Details of the Target
General Information of Target
| Target ID | LDTP02227 | |||||
|---|---|---|---|---|---|---|
| Target Name | Heterogeneous nuclear ribonucleoproteins C1/C2 (HNRNPC) | |||||
| Gene Name | HNRNPC | |||||
| Gene ID | 3183 | |||||
| Synonyms |
HNRPC; Heterogeneous nuclear ribonucleoproteins C1/C2; hnRNP C1/C2 |
|||||
| 3D Structure | ||||||
| Sequence |
MASNVTNKTDPRSMNSRVFIGNLNTLVVKKSDVEAIFSKYGKIVGCSVHKGFAFVQYVNE
RNARAAVAGEDGRMIAGQVLDINLAAEPKVNRGKAGVKRSAAEMYGSVTEHPSPSPLLSS SFDLDYDFQRDYYDRMYSYPARVPPPPPIARAVVPSKRQRVSGNTSRRGKSGFNSKSGQR GSSKSGKLKGDDLQAIKKELTQIKQKVDSLLENLEKIEKEQSKQAVEMKNDKSEEEQSSS SVKKDETNVKMESEGGADDSAEEGDLLDDDDNEDRGDDQLELIKDDEKEAEEGEDDRDSA NGEDDS |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
RRM HNRPC family, RALY subfamily
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
Binds pre-mRNA and nucleates the assembly of 40S hnRNP particles. Interacts with poly-U tracts in the 3'-UTR or 5'-UTR of mRNA and modulates the stability and the level of translation of bound mRNA molecules. Single HNRNPC tetramers bind 230-240 nucleotides. Trimers of HNRNPC tetramers bind 700 nucleotides. May play a role in the early steps of spliceosome assembly and pre-mRNA splicing. N6-methyladenosine (m6A) has been shown to alter the local structure in mRNAs and long non-coding RNAs (lncRNAs) via a mechanism named 'm(6)A-switch', facilitating binding of HNRNPC, leading to regulation of mRNA splicing.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
