Details of the Target
General Information of Target
| Target ID | LDTP03519 | |||||
|---|---|---|---|---|---|---|
| Target Name | UMP-CMP kinase (CMPK1) | |||||
| Gene Name | CMPK1 | |||||
| Gene ID | 51727 | |||||
| Synonyms |
CMK; CMPK; UCK; UMK; UMPK; UMP-CMP kinase; EC 2.7.4.14; Deoxycytidylate kinase; CK; dCMP kinase; Nucleoside-diphosphate kinase; EC 2.7.4.6; Uridine monophosphate/cytidine monophosphate kinase; UMP/CMP kinase; UMP/CMPK
|
|||||
| 3D Structure | ||||||
| Sequence |
MKPLVVFVLGGPGAGKGTQCARIVEKYGYTHLSAGELLRDERKNPDSQYGELIEKYIKEG
KIVPVEITISLLKREMDQTMAANAQKNKFLIDGFPRNQDNLQGWNKTMDGKADVSFVLFF DCNNEICIERCLERGKSSGRSDDNRESLEKRIQTYLQSTKPIIDLYEEMGKVKKIDASKS VDEVFDEVVQIFDKEG |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Adenylate kinase family, UMP-CMP kinase subfamily
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
Catalyzes the phosphorylation of pyrimidine nucleoside monophosphates at the expense of ATP. Plays an important role in de novo pyrimidine nucleotide biosynthesis. Has preference for UMP and CMP as phosphate acceptors. Also displays broad nucleoside diphosphate kinase activity. {|HAMAP-Rule:MF_03172}.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
AZ-9 Probe Info |
 |
4.29 | LDD0393 | [2] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [3] | |
|
STPyne Probe Info |
 |
K106(1.15); K2(9.22); K43(6.08); K55(7.01) | LDD0277 | [4] | |
|
Probe 1 Probe Info |
 |
Y27(7.19); Y49(13.37) | LDD3495 | [5] | |
|
BTD Probe Info |
 |
C20(1.01) | LDD1700 | [6] | |
|
Jackson_1 Probe Info |
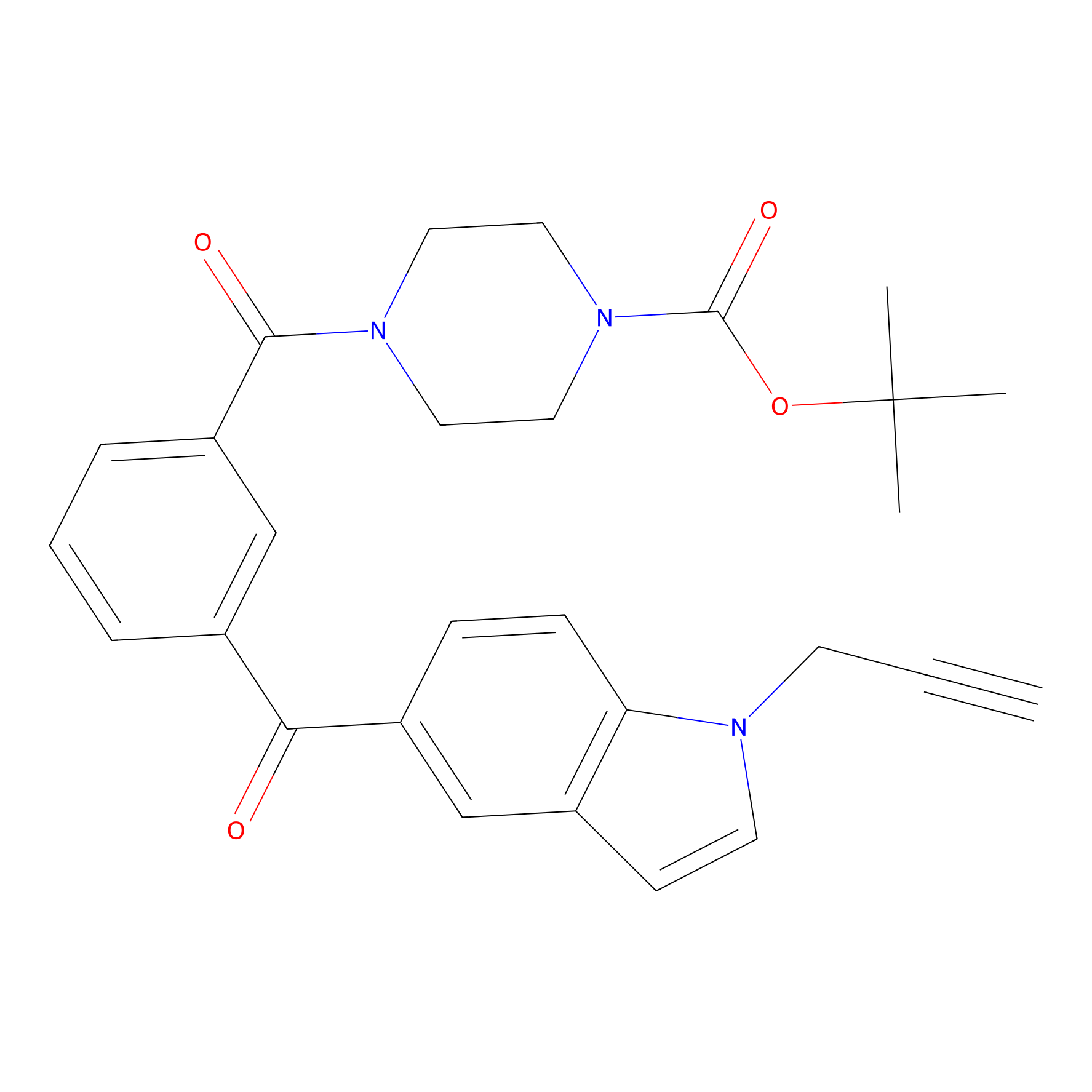 |
10.56 | LDD0120 | [7] | |
|
DBIA Probe Info |
 |
C20(332.45) | LDD0209 | [8] | |
|
HHS-482 Probe Info |
 |
Y49(0.91) | LDD0285 | [9] | |
|
HHS-475 Probe Info |
 |
Y49(0.87); Y155(0.99) | LDD0264 | [10] | |
|
HHS-465 Probe Info |
 |
Y49(5.18) | LDD2237 | [11] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0226 | [12] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [13] | |
|
ATP probe Probe Info |
 |
K16(0.00); K43(0.00) | LDD0199 | [14] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0162 | [15] | |
|
ATP probe Probe Info |
 |
K16(0.00); K2(0.00) | LDD0035 | [16] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [17] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [17] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [18] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [19] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [20] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [21] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FFF probe14 Probe Info |
 |
15.57 | LDD0477 | [22] | |
|
JN0003 Probe Info |
 |
20.00 | LDD0469 | [22] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0137 | [23] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C20(0.67) | LDD2142 | [6] |
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C20(0.89) | LDD2112 | [6] |
| LDCM0502 | 1-(Cyanoacetyl)piperidine | MDA-MB-231 | C20(0.52) | LDD2095 | [6] |
| LDCM0537 | 2-Cyano-N,N-dimethylacetamide | MDA-MB-231 | C20(0.87) | LDD2130 | [6] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C20(0.60) | LDD2117 | [6] |
