Details of the Target
General Information of Target
| Target ID | LDTP02815 | |||||
|---|---|---|---|---|---|---|
| Target Name | Desmoplakin (DSP) | |||||
| Gene Name | DSP | |||||
| Gene ID | 1832 | |||||
| Synonyms |
Desmoplakin; DP; 250/210 kDa paraneoplastic pemphigus antigen |
|||||
| 3D Structure | ||||||
| Sequence |
MSCNGGSHPRINTLGRMIRAESGPDLRYEVTSGGGGTSRMYYSRRGVITDQNSDGYCQTG
TMSRHQNQNTIQELLQNCSDCLMRAELIVQPELKYGDGIQLTRSRELDECFAQANDQMEI LDSLIREMRQMGQPCDAYQKRLLQLQEQMRALYKAISVPRVRRASSKGGGGYTCQSGSGW DEFTKHVTSECLGWMRQQRAEMDMVAWGVDLASVEQHINSHRGIHNSIGDYRWQLDKIKA DLREKSAIYQLEEEYENLLKASFERMDHLRQLQNIIQATSREIMWINDCEEEELLYDWSD KNTNIAQKQEAFSIRMSQLEVKEKELNKLKQESDQLVLNQHPASDKIEAYMDTLQTQWSW ILQITKCIDVHLKENAAYFQFFEEAQSTEAYLKGLQDSIRKKYPCDKNMPLQHLLEQIKE LEKEREKILEYKRQVQNLVNKSKKIVQLKPRNPDYRSNKPIILRALCDYKQDQKIVHKGD ECILKDNNERSKWYVTGPGGVDMLVPSVGLIIPPPNPLAVDLSCKIEQYYEAILALWNQL YINMKSLVSWHYCMIDIEKIRAMTIAKLKTMRQEDYMKTIADLELHYQEFIRNSQGSEMF GDDDKRKIQSQFTDAQKHYQTLVIQLPGYPQHQTVTTTEITHHGTCQDVNHNKVIETNRE NDKQETWMLMELQKIRRQIEHCEGRMTLKNLPLADQGSSHHITVKINELKSVQNDSQAIA EVLNQLKDMLANFRGSEKYCYLQNEVFGLFQKLENINGVTDGYLNSLCTVRALLQAILQT EDMLKVYEARLTEEETVCLDLDKVEAYRCGLKKIKNDLNLKKSLLATMKTELQKAQQIHS QTSQQYPLYDLDLGKFGEKVTQLTDRWQRIDKQIDFRLWDLEKQIKQLRNYRDNYQAFCK WLYDAKRRQDSLESMKFGDSNTVMRFLNEQKNLHSEISGKRDKSEEVQKIAELCANSIKD YELQLASYTSGLETLLNIPIKRTMIQSPSGVILQEAADVHARYIELLTRSGDYYRFLSEM