Details of the Target
General Information of Target
| Target ID | LDTP03953 | |||||
|---|---|---|---|---|---|---|
| Target Name | Eukaryotic translation initiation factor 1 (EIF1) | |||||
| Gene Name | EIF1 | |||||
| Gene ID | 10209 | |||||
| Synonyms |
SUI1; Eukaryotic translation initiation factor 1; eIF1; A121; Protein translation factor SUI1 homolog; Sui1iso1 |
|||||
| 3D Structure | ||||||
| Sequence |
MSAIQNLHSFDPFADASKGDDLLPAGTEDYIHIRIQQRNGRKTLTTVQGIADDYDKKKLV
KAFKKKFACNGTVIEHPEYGEVIQLQGDQRKNICQFLVEIGLAKDDQLKVHGF |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
SUI1 family
|
|||||
| Function |
Component of the 43S pre-initiation complex (43S PIC), which binds to the mRNA cap-proximal region, scans mRNA 5'-untranslated region, and locates the initiation codon. Together with eIF1A (EIF1AX), EIF1 facilitates scanning and is essential for start codon recognition on the basis of AUG nucleotide context and location relative to the 5'-cap. Participates to initiation codon selection by influencing the conformation of the 40S ribosomal subunit and the positions of bound mRNA and initiator tRNA; this is possible after its binding to the interface surface of the platform of the 40S ribosomal subunit close to the P-site. Together with eIF1A (EIF1AX), also regulates the opening and closing of the mRNA binding channel, which ensures mRNA recruitment, scanning and the fidelity of initiation codon selection. Continuously monitors and protects against premature and partial base-pairing of codons in the 5'-UTR with the anticodon of initiator tRNA. Together with eIF1A (EIF1AX), acts for ribosomal scanning, promotion of the assembly of 48S complex at the initiation codon (43S PIC becomes 48S PIC after the start codon is reached), and dissociation of aberrant complexes. Interacts with EIF4G1, which in a mutual exclusive interaction associates either with EIF1 or with EIF4E on a common binding site. EIF4G1-EIF1 complex promotes ribosome scanning (on both short and long 5'UTR), leaky scanning (on short 5'UTR) which is the bypass of the initial start codon, and discrimination against cap-proximal AUG. Is probably maintained within the 43S PIC in open conformation thanks to eIF1A-EIF5 interaction. Once the correct start codon is reached, EIF1 is physically excluded from the decoding site, shifting the PIC into the closed conformation and arresting it at the start codon.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
P8 Probe Info |
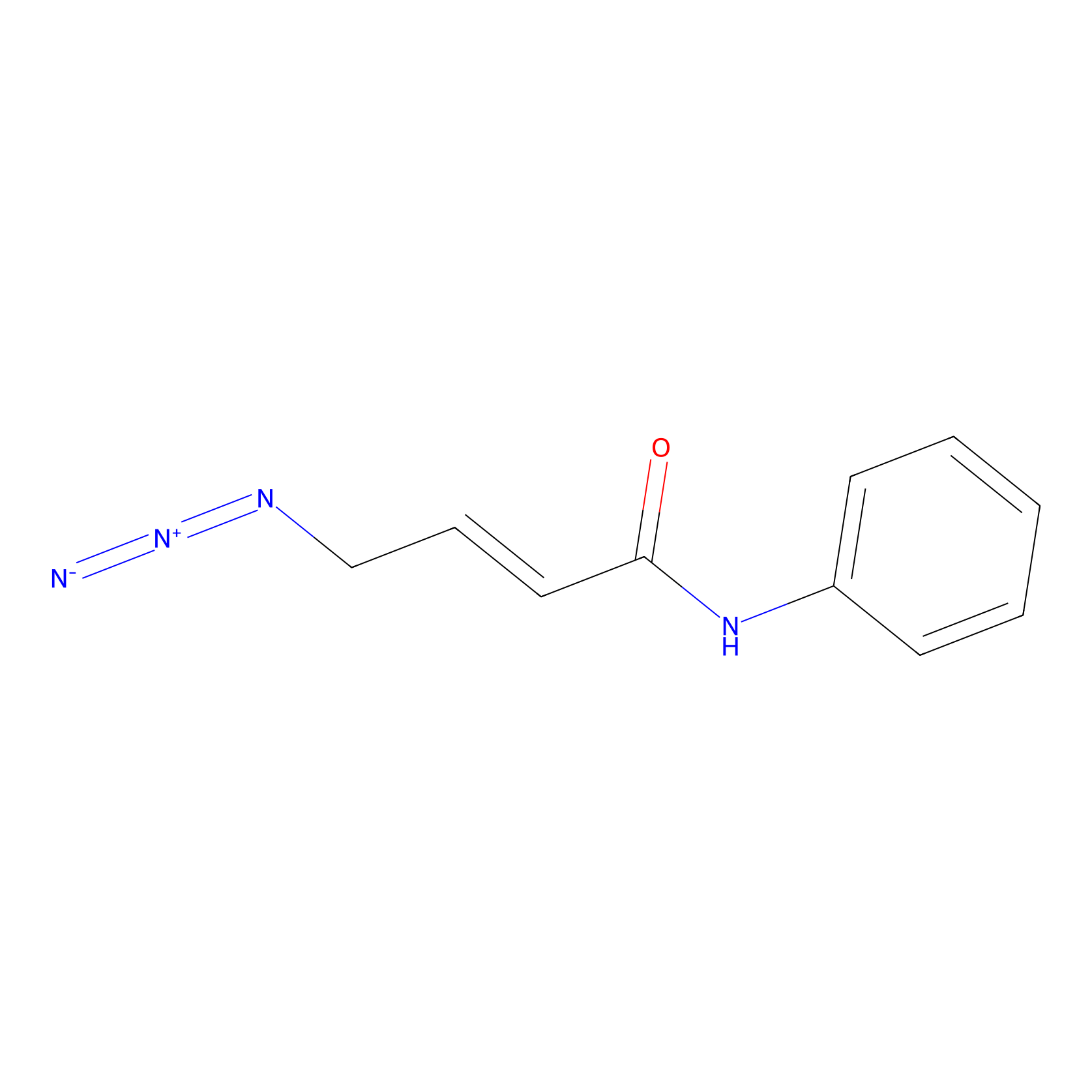 |
10.00 | LDD0451 | [1] | |
|
TH216 Probe Info |
 |
Y54(20.00) | LDD0259 | [2] | |
|
BTD Probe Info |
 |
C69(1.00) | LDD2117 | [3] | |
|
HHS-482 Probe Info |
 |
Y79(0.99) | LDD0285 | [4] | |
|
HHS-475 Probe Info |
 |
Y54(0.87); Y79(0.99); Y30(1.14) | LDD0264 | [5] | |
|
HHS-465 Probe Info |
 |
Y54(10.00); Y79(6.18) | LDD2237 | [6] | |
