Details of the Target
General Information of Target
| Target ID | LDTP11281 | |||||
|---|---|---|---|---|---|---|
| Target Name | Voltage-gated monoatomic cation channel TMEM109 (TMEM109) | |||||
| Gene Name | TMEM109 | |||||
| Gene ID | 79073 | |||||
| Synonyms |
Voltage-gated monoatomic cation channel TMEM109; Mitsugumin-23; Mg23; Transmembrane protein 109 |
|||||
| 3D Structure | ||||||
| Sequence |
MNTSPGTVGSDPVILATAGYDHTVRFWQAHSGICTRTVQHQDSQVNALEVTPDRSMIAAA
GYQHIRMYDLNSNNPNPIISYDGVNKNIASVGFHEDGRWMYTGGEDCTARIWDLRSRNLQ CQRIFQVNAPINCVCLHPNQAELIVGDQSGAIHIWDLKTDHNEQLIPEPEVSITSAHIDP DASYMAAVNSTGNCYVWNLTGGIGDEVTQLIPKTKIPAHTRYALQCRFSPDSTLLATCSA DQTCKIWRTSNFSLMTELSIKSGNPGESSRGWMWGCAFSGDSQYIVTASSDNLARLWCVE TGEIKREYGGHQKAVVCLAFNDSVLG |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Subcellular location |
Nucleus outer membrane
|
|||||
| Function | Functions as a voltage-gated monoatomic cation channel permeable to both potassium and calcium. Plays a role in the cellular response to DNA damage. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C-Sul Probe Info |
 |
3.16 | LDD0066 | [1] | |
|
ONAyne Probe Info |
 |
N.A. | LDD0273 | [2] | |
|
OPA-S-S-alkyne Probe Info |
 |
K46(3.24); K215(3.67) | LDD3494 | [3] | |
|
HPAP Probe Info |
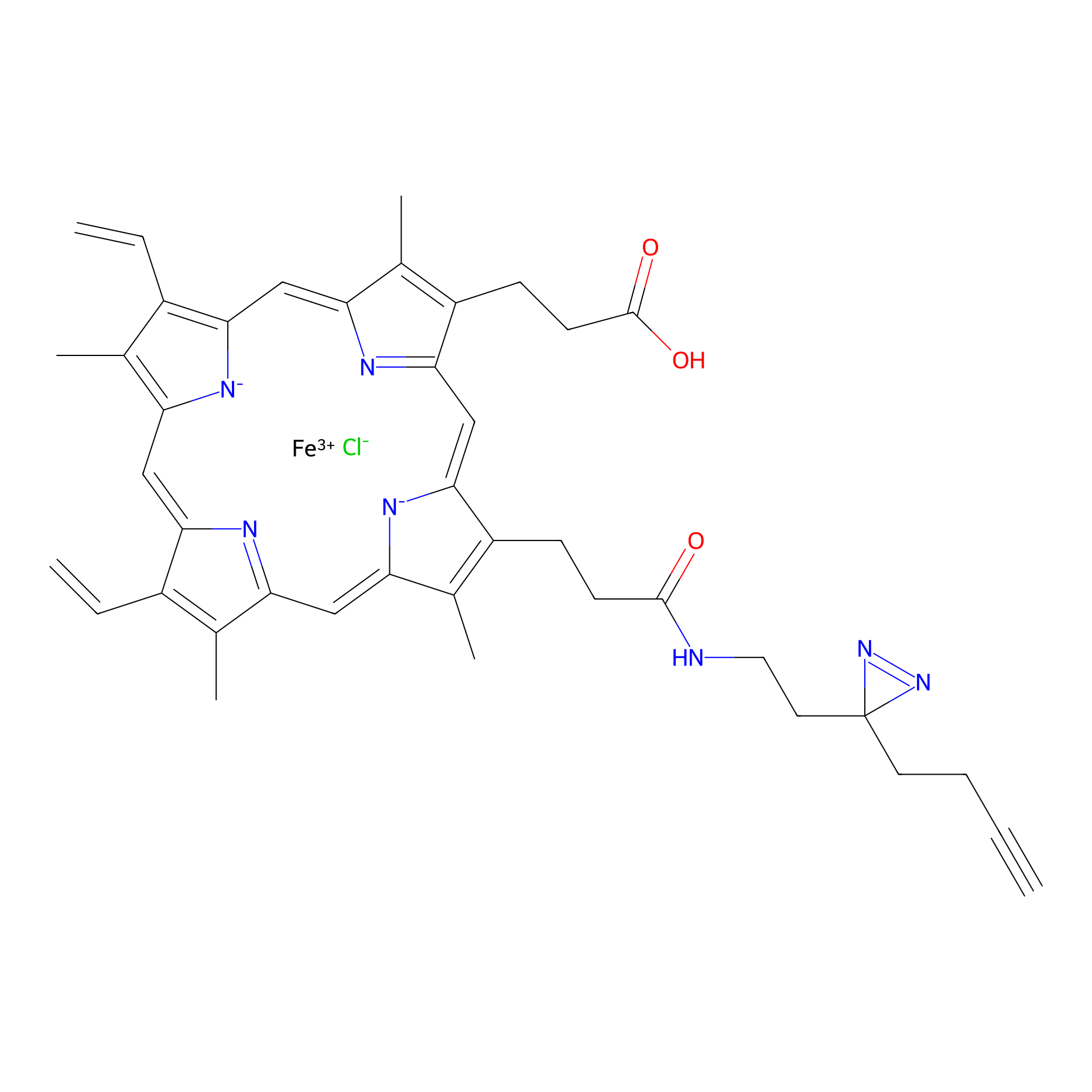 |
4.70 | LDD0062 | [4] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [5] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C040 Probe Info |
 |
11.39 | LDD1740 | [6] | |
|
C056 Probe Info |
 |
16.22 | LDD1753 | [6] | |
|
C094 Probe Info |
 |
32.67 | LDD1785 | [6] | |
|
C112 Probe Info |
 |
20.25 | LDD1799 | [6] | |
|
C218 Probe Info |
 |
11.47 | LDD1892 | [6] | |
|
C228 Probe Info |
 |
15.24 | LDD1901 | [6] | |
|
C231 Probe Info |
 |
37.27 | LDD1904 | [6] | |
|
C232 Probe Info |
 |
35.75 | LDD1905 | [6] | |
|
C235 Probe Info |
 |
39.67 | LDD1908 | [6] | |
|
C289 Probe Info |
 |
32.22 | LDD1959 | [6] | |
|
C349 Probe Info |
 |
9.71 | LDD2010 | [6] | |
|
C350 Probe Info |
 |
35.02 | LDD2011 | [6] | |
|
C362 Probe Info |
 |
31.12 | LDD2023 | [6] | |
|
OEA-DA Probe Info |
 |
5.94 | LDD0046 | [7] | |
Competitor(s) Related to This Target
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
Other
References
