Details of the Target
General Information of Target
| Target ID | LDTP11732 | |||||
|---|---|---|---|---|---|---|
| Target Name | Magnesium transporter protein 1 (MAGT1) | |||||
| Gene Name | MAGT1 | |||||
| Gene ID | 84061 | |||||
| Synonyms |
IAG2; Magnesium transporter protein 1; MagT1; Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit MAGT1; Oligosaccharyl transferase subunit MAGT1; Implantation-associated protein; IAP
|
|||||
| 3D Structure | ||||||
| Sequence |
MAQAHRTPQPRAAPSQPRVFKLVLLGSGSVGKSSLALRYVKNDFKSILPTVGCAFFTKVV
DVGATSLKLEIWDTAGQEKYHSVCHLYFRGANAALLVYDITRKDSFLKAQQWLKDLEEEL HPGEVLVMLVGNKTDLSQEREVTFQEGKEFADSQKLLFMETSAKLNHQVSEVFNTVAQEL LQRSDEEGQALRGDAAVALNKGPARQAKCCAH |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
OST3/OST6 family
|
|||||
| Subcellular location |
Cell membrane
|
|||||
| Function |
Accessory component of the STT3B-containing form of the N-oligosaccharyl transferase (OST) complex which catalyzes the transfer of a high mannose oligosaccharide from a lipid-linked oligosaccharide donor to an asparagine residue within an Asn-X-Ser/Thr consensus motif in nascent polypeptide chains. Involved in N-glycosylation