|
DBIA Probe Info |
 |
C69(0.88) | LDD0531 | [7] | |
|
5E-2FA Probe Info |
 |
H76(0.00); H8(0.00) | LDD2235 | [8] | |
|
AMP probe Probe Info |
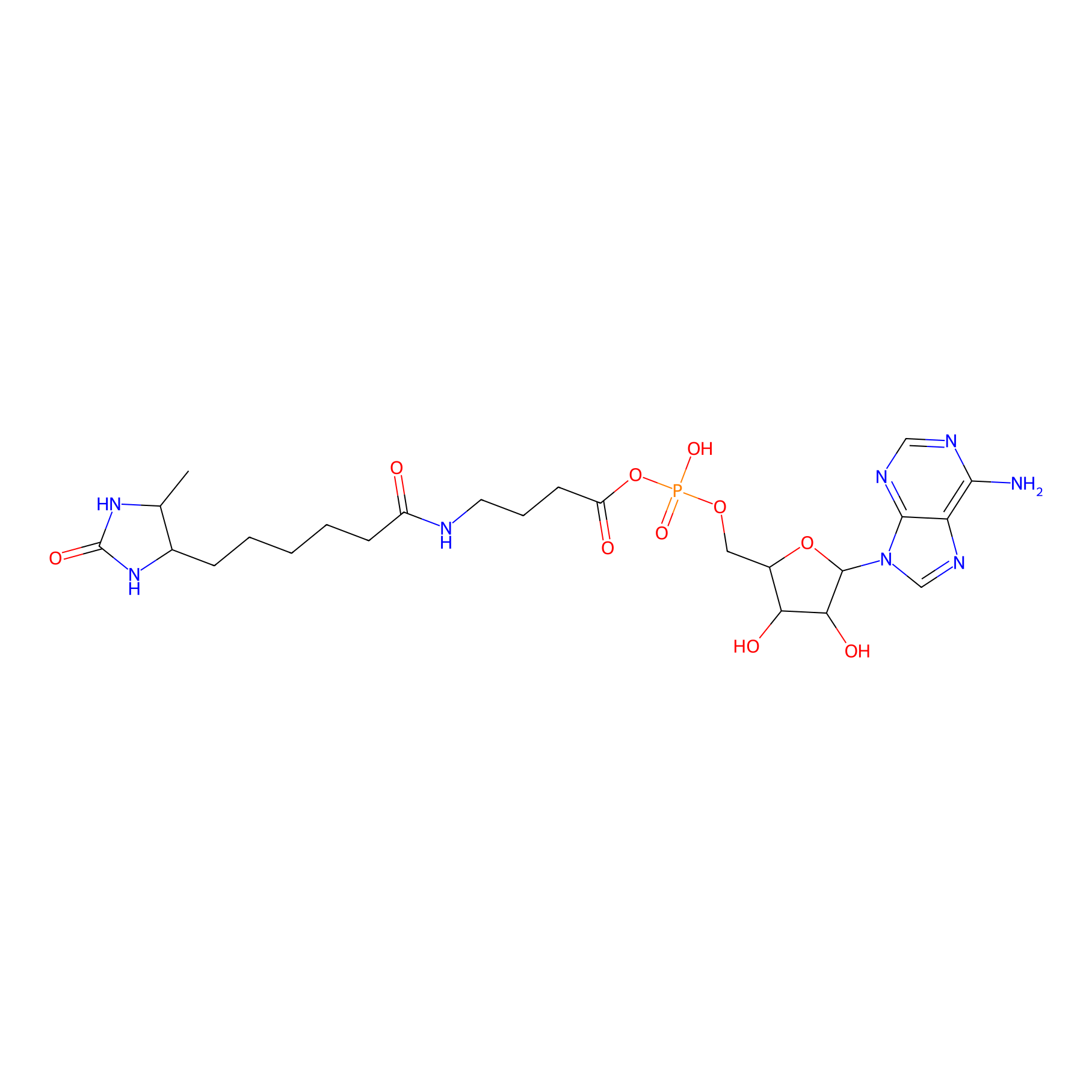 |
N.A. | LDD0200 | [9] | |
|
ATP probe Probe Info |
 |
K42(0.00); K109(0.00); K56(0.00); K57(0.00) | LDD0199 | [9] | |
|
m-APA Probe Info |
 |
H76(0.00); H32(0.00); H8(0.00) | LDD2231 | [8] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C69(0.00); C94(0.00) | LDD0038 | [10] | |
|
IA-alkyne Probe Info |
 |
C94(0.00); C69(0.00) | LDD0036 | [10] | |
|
IPIAA_L Probe Info |
 |
C94(0.00); C69(0.00) | LDD0031 | [11] | |
|
Lodoacetamide azide Probe Info |
 |
C69(0.00); C94(0.00) | LDD0037 | [10] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [12] | |
|
NAIA_4 Probe Info |
 |
C69(0.00); C94(0.00) | LDD2226 | [13] | |
|
IPM Probe Info |
 |
N.A. | LDD0147 | [12] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [14] | |
|
SF Probe Info |
 |
Y54(0.00); Y30(0.00) | LDD0028 | [15] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [14] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [12] | |
|
1c-yne Probe Info |
 |
K42(0.00); K91(0.00); K18(0.00) | LDD0228 | [16] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [17] | |
|
W1 Probe Info |
 |
N.A. | LDD0236 | [18] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [19] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [13] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C69(1.00) | LDD2117 | [3] |
| LDCM0563 | Abegg_cp(+)-11 | HeLa | C94(2.28) | LDD0314 | [12] |
| LDCM0214 | AC1 | HCT 116 | C69(0.88) | LDD0531 | [7] |
| LDCM0215 | AC10 | HCT 116 | C69(1.38) | LDD0532 | [7] |
| LDCM0216 | AC100 | HCT 116 | C69(1.61) | LDD0533 | [7] |
| LDCM0217 | AC101 | HCT 116 | C69(1.06) | LDD0534 | [7] |
| LDCM0218 | AC102 | HCT 116 | C69(1.06) | LDD0535 | [7] |
| LDCM0219 | AC103 | HCT 116 | C69(1.20) | LDD0536 | [7] |
| LDCM0220 | AC104 | HCT 116 | C69(1.08) | LDD0537 | [7] |
| LDCM0221 | AC105 | HCT 116 | C69(1.00) | LDD0538 | [7] |
| LDCM0222 | AC106 | HCT 116 | C69(1.04) | LDD0539 | [7] |
| LDCM0223 | AC107 | HCT 116 | C69(1.20) | LDD0540 | [7] |
| LDCM0224 | AC108 | HCT 116 | C69(0.96) | LDD0541 | [7] |
| LDCM0225 | AC109 | HCT 116 | C69(1.22) | LDD0542 | [7] |
| LDCM0226 | AC11 | HCT 116 | C69(1.22) | LDD0543 | [7] |
| LDCM0227 | AC110 | HCT 116 | C69(1.13) | LDD0544 | [7] |
| LDCM0228 | AC111 | HCT 116 | C69(0.91) | LDD0545 | [7] |
| LDCM0229 | AC112 | HCT 116 | C69(1.23) | LDD0546 | [7] |
| LDCM0230 | AC113 | HCT 116 | C69(0.80); C94(1.05) | LDD0547 | [7] |
| LDCM0231 | AC114 | HCT 116 | C69(1.17); C94(0.95) | LDD0548 | [7] |
| LDCM0232 | AC115 | HCT 116 | C69(1.19); C94(0.94) | LDD0549 | [7] |
| LDCM0233 | AC116 | HCT 116 | C69(0.97); C94(1.02) | LDD0550 | [7] |
| LDCM0234 | AC117 | HCT 116 | C69(1.26); C94(0.97) | LDD0551 | [7] |