P2 Probe Info |
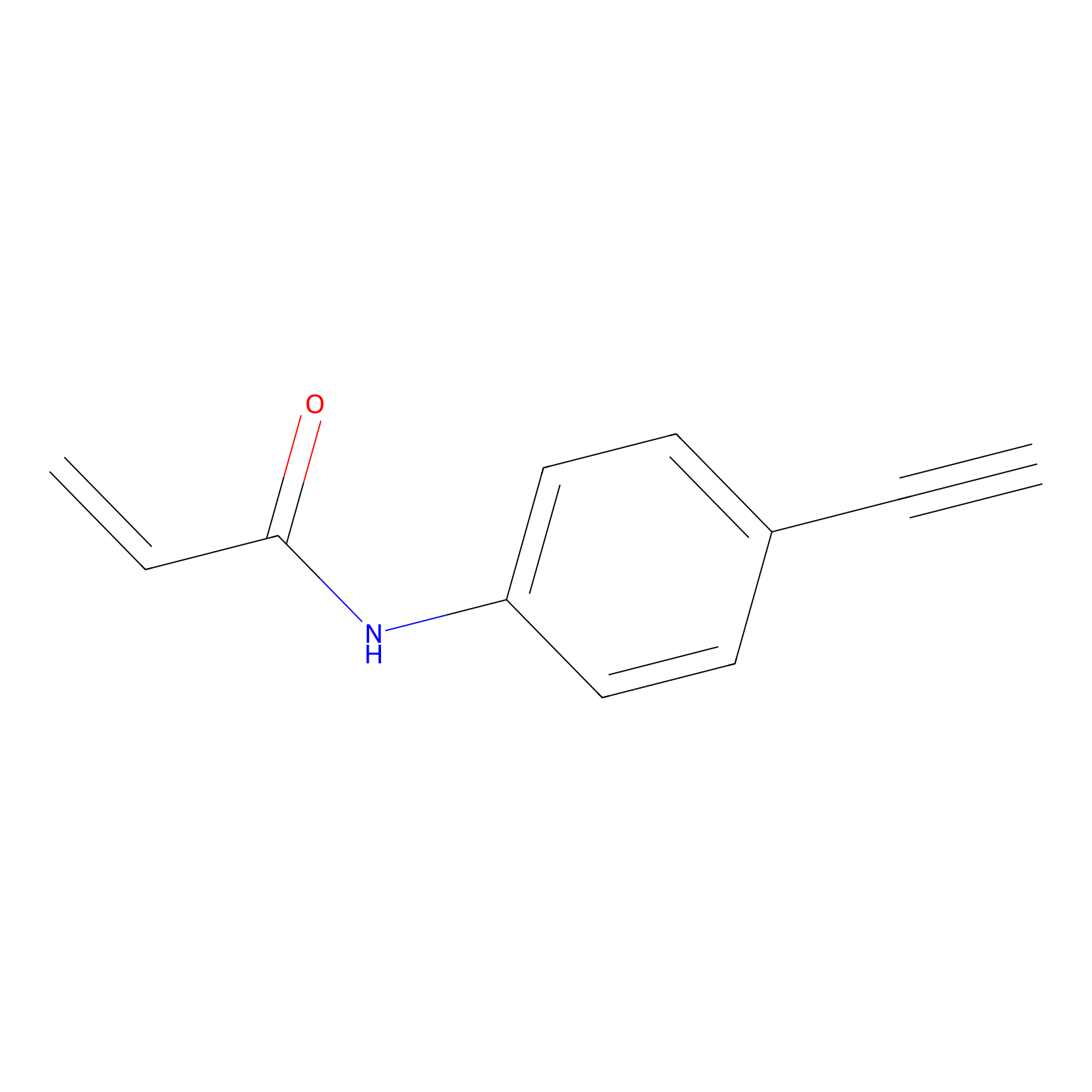 |
3.84 | LDD0449 | [1] | |
|
P3 Probe Info |
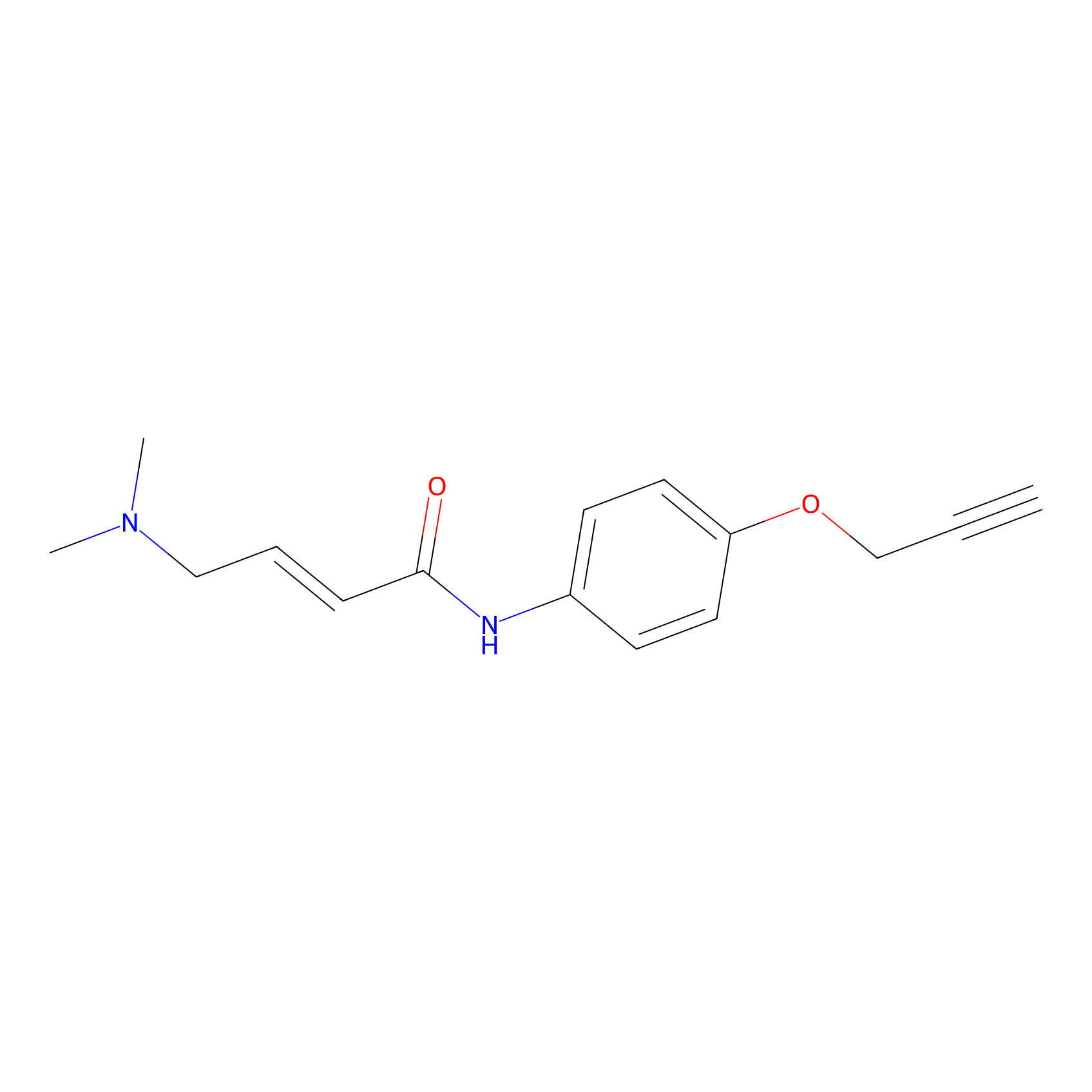 |
3.62 | LDD0450 | [1] | |
|
P8 Probe Info |
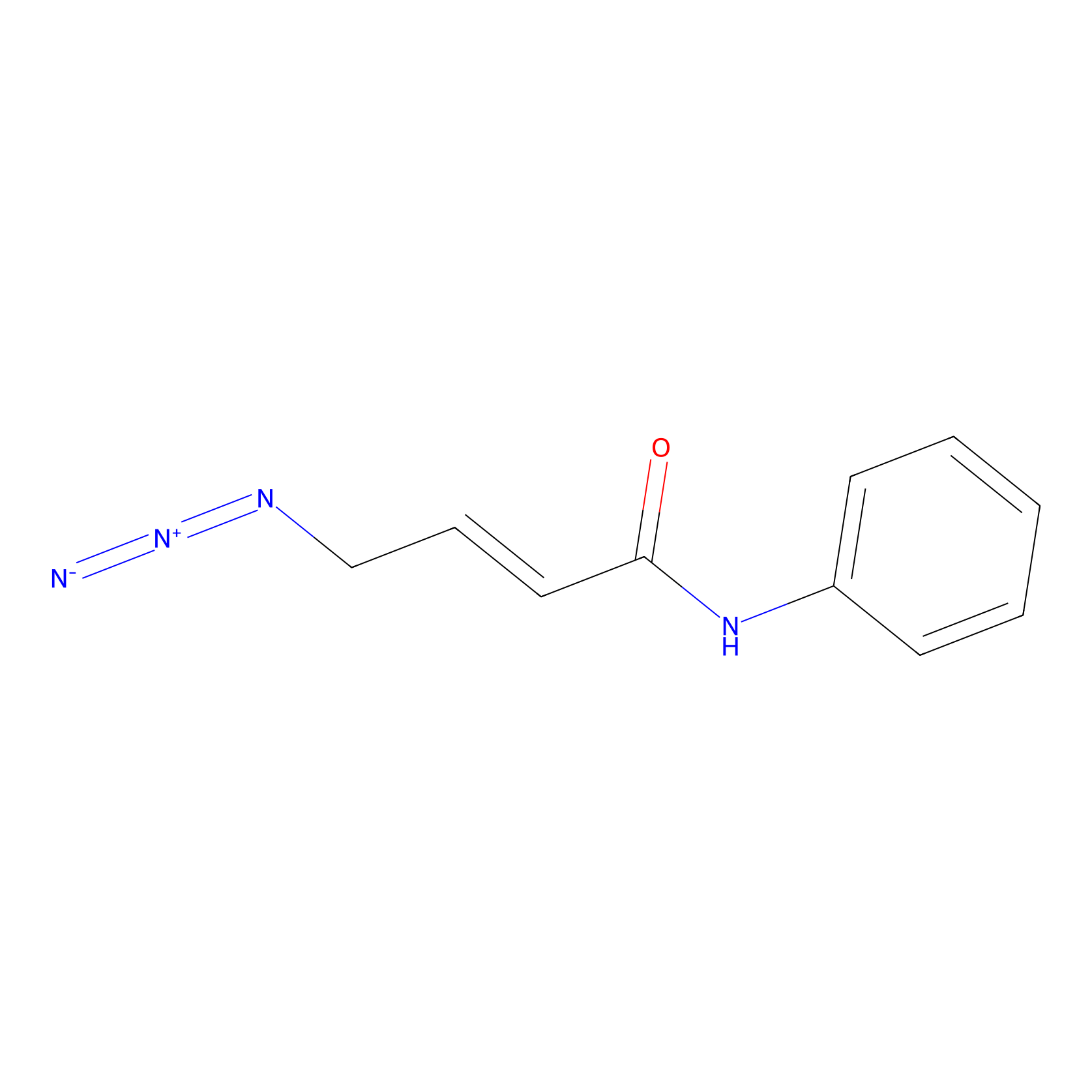 |
5.05 | LDD0451 | [1] | |
|
A-EBA Probe Info |
 |
4.52 | LDD0215 | [2] | |
|
CY-1 Probe Info |
 |
100.00 | LDD0243 | [3] | |
|
CY4 Probe Info |
 |
100.00 | LDD0244 | [3] | |
|
N1 Probe Info |
 |
100.00 | LDD0242 | [3] | |
|
C-Sul Probe Info |
 |
5.31 | LDD0066 | [4] | |
|
TH211 Probe Info |
 |
Y139(8.58) | LDD0257 | [5] | |
|
TH216 Probe Info |
 |
Y137(5.47) | LDD0259 | [5] | |
|
AZ-9 Probe Info |
 |
E60(1.11); E246(1.80) | LDD2208 | [6] | |
|
ONAyne Probe Info |
 |
K184(0.00); K189(0.00); K197(0.00); K198(0.00) | LDD0273 | [7] | |
|
OPA-S-S-alkyne Probe Info |
 |
K170(2.35); K176(3.10); K98(3.25); K94(3.25) | LDD3494 | [8] | |
|
Probe 1 Probe Info |
 |
Y139(65.94) | LDD3495 | [9] | |
|
BTD Probe Info |
 |
C46(1.83) | LDD1700 | [10] | |
|
ATP probe Probe Info |
 |
K198(0.00); K189(0.00); K170(0.00); K8(0.00) | LDD0199 | [11] | |
|
m-APA Probe Info |
 |
N.A. | LDD2231 | [12] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [13] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [13] | |
|
IPIAA_L Probe Info |
 |
N.A. | LDD0031 | [14] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [13] | |