| LDCM0538 | 4-(Cyanoacetyl)morpholine | MDA-MB-231 | C20(1.21) | LDD2131 | [6] |
| LDCM0214 | AC1 | PaTu 8988t | C122(1.77); C127(1.77) | LDD1093 | [24] |
| LDCM0215 | AC10 | PaTu 8988t | C122(1.47); C127(1.47) | LDD1094 | [24] |
| LDCM0216 | AC100 | PaTu 8988t | C122(1.44); C127(1.44) | LDD1095 | [24] |
| LDCM0217 | AC101 | PaTu 8988t | C122(1.60); C127(1.60) | LDD1096 | [24] |
| LDCM0218 | AC102 | PaTu 8988t | C122(1.03); C127(1.03) | LDD1097 | [24] |
| LDCM0219 | AC103 | PaTu 8988t | C122(1.83); C127(1.83) | LDD1098 | [24] |
| LDCM0220 | AC104 | PaTu 8988t | C122(0.92); C127(0.92) | LDD1099 | [24] |
| LDCM0221 | AC105 | PaTu 8988t | C122(1.18); C127(1.18) | LDD1100 | [24] |
| LDCM0222 | AC106 | PaTu 8988t | C122(1.48); C127(1.48) | LDD1101 | [24] |
| LDCM0223 | AC107 | PaTu 8988t | C122(2.10); C127(2.10) | LDD1102 | [24] |
| LDCM0224 | AC108 | PaTu 8988t | C122(1.20); C127(1.20) | LDD1103 | [24] |
| LDCM0225 | AC109 | PaTu 8988t | C122(1.13); C127(1.13) | LDD1104 | [24] |
| LDCM0226 | AC11 | PaTu 8988t | C122(1.34); C127(1.34) | LDD1105 | [24] |
| LDCM0227 | AC110 | PaTu 8988t | C122(1.90); C127(1.90) | LDD1106 | [24] |
| LDCM0228 | AC111 | PaTu 8988t | C122(1.42); C127(1.42) | LDD1107 | [24] |
| LDCM0229 | AC112 | PaTu 8988t | C122(1.27); C127(1.27) | LDD1108 | [24] |
| LDCM0230 | AC113 | PaTu 8988t | C122(0.70); C127(0.70) | LDD1109 | [24] |
| LDCM0231 | AC114 | PaTu 8988t | C122(0.67); C127(0.67) | LDD1110 | [24] |
| LDCM0232 | AC115 | PaTu 8988t | C122(0.81); C127(0.81) | LDD1111 | [24] |
| LDCM0233 | AC116 | PaTu 8988t | C122(0.85); C127(0.85) | LDD1112 | [24] |
| LDCM0234 | AC117 | PaTu 8988t | C122(0.65); C127(0.65) | LDD1113 | [24] |
| LDCM0235 | AC118 | PaTu 8988t | C122(0.92); C127(0.92) | LDD1114 | [24] |
| LDCM0236 | AC119 | PaTu 8988t | C122(0.82); C127(0.82) | LDD1115 | [24] |
| LDCM0237 | AC12 | PaTu 8988t | C122(1.36); C127(1.36) | LDD1116 | [24] |
| LDCM0238 | AC120 | PaTu 8988t | C122(0.67); C127(0.67) | LDD1117 | [24] |
| LDCM0239 | AC121 | PaTu 8988t | C122(0.70); C127(0.70) | LDD1118 | [24] |
| LDCM0240 | AC122 | PaTu 8988t | C122(1.03); C127(1.03) | LDD1119 | [24] |
| LDCM0241 | AC123 | PaTu 8988t | C122(1.28); C127(1.28) | LDD1120 | [24] |
| LDCM0242 | AC124 | PaTu 8988t | C122(1.72); C127(1.72) | LDD1121 | [24] |
| LDCM0243 | AC125 | PaTu 8988t | C122(1.65); C127(1.65) | LDD1122 | [24] |
| LDCM0244 | AC126 | PaTu 8988t | C122(1.11); C127(1.11) | LDD1123 | [24] |
| LDCM0245 | AC127 | PaTu 8988t | C122(1.09); C127(1.09) | LDD1124 | [24] |
| LDCM0246 | AC128 | PaTu 8988t | C122(0.41); C127(0.41) | LDD1125 | [24] |
| LDCM0247 | AC129 | PaTu 8988t | C122(0.63); C127(0.63) | LDD1126 | [24] |
| LDCM0249 | AC130 | PaTu 8988t | C122(0.58); C127(0.58) | LDD1128 | [24] |
| LDCM0250 | AC131 | PaTu 8988t | C122(0.61); C127(0.61) | LDD1129 | [24] |
| LDCM0251 | AC132 | PaTu 8988t | C122(0.62); C127(0.62) | LDD1130 | [24] |
| LDCM0252 | AC133 | PaTu 8988t | C122(0.61); C127(0.61) | LDD1131 | [24] |
| LDCM0253 | AC134 | PaTu 8988t | C122(0.60); C127(0.60) | LDD1132 | [24] |