| LDCM0235 | AC118 | HCT 116 | C69(1.06); C94(1.02) | LDD0552 | [7] |
| LDCM0236 | AC119 | HCT 116 | C69(1.07); C94(0.75) | LDD0553 | [7] |
| LDCM0237 | AC12 | HCT 116 | C69(1.11) | LDD0554 | [7] |
| LDCM0238 | AC120 | HCT 116 | C69(1.02); C94(0.85) | LDD0555 | [7] |
| LDCM0239 | AC121 | HCT 116 | C69(0.94); C94(1.03) | LDD0556 | [7] |
| LDCM0240 | AC122 | HCT 116 | C69(1.05); C94(0.89) | LDD0557 | [7] |
| LDCM0241 | AC123 | HCT 116 | C69(0.93); C94(0.87) | LDD0558 | [7] |
| LDCM0242 | AC124 | HCT 116 | C69(1.07); C94(1.21) | LDD0559 | [7] |
| LDCM0243 | AC125 | HCT 116 | C69(1.08); C94(1.00) | LDD0560 | [7] |
| LDCM0244 | AC126 | HCT 116 | C69(1.28); C94(0.96) | LDD0561 | [7] |
| LDCM0245 | AC127 | HCT 116 | C69(0.97); C94(0.98) | LDD0562 | [7] |
| LDCM0259 | AC14 | HCT 116 | C69(0.94) | LDD0576 | [7] |
| LDCM0263 | AC143 | HCT 116 | C69(0.98) | LDD0580 | [7] |
| LDCM0264 | AC144 | HCT 116 | C69(0.88) | LDD0581 | [7] |
| LDCM0265 | AC145 | HCT 116 | C69(1.30) | LDD0582 | [7] |
| LDCM0266 | AC146 | HCT 116 | C69(1.30) | LDD0583 | [7] |
| LDCM0267 | AC147 | HCT 116 | C69(0.92) | LDD0584 | [7] |
| LDCM0268 | AC148 | HCT 116 | C69(1.17) | LDD0585 | [7] |
| LDCM0269 | AC149 | HCT 116 | C69(1.21) | LDD0586 | [7] |
| LDCM0270 | AC15 | HCT 116 | C69(1.38) | LDD0587 | [7] |
| LDCM0271 | AC150 | HCT 116 | C69(0.98) | LDD0588 | [7] |
| LDCM0272 | AC151 | HCT 116 | C69(1.21) | LDD0589 | [7] |
| LDCM0273 | AC152 | HCT 116 | C69(1.11) | LDD0590 | [7] |
| LDCM0274 | AC153 | HCT 116 | C69(1.40) | LDD0591 | [7] |
| LDCM0621 | AC154 | HCT 116 | C69(1.43) | LDD2158 | [7] |
| LDCM0622 | AC155 | HCT 116 | C69(1.14) | LDD2159 | [7] |
| LDCM0623 | AC156 | HCT 116 | C69(1.12) | LDD2160 | [7] |
| LDCM0624 | AC157 | HCT 116 | C69(1.27) | LDD2161 | [7] |
| LDCM0276 | AC17 | HCT 116 | C69(0.85); C94(0.99) | LDD0593 | [7] |
| LDCM0277 | AC18 | HCT 116 | C94(0.74); C69(1.16) | LDD0594 | [7] |
| LDCM0278 | AC19 | HCT 116 | C69(0.86); C94(0.94) | LDD0595 | [7] |
| LDCM0279 | AC2 | HCT 116 | C69(0.93) | LDD0596 | [7] |
| LDCM0280 | AC20 | HCT 116 | C94(0.90); C69(0.91) | LDD0597 | [7] |
| LDCM0281 | AC21 | HCT 116 | C69(0.82); C94(0.95) | LDD0598 | [7] |
| LDCM0282 | AC22 | HCT 116 | C69(0.83); C94(0.95) | LDD0599 | [7] |
| LDCM0283 | AC23 | HCT 116 | C69(0.79); C94(0.80) | LDD0600 | [7] |
| LDCM0284 | AC24 | HCT 116 | C69(0.92); C94(0.97) | LDD0601 | [7] |
| LDCM0285 | AC25 | HCT 116 | C69(0.83) | LDD0602 | [7] |
| LDCM0286 | AC26 | HCT 116 | C69(0.90) | LDD0603 | [7] |
| LDCM0287 | AC27 | HCT 116 | C69(0.98) | LDD0604 | [7] |
| LDCM0288 | AC28 | HCT 116 | C69(1.00) | LDD0605 | [7] |
| LDCM0289 | AC29 | HCT 116 | C69(1.02) | LDD0606 | [7] |
| LDCM0290 | AC3 | HCT 116 | C69(1.04) | LDD0607 | [7] |
| LDCM0291 | AC30 | HCT 116 | C69(1.05) | LDD0608 | [7] |
| LDCM0292 | AC31 | HCT 116 | C69(1.03) | LDD0609 | [7] |
| LDCM0293 | AC32 | HCT 116 | C69(1.09) | LDD0610 | [7] |