LKSLEDLKLKNTKIEVLEEELRLARDANSENCNKNKFLDQNLQKYQAECSQFKAKLASLE ELKRQAELDGKSAKQNLDKCYGQIKELNEKITRLTYEIEDEKRRRKSVEDRFDQQKNDYD QLQKARQCEKENLGWQKLESEKAIKEKEYEIERLRVLLQEEGTRKREYENELAKVRNHYN EEMSNLRNKYETEINITKTTIKEISMQKEDDSKNLRNQLDRLSRENRDLKDEIVRLNDSI LQATEQRRRAEENALQQKACGSEIMQKKQHLEIELKQVMQQRSEDNARHKQSLEEAAKTI QDKNKEIERLKAEFQEEAKRRWEYENELSKVRNNYDEEIISLKNQFETEINITKTTIHQL TMQKEEDTSGYRAQIDNLTRENRSLSEEIKRLKNTLTQTTENLRRVEEDIQQQKATGSEV SQRKQQLEVELRQVTQMRTEESVRYKQSLDDAAKTIQDKNKEIERLKQLIDKETNDRKCL EDENARLQRVQYDLQKANSSATETINKLKVQEQELTRLRIDYERVSQERTVKDQDITRFQ NSLKELQLQKQKVEEELNRLKRTASEDSCKRKKLEEELEGMRRSLKEQAIKITNLTQQLE QASIVKKRSEDDLRQQRDVLDGHLREKQRTQEELRRLSSEVEALRRQLLQEQESVKQAHL RNEHFQKAIEDKSRSLNESKIEIERLQSLTENLTKEHLMLEEELRNLRLEYDDLRRGRSE ADSDKNATILELRSQLQISNNRTLELQGLINDLQRERENLRQEIEKFQKQALEASNRIQE SKNQCTQVVQERESLLVKIKVLEQDKARLQRLEDELNRAKSTLEAETRVKQRLECEKQQI QNDLNQWKTQYSRKEEAIRKIESEREKSEREKNSLRSEIERLQAEIKRIEERCRRKLEDS TRETQSQLETERSRYQREIDKLRQRPYGSHRETQTECEWTVDTSKLVFDGLRKKVTAMQL YECQLIDKTTLDKLLKGKKSVEEVASEIQPFLRGAGSIAGASASPKEKYSLVEAKRKKLI SPESTVMLLEAQAATGGIIDPHRNEKLTVDSAIARDLIDFDDRQQIYAAEKAITGFDDPF SGKTVSVSEAIKKNLIDRETGMRLLEAQIASGGVVDPVNSVFLPKDVALARGLIDRDLYR SLNDPRDSQKNFVDPVTKKKVSYVQLKERCRIEPHTGLLLLSVQKRSMSFQGIRQPVTVT ELVDSGILRPSTVNELESGQISYDEVGERIKDFLQGSSCIAGIYNETTKQKLGIYEAMKI