| LDCM0254 | AC135 | PaTu 8988t | C122(0.56); C127(0.56) | LDD1133 | [24] |
| LDCM0255 | AC136 | PaTu 8988t | C122(0.42); C127(0.42) | LDD1134 | [24] |
| LDCM0256 | AC137 | PaTu 8988t | C122(0.71); C127(0.71) | LDD1135 | [24] |
| LDCM0257 | AC138 | PaTu 8988t | C122(0.69); C127(0.69) | LDD1136 | [24] |
| LDCM0258 | AC139 | PaTu 8988t | C122(0.49); C127(0.49) | LDD1137 | [24] |
| LDCM0259 | AC14 | PaTu 8988t | C122(1.20); C127(1.20) | LDD1138 | [24] |
| LDCM0260 | AC140 | PaTu 8988t | C122(0.77); C127(0.77) | LDD1139 | [24] |
| LDCM0261 | AC141 | PaTu 8988t | C122(0.77); C127(0.77) | LDD1140 | [24] |
| LDCM0262 | AC142 | PaTu 8988t | C122(0.73); C127(0.73) | LDD1141 | [24] |
| LDCM0263 | AC143 | PaTu 8988t | C122(1.00); C127(1.00) | LDD1142 | [24] |
| LDCM0264 | AC144 | PaTu 8988t | C122(1.09); C127(1.09) | LDD1143 | [24] |
| LDCM0265 | AC145 | PaTu 8988t | C122(1.30); C127(1.30) | LDD1144 | [24] |
| LDCM0266 | AC146 | PaTu 8988t | C122(1.40); C127(1.40) | LDD1145 | [24] |
| LDCM0267 | AC147 | PaTu 8988t | C122(1.22); C127(1.22) | LDD1146 | [24] |
| LDCM0268 | AC148 | PaTu 8988t | C122(1.37); C127(1.37) | LDD1147 | [24] |
| LDCM0269 | AC149 | PaTu 8988t | C122(1.33); C127(1.33) | LDD1148 | [24] |
| LDCM0270 | AC15 | PaTu 8988t | C122(1.14); C127(1.14) | LDD1149 | [24] |
| LDCM0271 | AC150 | PaTu 8988t | C122(1.53); C127(1.53) | LDD1150 | [24] |
| LDCM0272 | AC151 | PaTu 8988t | C122(2.17); C127(2.17) | LDD1151 | [24] |
| LDCM0273 | AC152 | PaTu 8988t | C122(1.36); C127(1.36) | LDD1152 | [24] |
| LDCM0274 | AC153 | PaTu 8988t | C122(1.35); C127(1.35) | LDD1153 | [24] |
| LDCM0621 | AC154 | PaTu 8988t | C122(1.91); C127(1.91) | LDD2166 | [24] |
| LDCM0622 | AC155 | PaTu 8988t | C122(1.64); C127(1.64) | LDD2167 | [24] |
| LDCM0623 | AC156 | PaTu 8988t | C122(1.42); C127(1.42) | LDD2168 | [24] |
| LDCM0624 | AC157 | PaTu 8988t | C122(1.69); C127(1.69) | LDD2169 | [24] |
| LDCM0278 | AC19 | HEK-293T | C122(0.99) | LDD1517 | [25] |
| LDCM0279 | AC2 | PaTu 8988t | C122(1.52); C127(1.52) | LDD1158 | [24] |
| LDCM0283 | AC23 | HEK-293T | C122(0.80) | LDD1522 | [25] |
| LDCM0287 | AC27 | HEK-293T | C122(0.92) | LDD1526 | [25] |
| LDCM0290 | AC3 | PaTu 8988t | C122(1.25); C127(1.25) | LDD1169 | [24] |
| LDCM0292 | AC31 | HEK-293T | C122(1.07) | LDD1531 | [25] |
| LDCM0296 | AC35 | PaTu 8988t | C122(1.24); C127(1.24) | LDD1175 | [24] |
| LDCM0297 | AC36 | PaTu 8988t | C122(1.37); C127(1.37) | LDD1176 | [24] |
| LDCM0298 | AC37 | PaTu 8988t | C122(1.36); C127(1.36) | LDD1177 | [24] |
| LDCM0299 | AC38 | PaTu 8988t | C122(1.44); C127(1.44) | LDD1178 | [24] |
| LDCM0300 | AC39 | PaTu 8988t | C122(1.91); C127(1.91) | LDD1179 | [24] |
| LDCM0301 | AC4 | PaTu 8988t | C122(1.26); C127(1.26) | LDD1180 | [24] |
| LDCM0302 | AC40 | PaTu 8988t | C122(2.12); C127(2.12) | LDD1181 | [24] |
| LDCM0303 | AC41 | PaTu 8988t | C122(1.60); C127(1.60) | LDD1182 | [24] |
| LDCM0304 | AC42 | PaTu 8988t | C122(1.45); C127(1.45) | LDD1183 | [24] |
| LDCM0305 | AC43 | PaTu 8988t | C122(1.59); C127(1.59) | LDD1184 | [24] |
| LDCM0306 | AC44 | PaTu 8988t | C122(2.43); C127(2.43) | LDD1185 | [24] |