of STT3B-dependent substrates. Specifically required for the glycosylation of a subset of acceptor sites that are near cysteine residues; in this function seems to act redundantly with TUSC3. In its oxidized form proposed to form transient mixed disulfides with a glycoprotein substrate to facilitate access of STT3B to the unmodified acceptor site. Has also oxidoreductase-independent functions in the STT3B-containing OST complex possibly involving substrate recognition.; May be involved in Mg(2+) transport in epithelial cells.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
TH211 Probe Info |
 |
Y106(15.15) | LDD0260 | [1] | |
|
C-Sul Probe Info |
 |
6.35 | LDD0066 | [2] | |
|
STPyne Probe Info |
 |
K50(7.90); K60(4.32); K66(10.00) | LDD0277 | [3] | |
|
OPA-S-S-alkyne Probe Info |
 |
K66(4.14) | LDD3494 | [4] | |
|
DA-P3 Probe Info |
 |
4.40 | LDD0179 | [5] | |
|
IPM Probe Info |
 |
C87(1.37) | LDD1701 | [6] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [7] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [8] | |
|
AOyne Probe Info |
 |
7.20 | LDD0443 | [9] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C040 Probe Info |
 |
9.71 | LDD1740 | [10] | |
|
C092 Probe Info |
 |
17.63 | LDD1783 | [10] | |
|
C094 Probe Info |
 |
33.59 | LDD1785 | [10] | |
|
C134 Probe Info |
 |
20.97 | LDD1816 | [10] | |
|
C187 Probe Info |
 |
15.78 | LDD1865 | [10] | |
|
C218 Probe Info |
 |
12.13 | LDD1892 | [10] | |
|
C220 Probe Info |
 |
12.73 | LDD1894 | [10] | |
|
C228 Probe Info |
 |
15.45 | LDD1901 | [10] | |
|
C231 Probe Info |
 |
19.70 | LDD1904 | [10] | |
|
C232 Probe Info |
 |
40.79 | LDD1905 | [10] | |
|
C233 Probe Info |
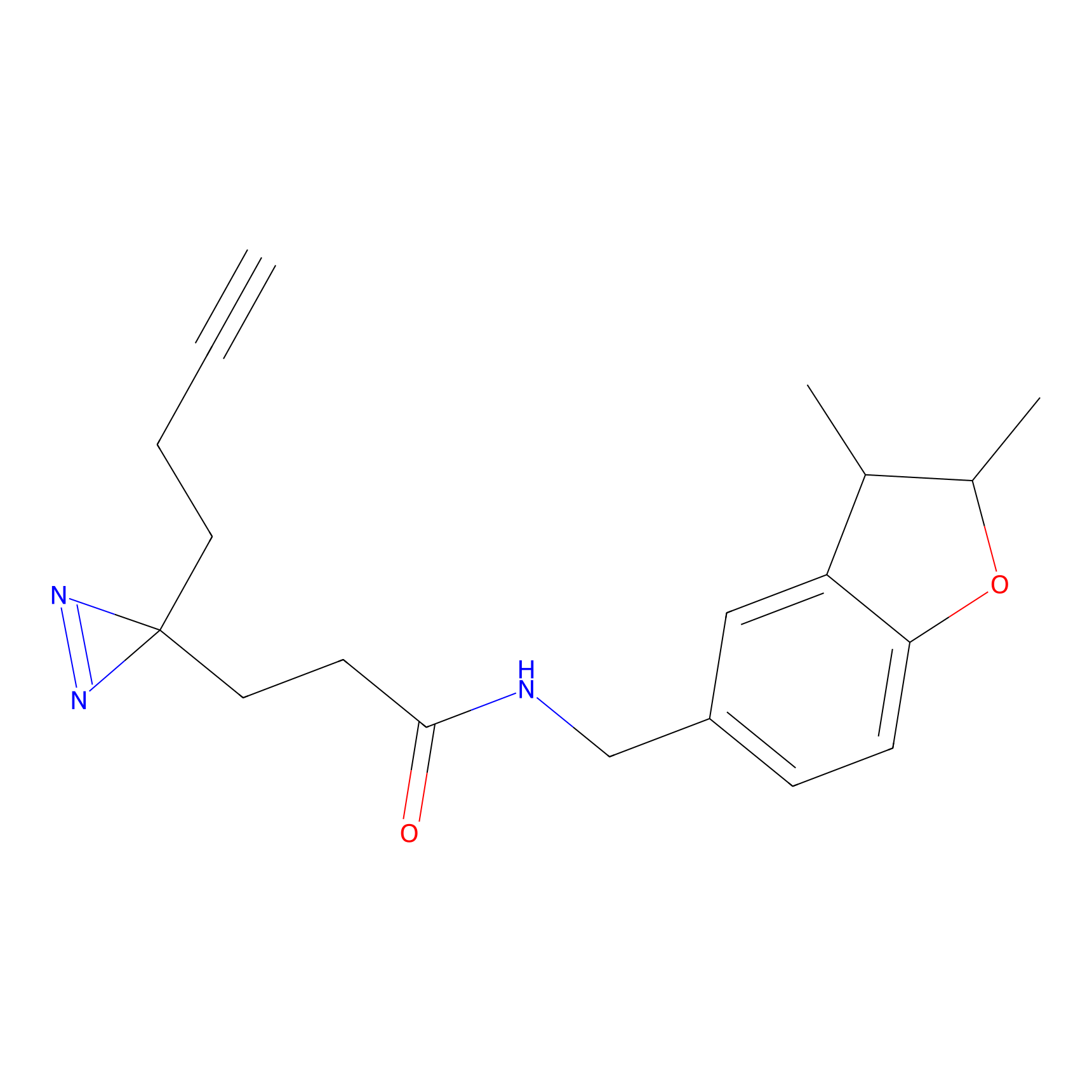 |
9.58 | LDD1906 | [10] | |
|
C234 Probe Info |
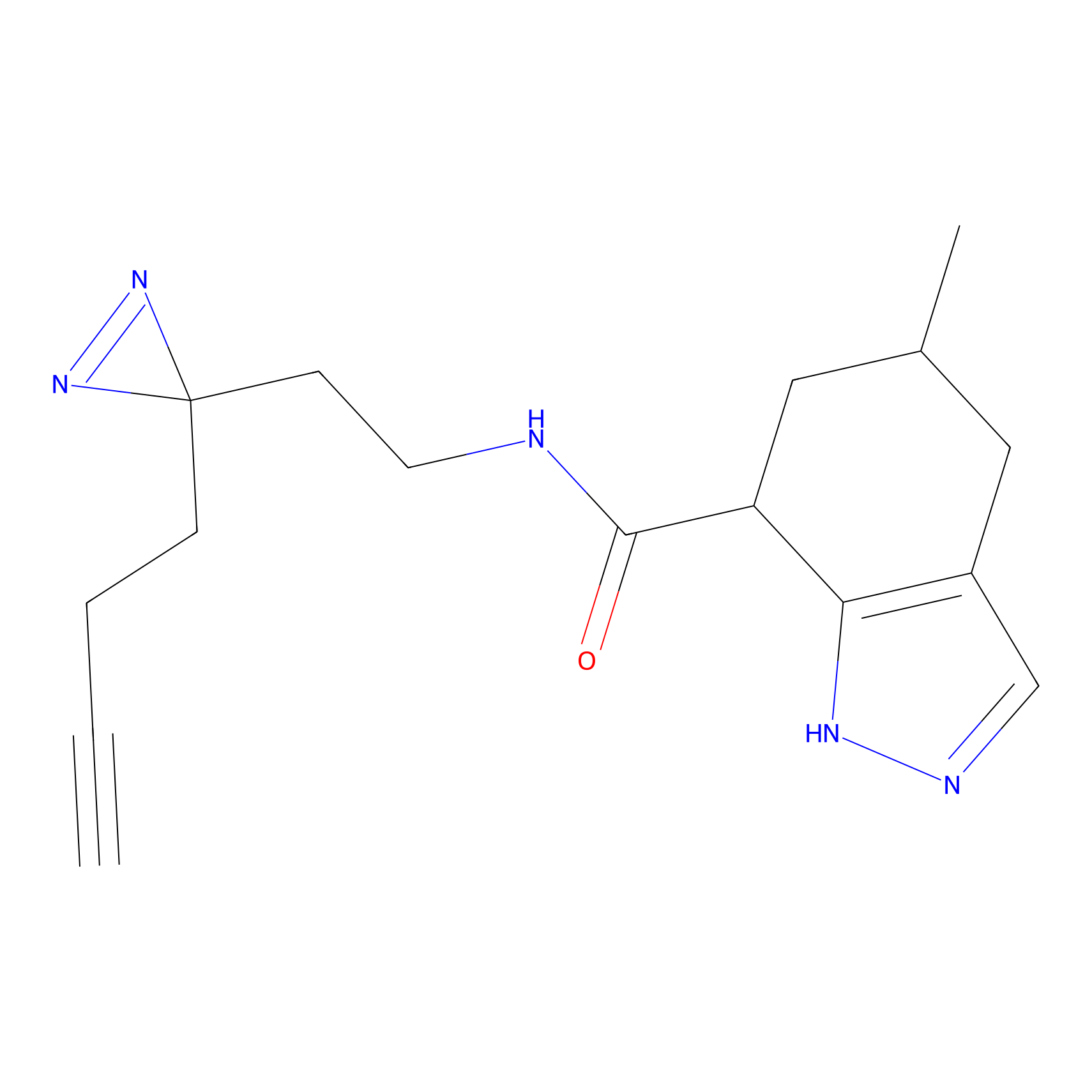 |
6.32 | LDD1907 | [10] | |
|
C235 Probe Info |
 |
26.72 | LDD1908 | [10] | |
|
C388 Probe Info |
 |
50.56 | LDD2047 | [10] | |
|
FFF probe11 Probe Info |
 |
20.00 | LDD0471 | [11] | |
|
FFF probe14 Probe Info |
 |
20.00 | LDD0477 | [11] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0136 | [12] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0139 | [12] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [13] | |
|
OEA-DA Probe Info |
 |
10.40 | LDD0046 | [14] | |
Competitor(s) Related to This Target
References
