD0805 | [6] |
| LDCM0489 | CL94 | HCT 116 | C380(1.29); C286(1.02); C407(1.01); C414(0.94) | LDD0806 | [6] |
| LDCM0490 | CL95 | HCT 116 | C380(1.75); C286(1.10); C407(1.07); C414(1.22) | LDD0807 | [6] |
| LDCM0491 | CL96 | HCT 116 | C380(1.58); C286(1.05); C407(0.96); C414(0.84) | LDD0808 | [6] |
| LDCM0492 | CL97 | HCT 116 | C380(1.18); C286(1.03); C407(0.77); C414(0.94) | LDD0809 | [6] |
| LDCM0493 | CL98 | HCT 116 | C380(1.05); C286(1.09); C407(1.03); C414(1.07) | LDD0810 | [6] |
| LDCM0494 | CL99 | HCT 116 | C380(1.57); C286(1.18); C407(0.97); C414(0.83) | LDD0811 | [6] |
| LDCM0634 | CY-0357 | Hep-G2 | C414(0.25) | LDD2228 | [8] |
| LDCM0097 | Dasatinib | K562 | N.A. | LDD0364 | [13] |
| LDCM0495 | E2913 | HEK-293T | C380(1.19) | LDD1698 | [14] |
| LDCM0625 | F8 | Ramos | C286(0.67) | LDD2187 | [15] |
| LDCM0576 | Fragment14 | Ramos | C286(0.77) | LDD2193 | [15] |
| LDCM0580 | Fragment21 | Ramos | C414(1.03) | LDD2195 | [15] |
| LDCM0578 | Fragment27 | Ramos | C414(1.57) | LDD2197 | [15] |
| LDCM0586 | Fragment28 | Ramos | C414(0.58) | LDD2198 | [15] |
| LDCM0588 | Fragment30 | Ramos | C414(1.58) | LDD2199 | [15] |
| LDCM0589 | Fragment31 | Ramos | C414(1.28) | LDD2200 | [15] |
| LDCM0468 | Fragment33 | HCT 116 | C380(0.82); C286(0.82); C407(0.81); C414(0.70) | LDD0785 | [6] |
| LDCM0596 | Fragment38 | Ramos | C414(1.73) | LDD2203 | [15] |
| LDCM0566 | Fragment4 | Ramos | C286(1.52) | LDD2184 | [15] |
| LDCM0427 | Fragment51 | HCT 116 | C380(0.99); C286(0.91) | LDD0744 | [6] |
| LDCM0614 | Fragment56 | Ramos | C414(1.28) | LDD2205 | [15] |
| LDCM0569 | Fragment7 | Ramos | C286(0.74) | LDD2186 | [15] |
| LDCM0179 | JZ128 | PC-3 | C324(0.00); C414(0.00); C346(0.00) | LDD0462 | [3] |
| LDCM0022 | KB02 | HCT 116 | C380(5.52) | LDD0080 | [6] |
| LDCM0023 | KB03 | HCT 116 | C380(2.13) | LDD0081 | [6] |
| LDCM0024 | KB05 | HCT 116 | C380(3.91) | LDD0082 | [6] |
| LDCM0032 | Oleacein | HEK-293T | 14.82 | LDD0184 | [4] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Other
The Drug(s) Related To This Target
Approved
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Fostamatinib | Small molecular drug | DB12010 | |||
Phase 1
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Isis-crp | Antisense drug | D06CIJ | |||
Investigative
References