| LDCM0307 | AC45 | PaTu 8988t | C122(1.59); C127(1.59) | LDD1186 | [24] |
| LDCM0309 | AC47 | HEK-293T | C122(0.86) | LDD1548 | [25] |
| LDCM0312 | AC5 | PaTu 8988t | C122(2.12); C127(2.12) | LDD1191 | [24] |
| LDCM0314 | AC51 | HEK-293T | C122(0.98) | LDD1553 | [25] |
| LDCM0318 | AC55 | HEK-293T | C122(0.77) | LDD1557 | [25] |
| LDCM0320 | AC57 | PaTu 8988t | C122(0.81); C127(0.81) | LDD1199 | [24] |
| LDCM0321 | AC58 | PaTu 8988t | C122(0.90); C127(0.90) | LDD1200 | [24] |
| LDCM0322 | AC59 | PaTu 8988t | C122(1.10); C127(1.10) | LDD1201 | [24] |
| LDCM0323 | AC6 | PaTu 8988t | C122(1.17); C127(1.17) | LDD1202 | [24] |
| LDCM0324 | AC60 | PaTu 8988t | C122(0.80); C127(0.80) | LDD1203 | [24] |
| LDCM0325 | AC61 | PaTu 8988t | C122(0.85); C127(0.85) | LDD1204 | [24] |
| LDCM0326 | AC62 | PaTu 8988t | C122(1.02); C127(1.02) | LDD1205 | [24] |
| LDCM0327 | AC63 | PaTu 8988t | C122(0.82); C127(0.82) | LDD1206 | [24] |
| LDCM0328 | AC64 | PaTu 8988t | C122(0.57); C127(0.57) | LDD1207 | [24] |
| LDCM0329 | AC65 | PaTu 8988t | C122(1.15); C127(1.15) | LDD1208 | [24] |
| LDCM0330 | AC66 | PaTu 8988t | C122(0.80); C127(0.80) | LDD1209 | [24] |
| LDCM0331 | AC67 | PaTu 8988t | C122(0.74); C127(0.74) | LDD1210 | [24] |
| LDCM0332 | AC68 | PaTu 8988t | C122(0.91); C127(0.91) | LDD1211 | [24] |
| LDCM0333 | AC69 | PaTu 8988t | C122(0.96); C127(0.96) | LDD1212 | [24] |
| LDCM0334 | AC7 | PaTu 8988t | C122(1.11); C127(1.11) | LDD1213 | [24] |
| LDCM0335 | AC70 | PaTu 8988t | C122(1.65); C127(1.65) | LDD1214 | [24] |
| LDCM0336 | AC71 | PaTu 8988t | C122(1.02); C127(1.02) | LDD1215 | [24] |
| LDCM0337 | AC72 | PaTu 8988t | C122(0.90); C127(0.90) | LDD1216 | [24] |
| LDCM0338 | AC73 | PaTu 8988t | C122(0.83); C127(0.83) | LDD1217 | [24] |
| LDCM0339 | AC74 | PaTu 8988t | C122(0.94); C127(0.94) | LDD1218 | [24] |
| LDCM0340 | AC75 | PaTu 8988t | C122(1.02); C127(1.02) | LDD1219 | [24] |
| LDCM0341 | AC76 | PaTu 8988t | C122(0.78); C127(0.78) | LDD1220 | [24] |
| LDCM0342 | AC77 | PaTu 8988t | C122(1.10); C127(1.10) | LDD1221 | [24] |
| LDCM0343 | AC78 | PaTu 8988t | C122(0.90); C127(0.90) | LDD1222 | [24] |
| LDCM0344 | AC79 | PaTu 8988t | C122(0.84); C127(0.84) | LDD1223 | [24] |
| LDCM0345 | AC8 | PaTu 8988t | C122(1.18); C127(1.18) | LDD1224 | [24] |
| LDCM0346 | AC80 | PaTu 8988t | C122(0.87); C127(0.87) | LDD1225 | [24] |
| LDCM0347 | AC81 | PaTu 8988t | C122(0.88); C127(0.88) | LDD1226 | [24] |
| LDCM0348 | AC82 | PaTu 8988t | C122(1.01); C127(1.01) | LDD1227 | [24] |
| LDCM0365 | AC98 | PaTu 8988t | C122(1.25); C127(1.25) | LDD1244 | [24] |
| LDCM0366 | AC99 | PaTu 8988t | C122(0.89); C127(0.89) | LDD1245 | [24] |
| LDCM0545 | Acetamide | MDA-MB-231 | C20(0.26) | LDD2138 | [6] |
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C20(0.71) | LDD2113 | [6] |
| LDCM0248 | AKOS034007472 | PaTu 8988t | C122(1.40); C127(1.40) | LDD1127 | [24] |
| LDCM0356 | AKOS034007680 | PaTu 8988t | C122(1.34); C127(1.34) | LDD1235 | [24] |
| LDCM0275 | AKOS034007705 | PaTu 8988t | C122(1.25); C127(1.25) | LDD1154 | [24] |