| LDCM0294 | AC33 | HCT 116 | C69(0.89) | LDD0611 | [7] |
| LDCM0295 | AC34 | HCT 116 | C69(1.01) | LDD0612 | [7] |
| LDCM0296 | AC35 | HCT 116 | C69(0.71) | LDD0613 | [7] |
| LDCM0297 | AC36 | HCT 116 | C69(0.69) | LDD0614 | [7] |
| LDCM0298 | AC37 | HCT 116 | C69(0.84) | LDD0615 | [7] |
| LDCM0299 | AC38 | HCT 116 | C69(0.82) | LDD0616 | [7] |
| LDCM0300 | AC39 | HCT 116 | C69(0.89) | LDD0617 | [7] |
| LDCM0301 | AC4 | HCT 116 | C69(1.15) | LDD0618 | [7] |
| LDCM0302 | AC40 | HCT 116 | C69(0.89) | LDD0619 | [7] |
| LDCM0303 | AC41 | HCT 116 | C69(0.77) | LDD0620 | [7] |
| LDCM0304 | AC42 | HCT 116 | C69(1.03) | LDD0621 | [7] |
| LDCM0305 | AC43 | HCT 116 | C69(0.71) | LDD0622 | [7] |
| LDCM0306 | AC44 | HCT 116 | C69(1.11) | LDD0623 | [7] |
| LDCM0307 | AC45 | HCT 116 | C69(0.99) | LDD0624 | [7] |
| LDCM0308 | AC46 | PaTu 8988t | C69(1.07); C94(1.08) | LDD1187 | [7] |
| LDCM0309 | AC47 | PaTu 8988t | C69(1.79); C94(1.10) | LDD1188 | [7] |
| LDCM0310 | AC48 | PaTu 8988t | C69(1.32); C94(1.14) | LDD1189 | [7] |
| LDCM0311 | AC49 | PaTu 8988t | C69(1.38); C94(1.25) | LDD1190 | [7] |
| LDCM0312 | AC5 | HCT 116 | C69(1.39) | LDD0629 | [7] |
| LDCM0313 | AC50 | PaTu 8988t | C69(1.34); C94(1.15) | LDD1192 | [7] |
| LDCM0314 | AC51 | PaTu 8988t | C69(1.25); C94(0.51) | LDD1193 | [7] |
| LDCM0315 | AC52 | PaTu 8988t | C69(1.40); C94(1.00) | LDD1194 | [7] |
| LDCM0316 | AC53 | PaTu 8988t | C69(1.74); C94(1.16) | LDD1195 | [7] |
| LDCM0317 | AC54 | PaTu 8988t | C69(2.03); C94(1.16) | LDD1196 | [7] |
| LDCM0318 | AC55 | PaTu 8988t | C69(1.48); C94(0.83) | LDD1197 | [7] |
| LDCM0319 | AC56 | PaTu 8988t | C69(1.11); C94(0.85) | LDD1198 | [7] |
| LDCM0320 | AC57 | PaTu 8988t | C94(0.90) | LDD1199 | [7] |
| LDCM0321 | AC58 | PaTu 8988t | C94(0.89) | LDD1200 | [7] |
| LDCM0322 | AC59 | PaTu 8988t | C94(0.97) | LDD1201 | [7] |
| LDCM0323 | AC6 | HCT 116 | C69(1.55) | LDD0640 | [7] |
| LDCM0324 | AC60 | PaTu 8988t | C94(0.79) | LDD1203 | [7] |
| LDCM0325 | AC61 | PaTu 8988t | C94(0.79) | LDD1204 | [7] |
| LDCM0326 | AC62 | PaTu 8988t | C94(0.74) | LDD1205 | [7] |
| LDCM0327 | AC63 | PaTu 8988t | C94(0.89) | LDD1206 | [7] |
| LDCM0328 | AC64 | PaTu 8988t | C94(0.91) | LDD1207 | [7] |
| LDCM0329 | AC65 | PaTu 8988t | C94(0.78) | LDD1208 | [7] |
| LDCM0330 | AC66 | PaTu 8988t | C94(0.75) | LDD1209 | [7] |
| LDCM0331 | AC67 | PaTu 8988t | C94(0.84) | LDD1210 | [7] |
| LDCM0332 | AC68 | HCT 116 | C69(0.87) | LDD0649 | [7] |
| LDCM0333 | AC69 | HCT 116 | C69(0.77) | LDD0650 | [7] |
| LDCM0334 | AC7 | HCT 116 | C69(0.79) | LDD0651 | [7] |
| LDCM0335 | AC70 | HCT 116 | C69(0.79) | LDD0652 | [7] |
| LDCM0336 | AC71 | HCT 116 | C69(1.00) | LDD0653 | [7] |
| LDCM0337 | AC72 | HCT 116 | C69(0.91) | LDD0654 | [7] |
| LDCM0338 | AC73 | HCT 116 | C69(0.86) | LDD0655 | [7] |
| LDCM0339 | AC74 | HCT 116 | C69(0.90) | LDD0656 | [7] |