GLVRPGTALELLEAQAATGFIVDPVSNLRLPVEEAYKRGLVGIEFKEKLLSAERAVTGYN DPETGNIISLFQAMNKELIEKGHGIRLLEAQIATGGIIDPKESHRLPVDIAYKRGYFNEE LSEILSDPSDDTKGFFDPNTEENLTYLQLKERCIKDEETGLCLLPLKEKKKQVQTSQKNT LRKRRVVIVDPETNKEMSVQEAYKKGLIDYETFKELCEQECEWEEITITGSDGSTRVVLV DRKTGSQYDIQDAIDKGLVDRKFFDQYRSGSLSLTQFADMISLKNGVGTSSSMGSGVSDD VFSSSRHESVSKISTISSVRNLTIRSSSFSDTLEESSPIAAIFDTENLEKISITEGIERG IVDSITGQRLLEAQACTGGIIHPTTGQKLSLQDAVSQGVIDQDMATRLKPAQKAFIGFEG VKGKKKMSAAEAVKEKWLPYEAGQRFLEFQYLTGGLVDPEVHGRISTEEAIRKGFIDGRA AQRLQDTSSYAKILTCPKTKLKISYKDAINRSMVEDITGLRLLEAASVSSKGLPSPYNMS SAPGSRSGSRSGSRSGSRSGSRSGSRRGSFDATGNSSYSYSYSFSSSSIGH |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
Plakin or cytolinker family
|
|||||
| Subcellular location |
Cell junction, desmosome
|
|||||
| Function |
Major high molecular weight protein of desmosomes. Regulates profibrotic gene expression in cardiomyocytes via activation of the MAPK14/p38 MAPK signaling cascade and increase in TGFB1 protein abundance.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| 22RV1 | SNV: p.A311T | DBIA Probe Info | |||
| A204 | SNV: p.E683D | DBIA Probe Info | |||
| CCK81 | SNV: p.W550Ter | DBIA Probe Info | |||
| CHL1 | SNV: p.G1154D | . | |||
| COLO792 | SNV: p.P24Q | DBIA Probe Info | |||
| DU145 | SNV: p.A951T | DBIA Probe Info | |||
| EGI1 | SNV: p.R1762W | DBIA Probe Info | |||