| LDCM0156 | Aniline | NCI-H1299 | 13.26 | LDD0403 | [1] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C20(0.33) | LDD2091 | [6] |
| LDCM0070 | Cisar_cp37 | MDA-MB-231 | 10.56 | LDD0120 | [7] |
| LDCM0632 | CL-Sc | Hep-G2 | C20(0.91); C20(0.72) | LDD2227 | [21] |
| LDCM0369 | CL100 | PaTu 8988t | C122(1.23); C127(1.23) | LDD1248 | [24] |
| LDCM0370 | CL101 | PaTu 8988t | C122(1.14); C127(1.14) | LDD1249 | [24] |
| LDCM0371 | CL102 | PaTu 8988t | C122(1.39); C127(1.39) | LDD1250 | [24] |
| LDCM0372 | CL103 | PaTu 8988t | C122(1.88); C127(1.88) | LDD1251 | [24] |
| LDCM0373 | CL104 | PaTu 8988t | C122(1.50); C127(1.50) | LDD1252 | [24] |
| LDCM0379 | CL11 | HEK-293T | C122(0.92) | LDD1583 | [25] |
| LDCM0387 | CL117 | PaTu 8988t | C122(1.30); C127(1.30) | LDD1266 | [24] |
| LDCM0388 | CL118 | PaTu 8988t | C122(1.52); C127(1.52) | LDD1267 | [24] |
| LDCM0389 | CL119 | PaTu 8988t | C122(1.51); C127(1.51) | LDD1268 | [24] |
| LDCM0391 | CL120 | PaTu 8988t | C122(1.52); C127(1.52) | LDD1270 | [24] |
| LDCM0396 | CL125 | PaTu 8988t | C122(0.89); C127(0.89) | LDD1275 | [24] |
| LDCM0397 | CL126 | PaTu 8988t | C122(0.82); C127(0.82) | LDD1276 | [24] |
| LDCM0398 | CL127 | PaTu 8988t | C122(1.06); C127(1.06) | LDD1277 | [24] |
| LDCM0399 | CL128 | PaTu 8988t | C122(0.75); C127(0.75) | LDD1278 | [24] |
| LDCM0403 | CL16 | PaTu 8988t | C122(1.35); C127(1.35) | LDD1282 | [24] |
| LDCM0404 | CL17 | PaTu 8988t | C122(1.39); C127(1.39) | LDD1283 | [24] |
| LDCM0405 | CL18 | PaTu 8988t | C122(1.24); C127(1.24) | LDD1284 | [24] |
| LDCM0406 | CL19 | PaTu 8988t | C122(1.72); C127(1.72) | LDD1285 | [24] |
| LDCM0408 | CL20 | PaTu 8988t | C122(2.67); C127(2.67) | LDD1287 | [24] |
| LDCM0409 | CL21 | PaTu 8988t | C122(2.02); C127(2.02) | LDD1288 | [24] |
| LDCM0410 | CL22 | PaTu 8988t | C122(2.05); C127(2.05) | LDD1289 | [24] |
| LDCM0411 | CL23 | PaTu 8988t | C122(1.67); C127(1.67) | LDD1290 | [24] |
| LDCM0412 | CL24 | PaTu 8988t | C122(1.02); C127(1.02) | LDD1291 | [24] |
| LDCM0413 | CL25 | PaTu 8988t | C122(0.99); C127(0.99) | LDD1292 | [24] |
| LDCM0414 | CL26 | PaTu 8988t | C122(1.58); C127(1.58) | LDD1293 | [24] |
| LDCM0415 | CL27 | PaTu 8988t | C122(1.21); C127(1.21) | LDD1294 | [24] |
| LDCM0416 | CL28 | PaTu 8988t | C122(2.45); C127(2.45) | LDD1295 | [24] |
| LDCM0417 | CL29 | PaTu 8988t | C122(2.45); C127(2.45) | LDD1296 | [24] |
| LDCM0419 | CL30 | PaTu 8988t | C122(1.35); C127(1.35) | LDD1298 | [24] |
| LDCM0420 | CL31 | PaTu 8988t | C122(0.78); C127(0.78) | LDD1299 | [24] |
| LDCM0421 | CL32 | PaTu 8988t | C122(0.79); C127(0.79) | LDD1300 | [24] |
| LDCM0422 | CL33 | PaTu 8988t | C122(1.15); C127(1.15) | LDD1301 | [24] |
| LDCM0423 | CL34 | PaTu 8988t | C122(1.02); C127(1.02) | LDD1302 | [24] |
| LDCM0424 | CL35 | PaTu 8988t | C122(0.99); C127(0.99) | LDD1303 | [24] |
| LDCM0425 | CL36 | PaTu 8988t | C122(1.66); C127(1.66) | LDD1304 | [24] |
| LDCM0426 | CL37 | PaTu 8988t | C122(1.40); C127(1.40) | LDD1305 | [24] |
| LDCM0428 | CL39 | PaTu 8988t | C122(0.79); C127(0.79) | LDD1307 | [24] |
| LDCM0430 | CL40 | PaTu 8988t | C122(0.83); C127(0.83) | LDD1309 | [24] |