| LDCM0340 | AC75 | HCT 116 | C69(0.63) | LDD0657 | [7] |
| LDCM0341 | AC76 | HCT 116 | C69(0.65) | LDD0658 | [7] |
| LDCM0342 | AC77 | HCT 116 | C69(0.74) | LDD0659 | [7] |
| LDCM0343 | AC78 | HCT 116 | C69(0.95) | LDD0660 | [7] |
| LDCM0344 | AC79 | HCT 116 | C69(0.99) | LDD0661 | [7] |
| LDCM0345 | AC8 | HCT 116 | C69(1.18) | LDD0662 | [7] |
| LDCM0346 | AC80 | HCT 116 | C69(1.03) | LDD0663 | [7] |
| LDCM0347 | AC81 | HCT 116 | C69(0.67) | LDD0664 | [7] |
| LDCM0348 | AC82 | HCT 116 | C69(0.80) | LDD0665 | [7] |
| LDCM0349 | AC83 | HCT 116 | C69(1.04) | LDD0666 | [7] |
| LDCM0350 | AC84 | HCT 116 | C69(1.08) | LDD0667 | [7] |
| LDCM0351 | AC85 | HCT 116 | C69(0.93) | LDD0668 | [7] |
| LDCM0352 | AC86 | HCT 116 | C69(0.91) | LDD0669 | [7] |
| LDCM0353 | AC87 | HCT 116 | C69(1.00) | LDD0670 | [7] |
| LDCM0354 | AC88 | HCT 116 | C69(0.97) | LDD0671 | [7] |
| LDCM0355 | AC89 | HCT 116 | C69(1.09) | LDD0672 | [7] |
| LDCM0357 | AC90 | HCT 116 | C69(1.06) | LDD0674 | [7] |
| LDCM0358 | AC91 | HCT 116 | C69(0.99) | LDD0675 | [7] |
| LDCM0359 | AC92 | HCT 116 | C69(0.92) | LDD0676 | [7] |
| LDCM0360 | AC93 | HCT 116 | C69(0.85) | LDD0677 | [7] |
| LDCM0361 | AC94 | HCT 116 | C69(0.84) | LDD0678 | [7] |
| LDCM0362 | AC95 | HCT 116 | C69(0.87) | LDD0679 | [7] |
| LDCM0363 | AC96 | HCT 116 | C69(0.88) | LDD0680 | [7] |
| LDCM0364 | AC97 | HCT 116 | C69(1.11) | LDD0681 | [7] |
| LDCM0365 | AC98 | HCT 116 | C69(1.08) | LDD0682 | [7] |
| LDCM0366 | AC99 | HCT 116 | C69(0.93) | LDD0683 | [7] |
| LDCM0248 | AKOS034007472 | HCT 116 | C69(1.13) | LDD0565 | [7] |
| LDCM0356 | AKOS034007680 | HCT 116 | C69(1.49) | LDD0673 | [7] |
| LDCM0275 | AKOS034007705 | HCT 116 | C69(1.21) | LDD0592 | [7] |
| LDCM0020 | ARS-1620 | HCC44 | C69(1.01) | LDD2171 | [7] |
| LDCM0108 | Chloroacetamide | HeLa | C69(0.00); H32(0.00) | LDD0222 | [17] |
| LDCM0632 | CL-Sc | Hep-G2 | C69(2.02) | LDD2227 | [13] |
| LDCM0367 | CL1 | HCT 116 | C94(0.81) | LDD0684 | [7] |
| LDCM0368 | CL10 | HCT 116 | C94(0.84) | LDD0685 | [7] |
| LDCM0369 | CL100 | HCT 116 | C69(0.86) | LDD0686 | [7] |
| LDCM0370 | CL101 | HCT 116 | C69(1.40) | LDD0687 | [7] |
| LDCM0371 | CL102 | HCT 116 | C69(1.24) | LDD0688 | [7] |
| LDCM0372 | CL103 | HCT 116 | C69(1.11) | LDD0689 | [7] |
| LDCM0373 | CL104 | HCT 116 | C69(0.84) | LDD0690 | [7] |
| LDCM0374 | CL105 | HCT 116 | C69(0.78); C94(0.99) | LDD0691 | [7] |
| LDCM0375 | CL106 | HCT 116 | C94(0.94); C69(1.01) | LDD0692 | [7] |
| LDCM0376 | CL107 | HCT 116 | C94(1.03); C69(1.07) | LDD0693 | [7] |
| LDCM0377 | CL108 | HCT 116 | C69(1.10); C94(1.11) | LDD0694 | [7] |
| LDCM0378 | CL109 | HCT 116 | C69(0.88); C94(1.01) | LDD0695 | [7] |
| LDCM0379 | CL11 | HCT 116 | C94(0.95) | LDD0696 | [7] |
| LDCM0380 | CL110 | HCT 116 | C69(0.80); C94(1.22) | LDD0697 | [7] |
| LDCM0381 | CL111 | HCT 116 | C69(0.75); C94(0.92) | LDD0698 | [7] |