|
ATP probe Probe Info |
 |
K206(0.00); K89(0.00); K8(0.00); K29(0.00) | LDD0035 | [15] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [16] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [17] | |
|
NHS Probe Info |
 |
K223(0.00); K189(0.00); K176(0.00); K232(0.00) | LDD0010 | [17] | |
|
OSF Probe Info |
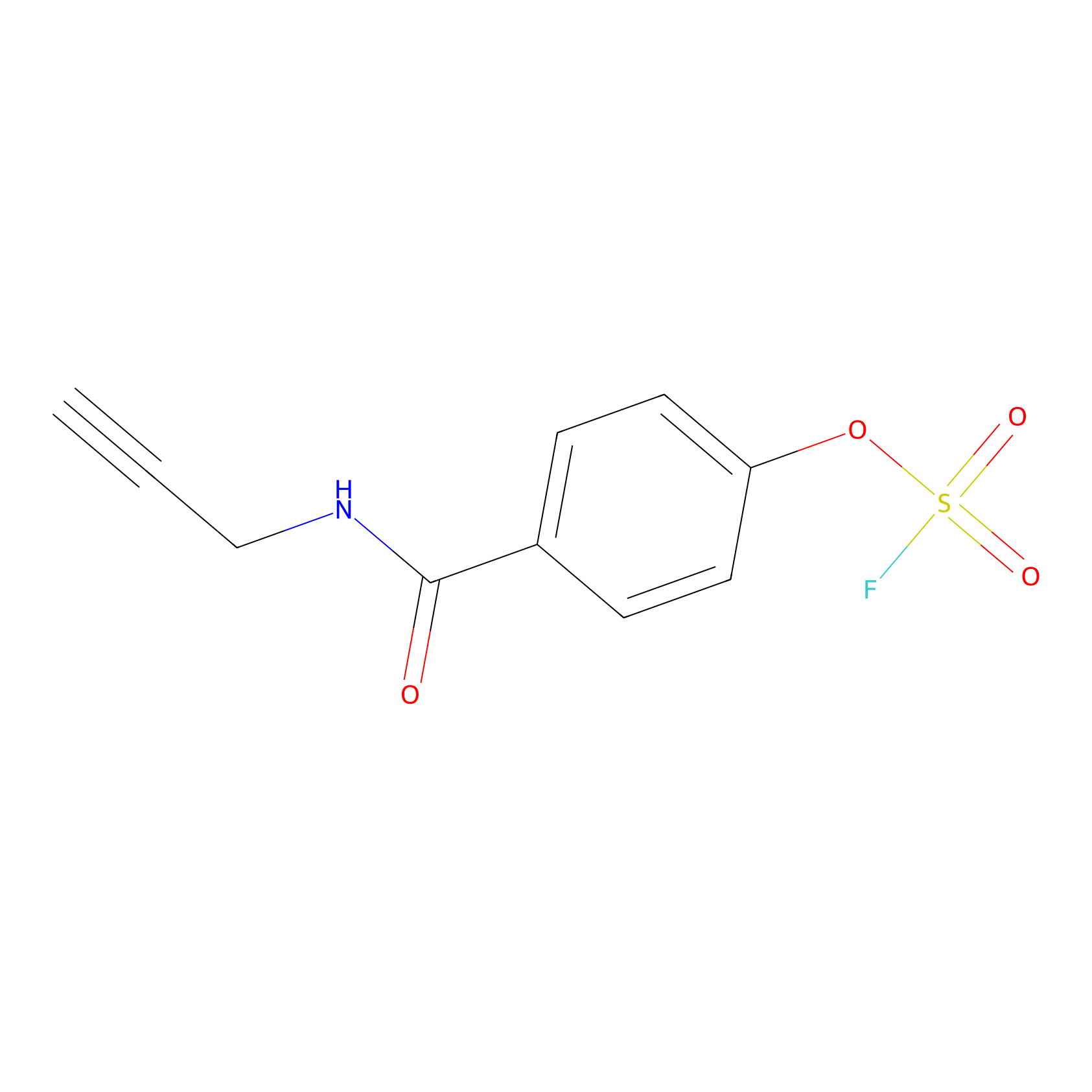 |
Y139(0.00); Y137(0.00) | LDD0029 | [18] | |
|
SF Probe Info |
 |
K8(0.00); Y137(0.00); Y139(0.00); K170(0.00) | LDD0028 | [18] | |
|
STPyne Probe Info |
 |
K30(0.00); K216(0.00); K206(0.00); K8(0.00) | LDD0009 | [17] | |
|
1c-yne Probe Info |
 |
K284(0.00); K50(0.00); K229(0.00); K39(0.00) | LDD0228 | [19] | |
|
AOyne Probe Info |
 |
7.00 | LDD0443 | [20] | |
|
MPP-AC Probe Info |
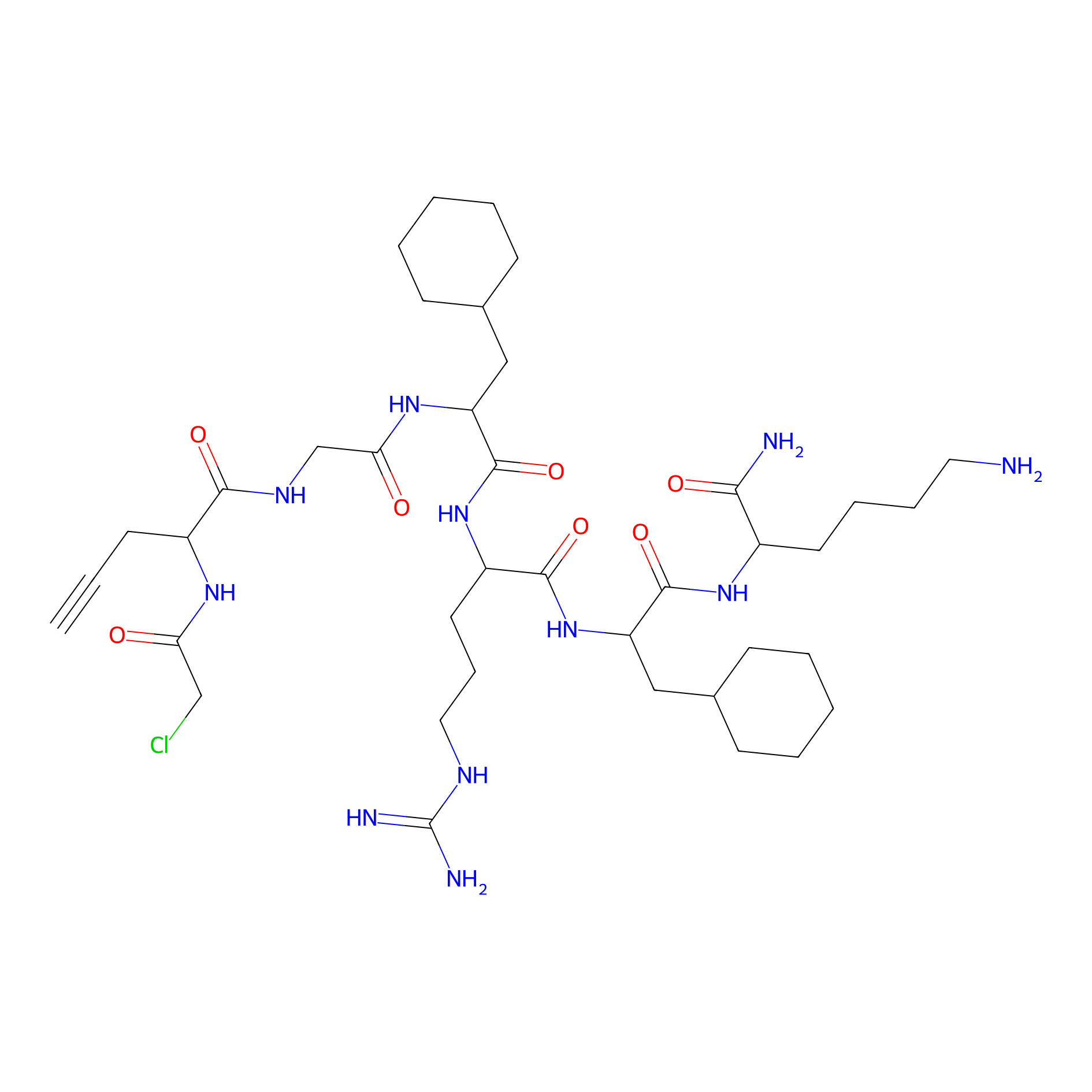 |
N.A. | LDD0428 | [21] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [22] | |
|
HHS-482 Probe Info |
 |
Y137(0.90) | LDD2239 | [23] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C338 Probe Info |
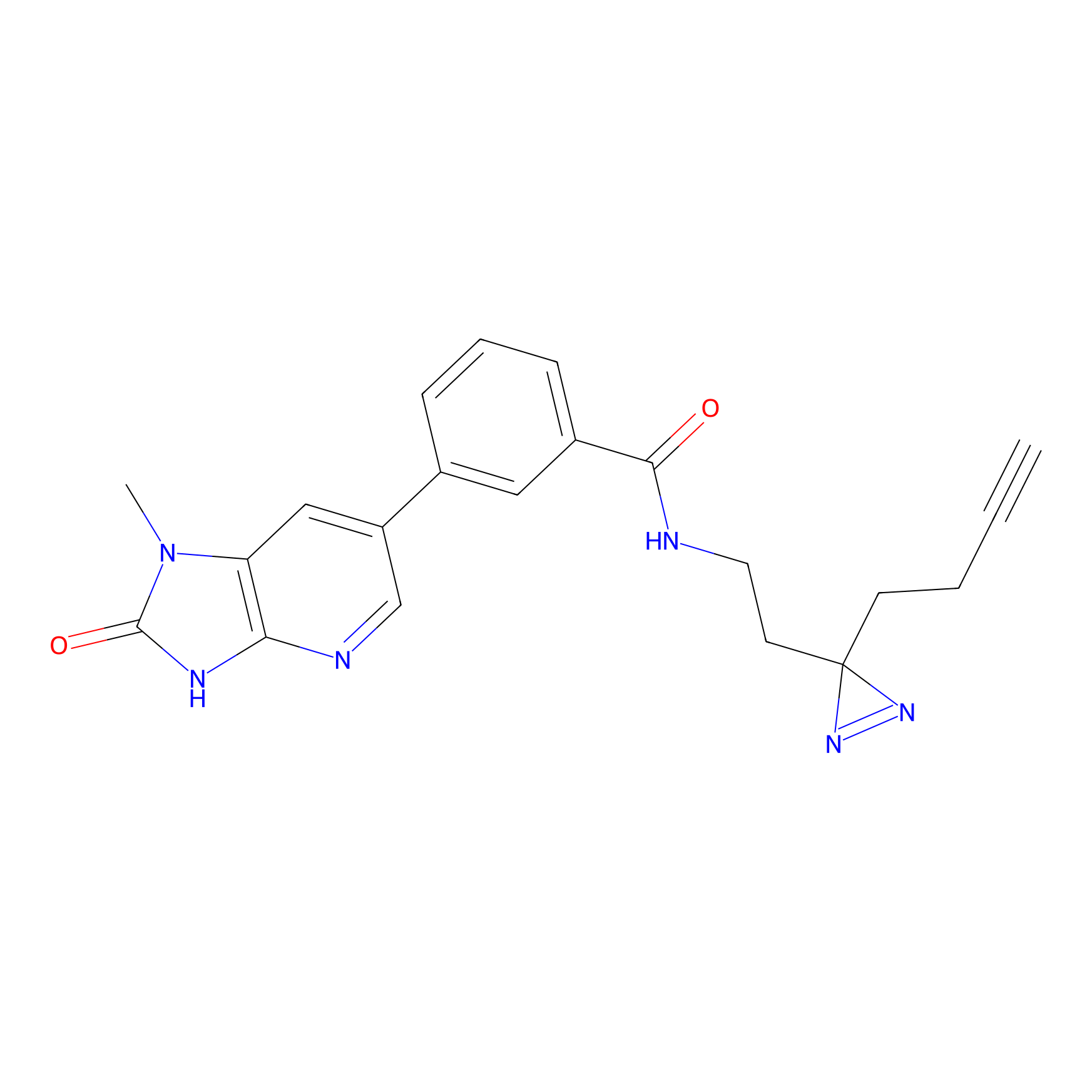 |
12.73 | LDD2001 | [24] | |
|
FFF probe11 Probe Info |
 |
10.41 | LDD0471 | [25] | |
|
FFF probe12 Probe Info |
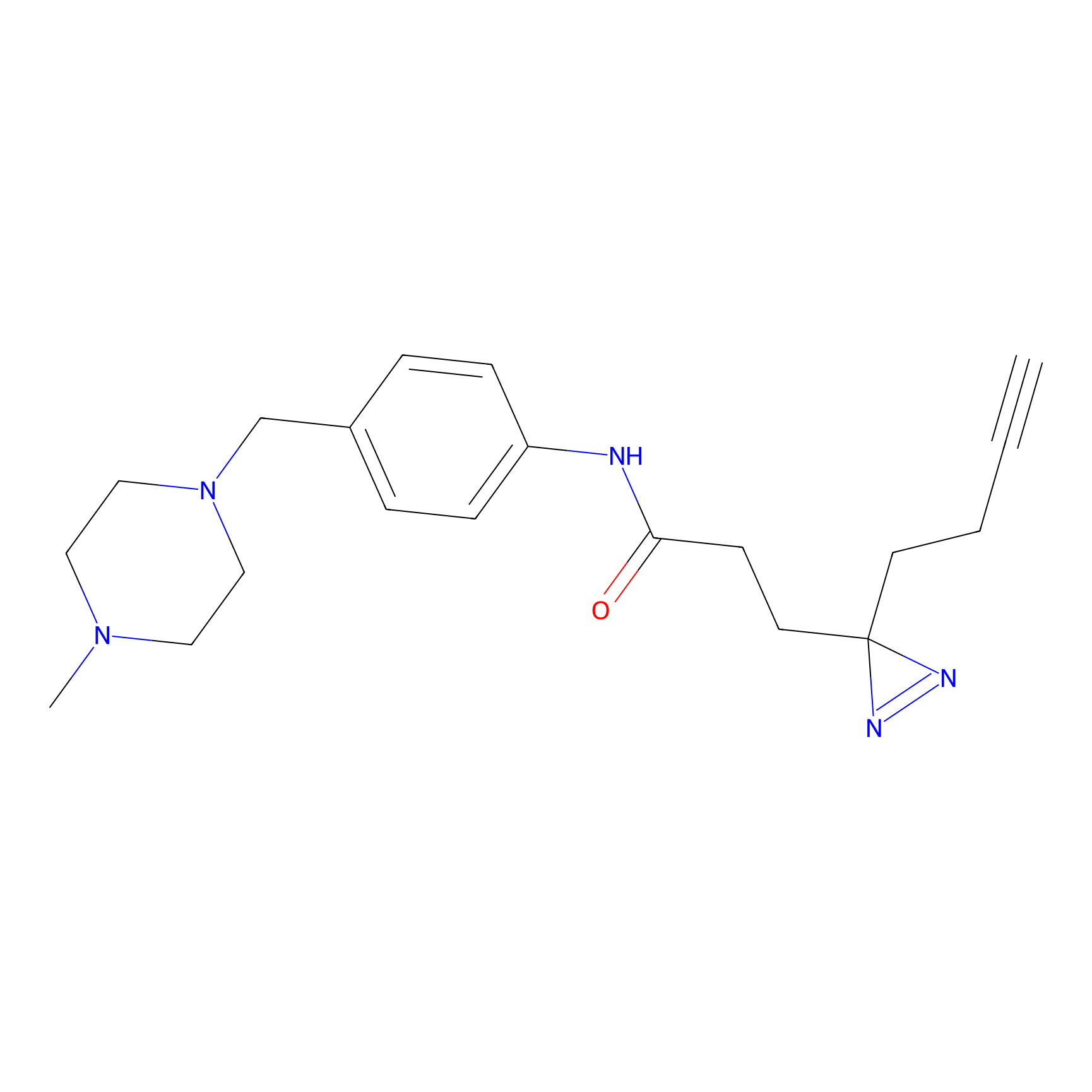 |
7.28 | LDD0473 | [25] | |
|
FFF probe13 Probe Info |
 |
14.38 | LDD0475 | [25] | |
|
FFF probe2 Probe Info |
 |
12.46 | LDD0463 | [25] | |
|
FFF probe3 Probe Info |
 |
7.95 | LDD0464 | [25] | |
|
FFF probe4 Probe Info |