| LDCM0431 | CL41 | PaTu 8988t | C122(0.92); C127(0.92) | LDD1310 | [24] |
| LDCM0432 | CL42 | PaTu 8988t | C122(1.66); C127(1.66) | LDD1311 | [24] |
| LDCM0433 | CL43 | PaTu 8988t | C122(1.83); C127(1.83) | LDD1312 | [24] |
| LDCM0434 | CL44 | PaTu 8988t | C122(0.91); C127(0.91) | LDD1313 | [24] |
| LDCM0435 | CL45 | PaTu 8988t | C122(1.65); C127(1.65) | LDD1314 | [24] |
| LDCM0436 | CL46 | PaTu 8988t | C122(0.85); C127(0.85) | LDD1315 | [24] |
| LDCM0437 | CL47 | PaTu 8988t | C122(0.86); C127(0.86) | LDD1316 | [24] |
| LDCM0438 | CL48 | PaTu 8988t | C122(0.98); C127(0.98) | LDD1317 | [24] |
| LDCM0439 | CL49 | PaTu 8988t | C122(0.90); C127(0.90) | LDD1318 | [24] |
| LDCM0441 | CL50 | PaTu 8988t | C122(1.02); C127(1.02) | LDD1320 | [24] |
| LDCM0442 | CL51 | PaTu 8988t | C122(0.83); C127(0.83) | LDD1321 | [24] |
| LDCM0443 | CL52 | PaTu 8988t | C122(1.14); C127(1.14) | LDD1322 | [24] |
| LDCM0444 | CL53 | PaTu 8988t | C122(1.69); C127(1.69) | LDD1323 | [24] |
| LDCM0445 | CL54 | PaTu 8988t | C122(1.14); C127(1.14) | LDD1324 | [24] |
| LDCM0446 | CL55 | PaTu 8988t | C122(1.00); C127(1.00) | LDD1325 | [24] |
| LDCM0447 | CL56 | PaTu 8988t | C122(0.71); C127(0.71) | LDD1326 | [24] |
| LDCM0448 | CL57 | PaTu 8988t | C122(2.09); C127(2.09) | LDD1327 | [24] |
| LDCM0449 | CL58 | PaTu 8988t | C122(1.37); C127(1.37) | LDD1328 | [24] |
| LDCM0450 | CL59 | PaTu 8988t | C122(2.53); C127(2.53) | LDD1329 | [24] |
| LDCM0452 | CL60 | PaTu 8988t | C122(1.46); C127(1.46) | LDD1331 | [24] |
| LDCM0459 | CL67 | HEK-293T | C122(1.35) | LDD1662 | [25] |
| LDCM0462 | CL7 | HEK-293T | C122(0.77) | LDD1665 | [25] |
| LDCM0464 | CL71 | HEK-293T | C122(1.04) | LDD1667 | [25] |
| LDCM0469 | CL76 | PaTu 8988t | C122(1.22); C127(1.22) | LDD1348 | [24] |
| LDCM0470 | CL77 | PaTu 8988t | C122(0.88); C127(0.88) | LDD1349 | [24] |
| LDCM0471 | CL78 | PaTu 8988t | C122(1.34); C127(1.34) | LDD1350 | [24] |
| LDCM0472 | CL79 | PaTu 8988t | C122(0.96); C127(0.96) | LDD1351 | [24] |
| LDCM0474 | CL80 | PaTu 8988t | C122(0.86); C127(0.86) | LDD1353 | [24] |
| LDCM0475 | CL81 | PaTu 8988t | C122(1.25); C127(1.25) | LDD1354 | [24] |
| LDCM0476 | CL82 | PaTu 8988t | C122(1.30); C127(1.30) | LDD1355 | [24] |
| LDCM0477 | CL83 | PaTu 8988t | C122(1.16); C127(1.16) | LDD1356 | [24] |
| LDCM0478 | CL84 | PaTu 8988t | C122(1.36); C127(1.36) | LDD1357 | [24] |
| LDCM0479 | CL85 | PaTu 8988t | C122(0.96); C127(0.96) | LDD1358 | [24] |
| LDCM0480 | CL86 | PaTu 8988t | C122(1.07); C127(1.07) | LDD1359 | [24] |
| LDCM0481 | CL87 | PaTu 8988t | C122(1.32); C127(1.32) | LDD1360 | [24] |
| LDCM0482 | CL88 | PaTu 8988t | C122(1.75); C127(1.75) | LDD1361 | [24] |
| LDCM0483 | CL89 | PaTu 8988t | C122(1.35); C127(1.35) | LDD1362 | [24] |
| LDCM0485 | CL90 | PaTu 8988t | C122(1.19); C127(1.19) | LDD1364 | [24] |
| LDCM0486 | CL91 | PaTu 8988t | C122(1.32); C127(1.32) | LDD1365 | [24] |
| LDCM0487 | CL92 | PaTu 8988t | C122(1.51); C127(1.51) | LDD1366 | [24] |
| LDCM0488 | CL93 | PaTu 8988t | C122(1.59); C127(1.59) | LDD1367 | [24] |
| LDCM0489 | CL94 | PaTu 8988t | C122(2.28); C127(2.28) | LDD1368 | [24] |