| LDCM0382 | CL112 | HCT 116 | C69(1.51) | LDD0699 | [7] |
| LDCM0383 | CL113 | HCT 116 | C69(1.10) | LDD0700 | [7] |
| LDCM0384 | CL114 | HCT 116 | C69(0.91) | LDD0701 | [7] |
| LDCM0385 | CL115 | HCT 116 | C69(1.05) | LDD0702 | [7] |
| LDCM0386 | CL116 | HCT 116 | C69(0.92) | LDD0703 | [7] |
| LDCM0387 | CL117 | HCT 116 | C69(1.14) | LDD0704 | [7] |
| LDCM0388 | CL118 | HCT 116 | C69(0.80) | LDD0705 | [7] |
| LDCM0389 | CL119 | HCT 116 | C69(1.01) | LDD0706 | [7] |
| LDCM0390 | CL12 | HCT 116 | C94(1.08) | LDD0707 | [7] |
| LDCM0391 | CL120 | HCT 116 | C69(0.86) | LDD0708 | [7] |
| LDCM0392 | CL121 | PaTu 8988t | C69(1.16); C94(0.98) | LDD1271 | [7] |
| LDCM0393 | CL122 | PaTu 8988t | C69(1.16); C94(1.05) | LDD1272 | [7] |
| LDCM0394 | CL123 | PaTu 8988t | C69(2.00); C94(1.05) | LDD1273 | [7] |
| LDCM0395 | CL124 | PaTu 8988t | C69(1.34); C94(1.12) | LDD1274 | [7] |
| LDCM0396 | CL125 | PaTu 8988t | C94(0.92) | LDD1275 | [7] |
| LDCM0397 | CL126 | PaTu 8988t | C94(0.92) | LDD1276 | [7] |
| LDCM0398 | CL127 | PaTu 8988t | C94(0.93) | LDD1277 | [7] |
| LDCM0399 | CL128 | PaTu 8988t | C94(0.83) | LDD1278 | [7] |
| LDCM0400 | CL13 | HCT 116 | C94(0.94) | LDD0717 | [7] |
| LDCM0401 | CL14 | HCT 116 | C94(0.91) | LDD0718 | [7] |
| LDCM0402 | CL15 | HCT 116 | C94(0.52) | LDD0719 | [7] |
| LDCM0403 | CL16 | HCT 116 | C69(1.29) | LDD0720 | [7] |
| LDCM0404 | CL17 | HCT 116 | C69(0.98) | LDD0721 | [7] |
| LDCM0405 | CL18 | HCT 116 | C69(0.80) | LDD0722 | [7] |
| LDCM0406 | CL19 | HCT 116 | C69(1.00) | LDD0723 | [7] |
| LDCM0407 | CL2 | HCT 116 | C94(0.78) | LDD0724 | [7] |
| LDCM0408 | CL20 | HCT 116 | C69(0.73) | LDD0725 | [7] |
| LDCM0409 | CL21 | HCT 116 | C69(0.88) | LDD0726 | [7] |
| LDCM0410 | CL22 | HCT 116 | C69(1.08) | LDD0727 | [7] |
| LDCM0411 | CL23 | HCT 116 | C69(0.99) | LDD0728 | [7] |
| LDCM0412 | CL24 | HCT 116 | C69(0.89) | LDD0729 | [7] |
| LDCM0413 | CL25 | HCT 116 | C69(0.72) | LDD0730 | [7] |
| LDCM0414 | CL26 | HCT 116 | C69(0.67) | LDD0731 | [7] |
| LDCM0415 | CL27 | HCT 116 | C69(1.06) | LDD0732 | [7] |
| LDCM0416 | CL28 | HCT 116 | C69(0.76) | LDD0733 | [7] |
| LDCM0417 | CL29 | HCT 116 | C69(0.93) | LDD0734 | [7] |
| LDCM0418 | CL3 | HCT 116 | C94(0.82) | LDD0735 | [7] |
| LDCM0419 | CL30 | HCT 116 | C69(0.84) | LDD0736 | [7] |
| LDCM0420 | CL31 | HCT 116 | C69(0.73); C94(0.86) | LDD0737 | [7] |
| LDCM0421 | CL32 | HCT 116 | C69(1.29); C94(0.77) | LDD0738 | [7] |
| LDCM0422 | CL33 | HCT 116 | C69(1.17); C94(0.89) | LDD0739 | [7] |
| LDCM0423 | CL34 | HCT 116 | C69(1.13); C94(1.07) | LDD0740 | [7] |
| LDCM0424 | CL35 | HCT 116 | C69(1.14); C94(1.14) | LDD0741 | [7] |
| LDCM0425 | CL36 | HCT 116 | C69(1.09); C94(1.11) | LDD0742 | [7] |
| LDCM0426 | CL37 | HCT 116 | C69(1.57); C94(1.34) | LDD0743 | [7] |
| LDCM0428 | CL39 | HCT 116 | C69(0.93); C94(1.18) | LDD0745 | [7] |