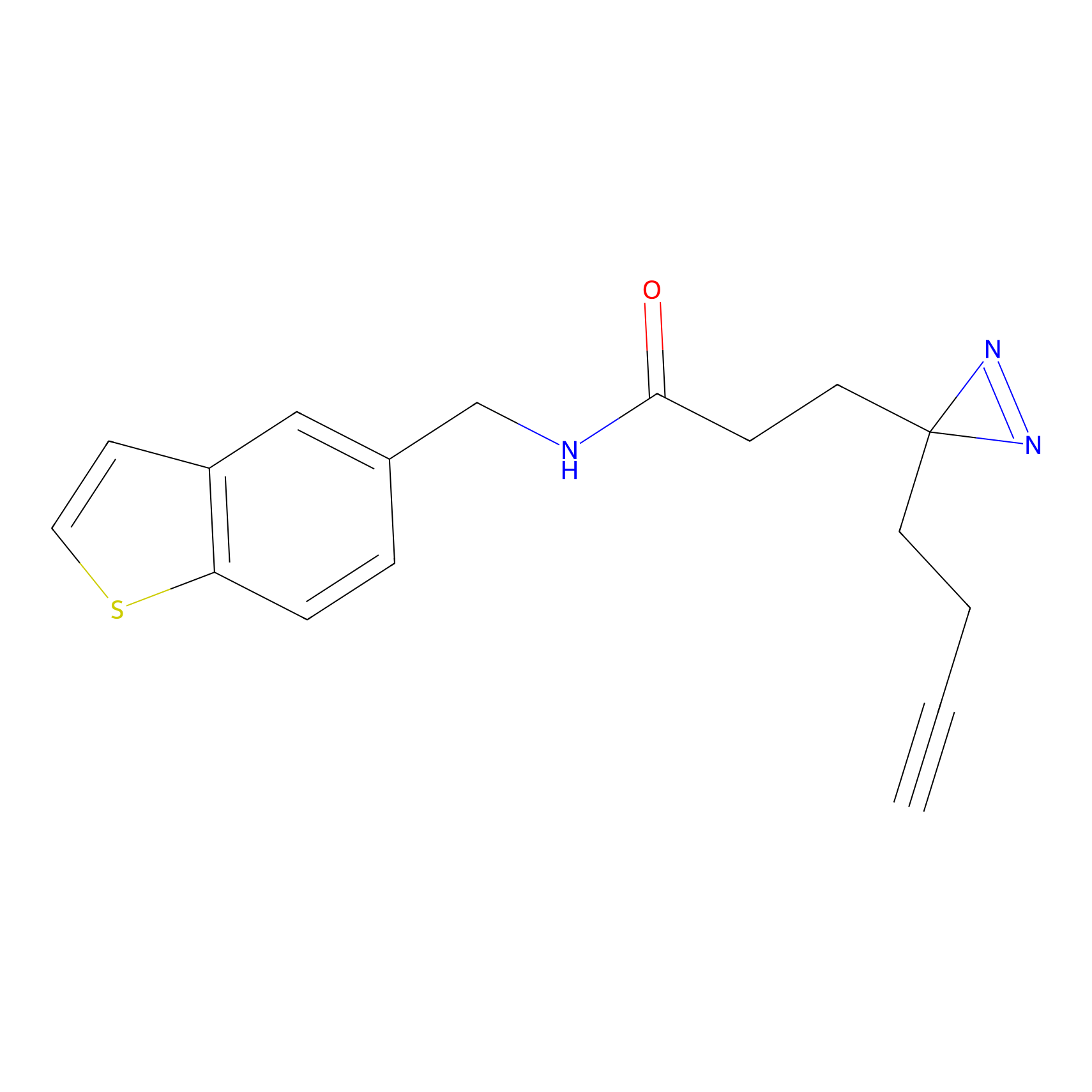 |
6.36 | LDD0466 | [25] | |
|
JN0003 Probe Info |
 |
11.73 | LDD0469 | [25] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0138 | [26] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [27] | |
|
DA-2 Probe Info |
 |
N.A. | LDD0070 | [28] | |
|
OEA-DA Probe Info |
 |
3.65 | LDD0046 | [29] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0069 | [30] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C46(0.44) | LDD2142 | [10] |
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C46(0.76) | LDD2112 | [10] |
| LDCM0502 | 1-(Cyanoacetyl)piperidine | MDA-MB-231 | C46(0.45) | LDD2095 | [10] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C46(0.58) | LDD2117 | [10] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C46(1.18) | LDD2152 | [10] |
| LDCM0510 | 3-(4-(Hydroxydiphenylmethyl)piperidin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C46(1.26) | LDD2103 | [10] |
| LDCM0539 | 3-(4-Isopropylpiperazin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C46(0.42) | LDD2132 | [10] |
| LDCM0538 | 4-(Cyanoacetyl)morpholine | MDA-MB-231 | C46(0.88) | LDD2131 | [10] |
| LDCM0545 | Acetamide | MDA-MB-231 | C46(0.37) | LDD2138 | [10] |
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C46(0.72) | LDD2113 | [10] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C46(0.55) | LDD2091 | [10] |
| LDCM0625 | F8 | Ramos | C46(0.59) | LDD2187 | [31] |