| LDCM0490 | CL95 | PaTu 8988t | C122(2.02); C127(2.02) | LDD1369 | [24] |
| LDCM0491 | CL96 | PaTu 8988t | C122(1.31); C127(1.31) | LDD1370 | [24] |
| LDCM0492 | CL97 | PaTu 8988t | C122(1.23); C127(1.23) | LDD1371 | [24] |
| LDCM0493 | CL98 | PaTu 8988t | C122(1.80); C127(1.80) | LDD1372 | [24] |
| LDCM0494 | CL99 | PaTu 8988t | C122(1.38); C127(1.38) | LDD1373 | [24] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C20(1.36) | LDD1702 | [6] |
| LDCM0625 | F8 | Ramos | C20(5.14) | LDD2187 | [26] |
| LDCM0576 | Fragment14 | Ramos | C20(0.23) | LDD2193 | [26] |
| LDCM0586 | Fragment28 | Ramos | C20(0.70) | LDD2198 | [26] |
| LDCM0566 | Fragment4 | Ramos | C20(1.15) | LDD2184 | [26] |
| LDCM0427 | Fragment51 | PaTu 8988t | C122(1.82); C127(1.82) | LDD1306 | [24] |
| LDCM0569 | Fragment7 | Ramos | C20(1.04) | LDD2186 | [26] |
| LDCM0116 | HHS-0101 | DM93 | Y49(0.87); Y155(0.99) | LDD0264 | [10] |
| LDCM0117 | HHS-0201 | DM93 | Y49(0.72) | LDD0265 | [10] |
| LDCM0118 | HHS-0301 | DM93 | Y49(0.79); Y155(1.22) | LDD0266 | [10] |
| LDCM0119 | HHS-0401 | DM93 | Y49(0.82); Y155(0.83) | LDD0267 | [10] |
| LDCM0120 | HHS-0701 | DM93 | Y49(0.99) | LDD0268 | [10] |
| LDCM0123 | JWB131 | DM93 | Y49(0.91) | LDD0285 | [9] |
| LDCM0124 | JWB142 | DM93 | Y49(0.60) | LDD0286 | [9] |
| LDCM0125 | JWB146 | DM93 | Y49(0.90) | LDD0287 | [9] |
| LDCM0126 | JWB150 | DM93 | Y49(1.28) | LDD0288 | [9] |
| LDCM0127 | JWB152 | DM93 | Y49(0.89) | LDD0289 | [9] |
| LDCM0128 | JWB198 | DM93 | Y49(0.56) | LDD0290 | [9] |
| LDCM0129 | JWB202 | DM93 | Y49(0.73) | LDD0291 | [9] |
| LDCM0130 | JWB211 | DM93 | Y49(1.01) | LDD0292 | [9] |
| LDCM0022 | KB02 | Ramos | C20(1.69) | LDD2182 | [26] |
| LDCM0023 | KB03 | Jurkat | C20(332.45) | LDD0209 | [8] |
| LDCM0024 | KB05 | MKN45 | C52(4.00) | LDD3332 | [27] |
| LDCM0509 | N-(4-bromo-3,5-dimethylphenyl)-2-nitroacetamide | MDA-MB-231 | C20(1.13) | LDD2102 | [6] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0226 | [12] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C20(1.35) | LDD2090 | [6] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C20(0.78) | LDD2093 | [6] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C20(0.96) | LDD2094 | [6] |
| LDCM0503 | Nucleophilic fragment 14b | MDA-MB-231 | C20(0.54) | LDD2096 | [6] |
| LDCM0504 | Nucleophilic fragment 15a | MDA-MB-231 | C20(0.78) | LDD2097 | [6] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C20(0.60) | LDD2099 | [6] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C20(0.50) | LDD2100 | [6] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C20(1.03) | LDD2101 | [6] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C20(0.63) | LDD2104 | [6] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C20(1.61) | LDD2105 | [6] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C20(0.60) | LDD2106 | [6] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C20(0.83) | LDD2107 | [6] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C20(0.71) | LDD2108 | [6] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C20(0.44) | LDD2109 | [6] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C20(0.51) | LDD2115 | [6] |
