Details of the Probe
General Information of Probe
The Probe Interaction Atlas
Target(s) List of this Probe
|
34 Enzyme Labeled by This Probe
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
18 Transporter and channel Labeled by This Probe
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
8 Other Labeled by This Probe
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Full Information of The Labelling Profiles of This Probe
Quantification: AfBPP Probe vs Negative Probe
Experiment 1 Reporting the Labelling Profiles of This Probe
Probe concentration
Quantitative Method
In Vitro Experiment Model
Note

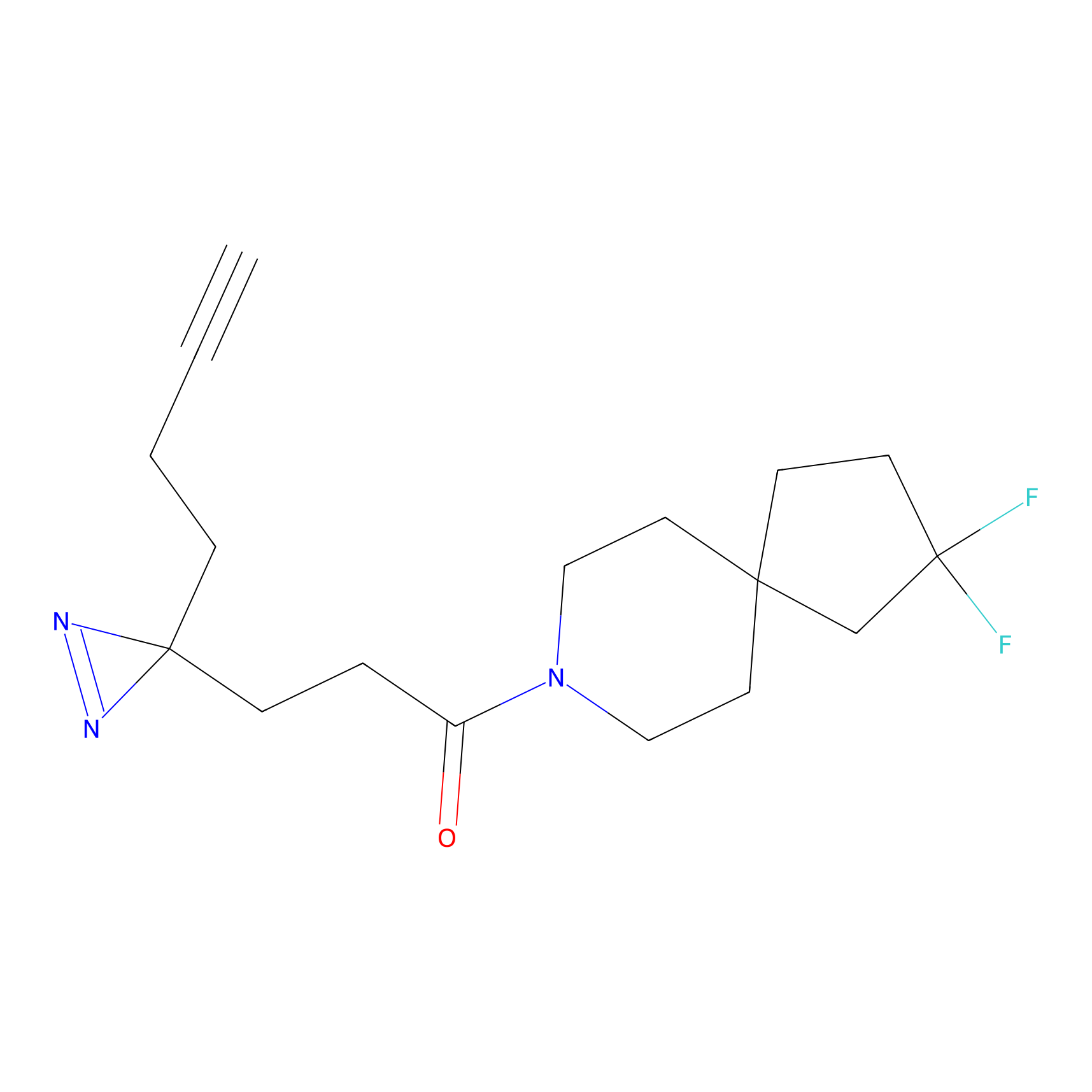


 Download The Altas
Download The Altas