| LDCM0572 | Fragment10 | Ramos | C46(0.60) | LDD2189 | [31] |
| LDCM0573 | Fragment11 | Ramos | C46(3.42) | LDD2190 | [31] |
| LDCM0574 | Fragment12 | Ramos | C46(0.66) | LDD2191 | [31] |
| LDCM0575 | Fragment13 | Ramos | C46(1.12) | LDD2192 | [31] |
| LDCM0576 | Fragment14 | Ramos | C46(0.88) | LDD2193 | [31] |
| LDCM0579 | Fragment20 | Ramos | C46(0.59) | LDD2194 | [31] |
| LDCM0580 | Fragment21 | Ramos | C46(0.97) | LDD2195 | [31] |
| LDCM0582 | Fragment23 | Ramos | C46(1.30) | LDD2196 | [31] |
| LDCM0578 | Fragment27 | Ramos | C46(1.06) | LDD2197 | [31] |
| LDCM0586 | Fragment28 | Ramos | C46(0.61) | LDD2198 | [31] |
| LDCM0588 | Fragment30 | Ramos | C46(1.41) | LDD2199 | [31] |
| LDCM0589 | Fragment31 | Ramos | C46(1.32) | LDD2200 | [31] |
| LDCM0590 | Fragment32 | Ramos | C46(0.77) | LDD2201 | [31] |
| LDCM0468 | Fragment33 | Ramos | C46(1.21) | LDD2202 | [31] |
| LDCM0596 | Fragment38 | Ramos | C46(1.31) | LDD2203 | [31] |
| LDCM0566 | Fragment4 | Ramos | C46(0.72) | LDD2184 | [31] |
| LDCM0610 | Fragment52 | Ramos | C46(1.61) | LDD2204 | [31] |
| LDCM0614 | Fragment56 | Ramos | C46(1.65) | LDD2205 | [31] |
| LDCM0569 | Fragment7 | Ramos | C46(0.53) | LDD2186 | [31] |
| LDCM0571 | Fragment9 | Ramos | C46(0.57) | LDD2188 | [31] |
| LDCM0022 | KB02 | Ramos | C46(0.54) | LDD2182 | [31] |
| LDCM0023 | KB03 | MDA-MB-231 | C46(1.82) | LDD1701 | [10] |
| LDCM0024 | KB05 | Ramos | C46(0.47) | LDD2185 | [31] |