| LDCM0523 | Nucleophilic fragment 24b | MDA-MB-231 | C20(0.98) | LDD2116 | [6] |
| LDCM0525 | Nucleophilic fragment 25b | MDA-MB-231 | C20(0.87) | LDD2118 | [6] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C20(0.90) | LDD2120 | [6] |
| LDCM0529 | Nucleophilic fragment 27b | MDA-MB-231 | C20(1.04) | LDD2122 | [6] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C20(0.53) | LDD2123 | [6] |
| LDCM0531 | Nucleophilic fragment 28b | MDA-MB-231 | C20(1.00) | LDD2124 | [6] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C20(0.50) | LDD2125 | [6] |
| LDCM0533 | Nucleophilic fragment 29b | MDA-MB-231 | C20(1.76) | LDD2126 | [6] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C20(0.86) | LDD2128 | [6] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C20(0.70) | LDD2129 | [6] |
| LDCM0540 | Nucleophilic fragment 35 | MDA-MB-231 | C20(0.62) | LDD2133 | [6] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C20(0.51) | LDD2134 | [6] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C20(0.83) | LDD2135 | [6] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C20(0.69) | LDD2136 | [6] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C20(0.83) | LDD2137 | [6] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C20(1.01) | LDD1700 | [6] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C20(0.65) | LDD2140 | [6] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C20(0.52) | LDD2141 | [6] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C20(1.36) | LDD2143 | [6] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C20(4.31) | LDD2144 | [6] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C20(0.59) | LDD2146 | [6] |
| LDCM0553 | Nucleophilic fragment 6b | MDA-MB-231 | C20(4.13) | LDD2147 | [6] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C20(0.78) | LDD2148 | [6] |
| LDCM0555 | Nucleophilic fragment 7b | MDA-MB-231 | C20(1.00) | LDD2149 | [6] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C20(0.29) | LDD2150 | [6] |
| LDCM0557 | Nucleophilic fragment 8b | MDA-MB-231 | C20(1.76) | LDD2151 | [6] |
| LDCM0559 | Nucleophilic fragment 9b | MDA-MB-231 | C20(2.07) | LDD2153 | [6] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Calpain-1 catalytic subunit (CAPN1) | Peptidase C2 family | P07384 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Lipid transferase CIDEB (CIDEB) | CIDE family | Q9UHD4 | |||
| Small glutamine-rich tetratricopeptide repeat-containing protein beta (SGTB) | SGT family | Q96EQ0 | |||
The Drug(s) Related To This Target
Approved
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Decitabine | Small molecular drug | DB01262 | |||
| Gemcitabine | Small molecular drug | DB00441 | |||
| Lamivudine | Small molecular drug | DB00709 | |||
| Sofosbuvir | Small molecular drug | DB08934 | |||
Investigative
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Cytidine-5'-monophosphate | Small molecular drug | DB03403 | |||
| Uridine-5'-diphosphate | Small molecular drug | DB03435 | |||
| Tetrafluoroaluminate Ion | . | DB04444 | |||
References