| LDCM0429 | CL4 | HCT 116 | C94(0.94) | LDD0746 | [7] |
| LDCM0430 | CL40 | HCT 116 | C69(1.28); C94(1.75) | LDD0747 | [7] |
| LDCM0431 | CL41 | HCT 116 | C69(1.38); C94(1.08) | LDD0748 | [7] |
| LDCM0432 | CL42 | HCT 116 | C69(2.46); C94(1.41) | LDD0749 | [7] |
| LDCM0433 | CL43 | HCT 116 | C69(1.08); C94(0.94) | LDD0750 | [7] |
| LDCM0434 | CL44 | HCT 116 | C69(1.70); C94(0.87) | LDD0751 | [7] |
| LDCM0435 | CL45 | HCT 116 | C69(1.06); C94(0.80) | LDD0752 | [7] |
| LDCM0436 | CL46 | HCT 116 | C69(0.70) | LDD0753 | [7] |
| LDCM0437 | CL47 | HCT 116 | C69(0.71) | LDD0754 | [7] |
| LDCM0438 | CL48 | HCT 116 | C69(0.75) | LDD0755 | [7] |
| LDCM0439 | CL49 | HCT 116 | C69(0.80) | LDD0756 | [7] |
| LDCM0440 | CL5 | HCT 116 | C94(0.90) | LDD0757 | [7] |
| LDCM0441 | CL50 | HCT 116 | C69(0.76) | LDD0758 | [7] |
| LDCM0442 | CL51 | HCT 116 | C69(0.72) | LDD0759 | [7] |
| LDCM0443 | CL52 | HCT 116 | C69(0.86) | LDD0760 | [7] |
| LDCM0444 | CL53 | HCT 116 | C69(0.84) | LDD0761 | [7] |
| LDCM0445 | CL54 | HCT 116 | C69(0.93) | LDD0762 | [7] |
| LDCM0446 | CL55 | HCT 116 | C69(0.81) | LDD0763 | [7] |
| LDCM0447 | CL56 | HCT 116 | C69(0.88) | LDD0764 | [7] |
| LDCM0448 | CL57 | HCT 116 | C69(0.92) | LDD0765 | [7] |
| LDCM0449 | CL58 | HCT 116 | C69(0.89) | LDD0766 | [7] |
| LDCM0450 | CL59 | HCT 116 | C69(0.71) | LDD0767 | [7] |
| LDCM0451 | CL6 | HCT 116 | C94(0.84) | LDD0768 | [7] |
| LDCM0452 | CL60 | HCT 116 | C69(0.62) | LDD0769 | [7] |
| LDCM0453 | CL61 | HCT 116 | C69(0.99); C94(1.10) | LDD0770 | [7] |
| LDCM0454 | CL62 | HCT 116 | C69(0.66); C94(1.09) | LDD0771 | [7] |
| LDCM0455 | CL63 | HCT 116 | C69(0.98); C94(1.07) | LDD0772 | [7] |
| LDCM0456 | CL64 | HCT 116 | C69(0.84); C94(1.15) | LDD0773 | [7] |
| LDCM0457 | CL65 | HCT 116 | C69(1.21); C94(1.08) | LDD0774 | [7] |
| LDCM0458 | CL66 | HCT 116 | C69(1.05); C94(1.26) | LDD0775 | [7] |
| LDCM0459 | CL67 | HCT 116 | C69(1.35); C94(1.41) | LDD0776 | [7] |
| LDCM0460 | CL68 | HCT 116 | C69(1.00); C94(1.09) | LDD0777 | [7] |
| LDCM0461 | CL69 | HCT 116 | C69(0.81); C94(0.93) | LDD0778 | [7] |
| LDCM0462 | CL7 | HCT 116 | C94(0.89) | LDD0779 | [7] |
| LDCM0463 | CL70 | HCT 116 | C69(0.88); C94(0.92) | LDD0780 | [7] |
| LDCM0464 | CL71 | HCT 116 | C69(1.74); C94(1.08) | LDD0781 | [7] |
| LDCM0465 | CL72 | HCT 116 | C69(1.20); C94(1.07) | LDD0782 | [7] |
| LDCM0466 | CL73 | HCT 116 | C69(0.75); C94(1.05) | LDD0783 | [7] |
| LDCM0467 | CL74 | HCT 116 | C69(1.08); C94(1.15) | LDD0784 | [7] |
| LDCM0469 | CL76 | HCT 116 | C69(1.09) | LDD0786 | [7] |
| LDCM0470 | CL77 | HCT 116 | C69(1.75) | LDD0787 | [7] |
| LDCM0471 | CL78 | HCT 116 | C69(1.40) | LDD0788 | [7] |
| LDCM0472 | CL79 | HCT 116 | C69(1.43) | LDD0789 | [7] |
| LDCM0473 | CL8 | HCT 116 | C94(1.07) | LDD0790 | [7] |
| LDCM0474 | CL80 | HCT 116 | C69(1.11) | LDD0791 | [7] |