| LDCM0509 | N-(4-bromo-3,5-dimethylphenyl)-2-nitroacetamide | MDA-MB-231 | C46(1.07) | LDD2102 | [10] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C46(0.61) | LDD2121 | [10] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C46(0.56) | LDD2089 | [10] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C46(0.90) | LDD2090 | [10] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C46(1.16) | LDD2092 | [10] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C46(0.99) | LDD2093 | [10] |
| LDCM0503 | Nucleophilic fragment 14b | MDA-MB-231 | C46(0.12) | LDD2096 | [10] |
| LDCM0504 | Nucleophilic fragment 15a | MDA-MB-231 | C46(0.88) | LDD2097 | [10] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C46(0.81) | LDD2098 | [10] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C46(0.97) | LDD2099 | [10] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C46(0.45) | LDD2100 | [10] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C46(0.64) | LDD2101 | [10] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C46(0.45) | LDD2104 | [10] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C46(1.49) | LDD2105 | [10] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C46(0.37) | LDD2106 | [10] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C46(0.92) | LDD2107 | [10] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C46(0.47) | LDD2108 | [10] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C46(0.45) | LDD2109 | [10] |
| LDCM0517 | Nucleophilic fragment 21b | MDA-MB-231 | C46(0.50) | LDD2110 | [10] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C46(0.81) | LDD2111 | [10] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C46(0.55) | LDD2114 | [10] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C46(0.36) | LDD2115 | [10] |
| LDCM0523 | Nucleophilic fragment 24b | MDA-MB-231 | C46(0.17) | LDD2116 | [10] |
| LDCM0525 | Nucleophilic fragment 25b | MDA-MB-231 | C46(0.23) | LDD2118 | [10] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C46(1.88) | LDD2119 | [10] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C46(0.76) | LDD2120 | [10] |
| LDCM0529 | Nucleophilic fragment 27b | MDA-MB-231 | C46(0.18) | LDD2122 | [10] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C46(0.68) | LDD2123 | [10] |
| LDCM0531 | Nucleophilic fragment 28b | MDA-MB-231 | C46(0.09) | LDD2124 | [10] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C46(0.64) | LDD2125 | [10] |
| LDCM0533 | Nucleophilic fragment 29b | MDA-MB-231 | C46(0.09) | LDD2126 | [10] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C46(0.66) | LDD2127 | [10] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C46(0.88) | LDD2128 | [10] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C46(1.13) | LDD2129 | [10] |