| LDCM0475 | CL81 | HCT 116 | C69(1.49) | LDD0792 | [7] |
| LDCM0476 | CL82 | HCT 116 | C69(1.66) | LDD0793 | [7] |
| LDCM0477 | CL83 | HCT 116 | C69(1.36) | LDD0794 | [7] |
| LDCM0478 | CL84 | HCT 116 | C69(1.53) | LDD0795 | [7] |
| LDCM0479 | CL85 | HCT 116 | C69(1.25) | LDD0796 | [7] |
| LDCM0480 | CL86 | HCT 116 | C69(1.39) | LDD0797 | [7] |
| LDCM0481 | CL87 | HCT 116 | C69(1.41) | LDD0798 | [7] |
| LDCM0482 | CL88 | HCT 116 | C69(1.46) | LDD0799 | [7] |
| LDCM0483 | CL89 | HCT 116 | C69(1.83) | LDD0800 | [7] |
| LDCM0484 | CL9 | HCT 116 | C94(0.94) | LDD0801 | [7] |
| LDCM0485 | CL90 | HCT 116 | C69(0.78) | LDD0802 | [7] |
| LDCM0486 | CL91 | HCT 116 | C69(1.28) | LDD0803 | [7] |
| LDCM0487 | CL92 | HCT 116 | C69(1.03) | LDD0804 | [7] |
| LDCM0488 | CL93 | HCT 116 | C69(0.90) | LDD0805 | [7] |
| LDCM0489 | CL94 | HCT 116 | C69(1.02) | LDD0806 | [7] |
| LDCM0490 | CL95 | HCT 116 | C69(1.17) | LDD0807 | [7] |
| LDCM0491 | CL96 | HCT 116 | C69(1.07) | LDD0808 | [7] |
| LDCM0492 | CL97 | HCT 116 | C69(1.03) | LDD0809 | [7] |
| LDCM0493 | CL98 | HCT 116 | C69(1.03) | LDD0810 | [7] |
| LDCM0494 | CL99 | HCT 116 | C69(1.00) | LDD0811 | [7] |
| LDCM0495 | E2913 | HEK-293T | C94(0.84) | LDD1698 | [20] |
| LDCM0468 | Fragment33 | HCT 116 | C69(1.04); C94(1.04) | LDD0785 | [7] |
| LDCM0427 | Fragment51 | HCT 116 | C69(1.05); C94(1.37) | LDD0744 | [7] |
| LDCM0116 | HHS-0101 | DM93 | Y54(0.87); Y79(0.99); Y30(1.14) | LDD0264 | [5] |
| LDCM0117 | HHS-0201 | DM93 | Y79(0.66); Y54(0.79); Y30(4.25) | LDD0265 | [5] |
| LDCM0118 | HHS-0301 | DM93 | Y54(0.51); Y79(0.78); Y30(1.66) | LDD0266 | [5] |
| LDCM0119 | HHS-0401 | DM93 | Y54(0.50); Y79(0.71); Y30(2.10) | LDD0267 | [5] |
| LDCM0120 | HHS-0701 | DM93 | Y79(0.75); Y30(1.01); Y54(1.19) | LDD0268 | [5] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [17] |
| LDCM0123 | JWB131 | DM93 | Y79(0.99) | LDD0285 | [4] |
| LDCM0124 | JWB142 | DM93 | Y79(0.70) | LDD0286 | [4] |
| LDCM0125 | JWB146 | DM93 | Y79(1.03) | LDD0287 | [4] |
| LDCM0126 | JWB150 | DM93 | Y79(4.58) | LDD0288 | [4] |
| LDCM0127 | JWB152 | DM93 | Y79(2.69) | LDD0289 | [4] |
| LDCM0128 | JWB198 | DM93 | Y79(1.63) | LDD0290 | [4] |
| LDCM0129 | JWB202 | DM93 | Y79(0.65) | LDD0291 | [4] |
| LDCM0130 | JWB211 | DM93 | Y79(1.51) | LDD0292 | [4] |
| LDCM0022 | KB02 | HEK-293T | C94(1.01) | LDD1492 | [20] |
| LDCM0023 | KB03 | HEK-293T | C94(0.90) | LDD1497 | [20] |
| LDCM0024 | KB05 | UACC257 | C94(1.73) | LDD3325 | [21] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [17] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C69(1.08) | LDD2134 | [3] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C69(1.22) | LDD2137 | [3] |
| LDCM0131 | RA190 | MM1.R | C94(1.21) | LDD0304 | [22] |
| LDCM0021 | THZ1 | HCT 116 | C69(1.01) | LDD2173 | [7] |
The Interaction Atlas With This Target
References