| LDCM0540 | Nucleophilic fragment 35 | MDA-MB-231 | C46(0.50) | LDD2133 | [10] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C46(0.48) | LDD2134 | [10] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C46(1.15) | LDD2135 | [10] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C46(0.97) | LDD2136 | [10] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C46(0.80) | LDD2137 | [10] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C46(1.83) | LDD1700 | [10] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C46(0.63) | LDD2140 | [10] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C46(0.45) | LDD2141 | [10] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C46(0.83) | LDD2143 | [10] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C46(1.84) | LDD2144 | [10] |
| LDCM0551 | Nucleophilic fragment 5b | MDA-MB-231 | C46(1.99) | LDD2145 | [10] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C46(0.86) | LDD2146 | [10] |
| LDCM0553 | Nucleophilic fragment 6b | MDA-MB-231 | C46(2.39) | LDD2147 | [10] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C46(0.38) | LDD2148 | [10] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C46(0.42) | LDD2150 | [10] |
| LDCM0559 | Nucleophilic fragment 9b | MDA-MB-231 | C46(2.03) | LDD2153 | [10] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Cell division cycle 5-like protein (CDC5L) | CEF1 family | Q99459 | |||
| Paired box protein Pax-6 (PAX6) | Paired homeobox family | P26367 | |||
Other
References