| HCC44 | SNV: p.Q175H | DBIA Probe Info | |||
| HCT15 | SNV: p.I120T; p.W901Ter; p.I1610T | DBIA Probe Info | |||
| HEC1 | SNV: p.Q1179R | DBIA Probe Info | |||
| HEC1B | SNV: p.L411M; p.Q1179R | IA-alkyne Probe Info | |||
| HT | SNV: p.L510P | . | |||
| HT115 | SNV: p.E2193K | DBIA Probe Info | |||
| ICC137 | SNV: p.D406N | DBIA Probe Info | |||
| ICC4 | SNV: p.A200V | DBIA Probe Info | |||
| KYSE510 | SNV: p.Y391N | DBIA Probe Info | |||
| LN18 | SNV: p.F917C | . | |||
| MDST8 | SNV: p.E1294Ter | DBIA Probe Info | |||
| MEC1 | SNV: p.G697Ter | . | |||
| MFE319 | SNV: p.L392P | DBIA Probe Info | |||
| NCIH446 | SNV: p.E1716Ter | DBIA Probe Info | |||
| OVTOKO | SNV: p.Q841E | DBIA Probe Info | |||
| P31FUJ | SNV: p.T184A; p.C2259G | DBIA Probe Info | |||
| REH | SNV: p.K432R | . | |||
| RKO | Insertion: p.K1892RfsTer38 SNV: . |
DBIA Probe Info | |||
| RL952 | Deletion: p.K1054del | DBIA Probe Info | |||
| SAOS2 | SNV: p.Q58Ter | . | |||
| SF295 | SNV: p.V1530I | DBIA Probe Info | |||
| SKMEL5 | SNV: p.V1808M | DBIA Probe Info | |||
| SW1783 | SNV: p.R1738Ter | . | |||
| U118MG | SNV: p.K906Ter | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
P1 Probe Info |
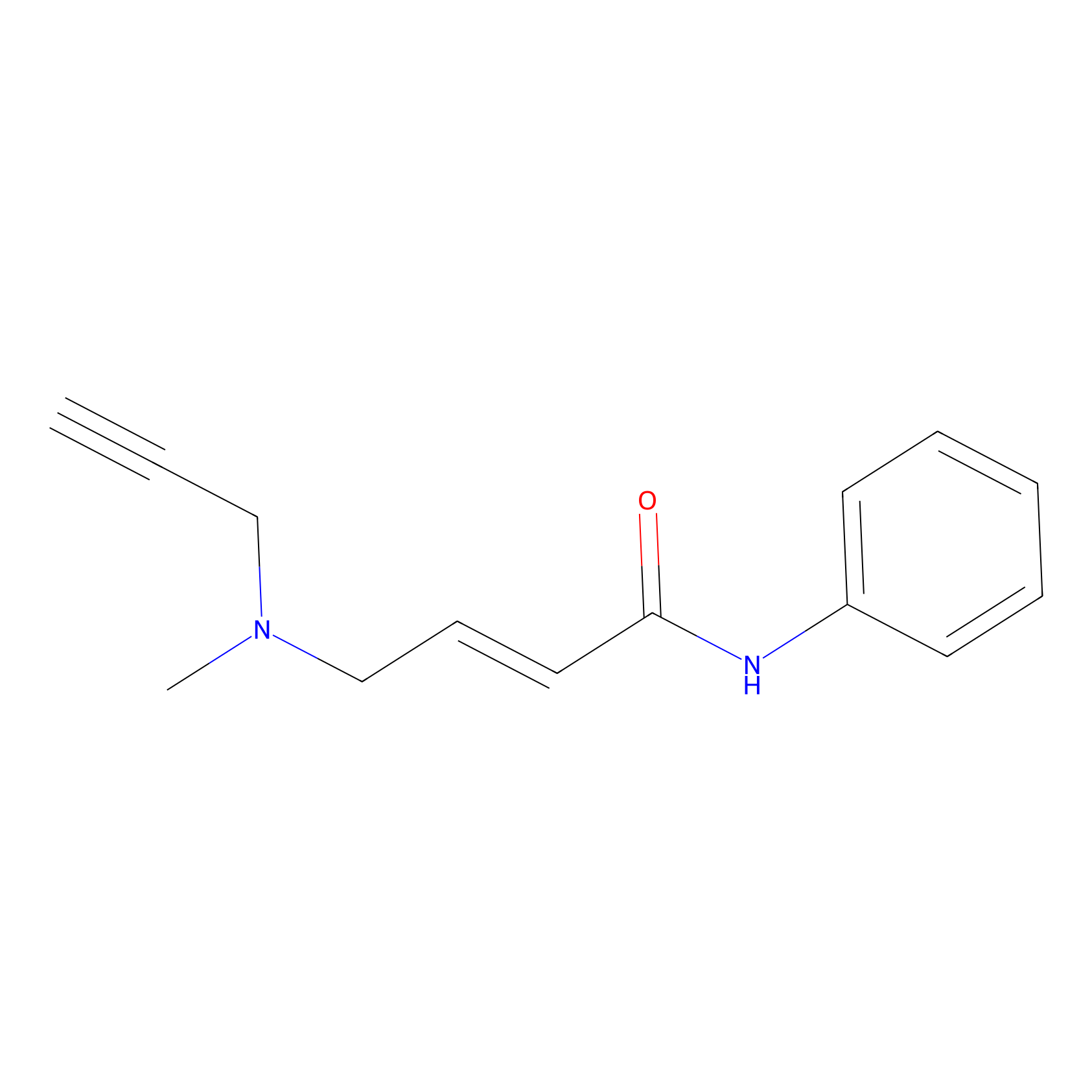 |
7.69 | LDD0452 | [1] | |
|
P2 Probe Info |
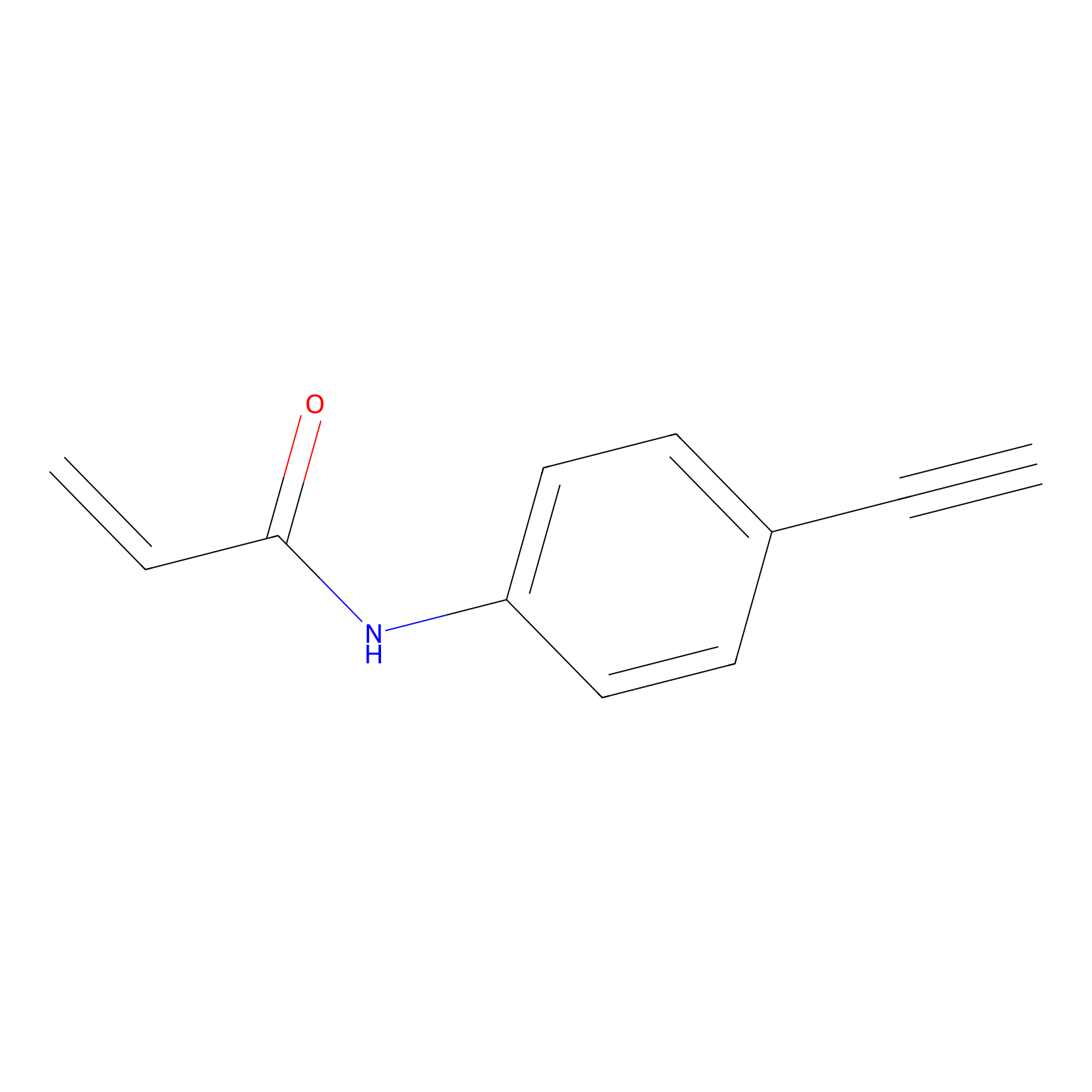 |
10.00 | LDD0453 | [1] | |
|
P3 Probe Info |
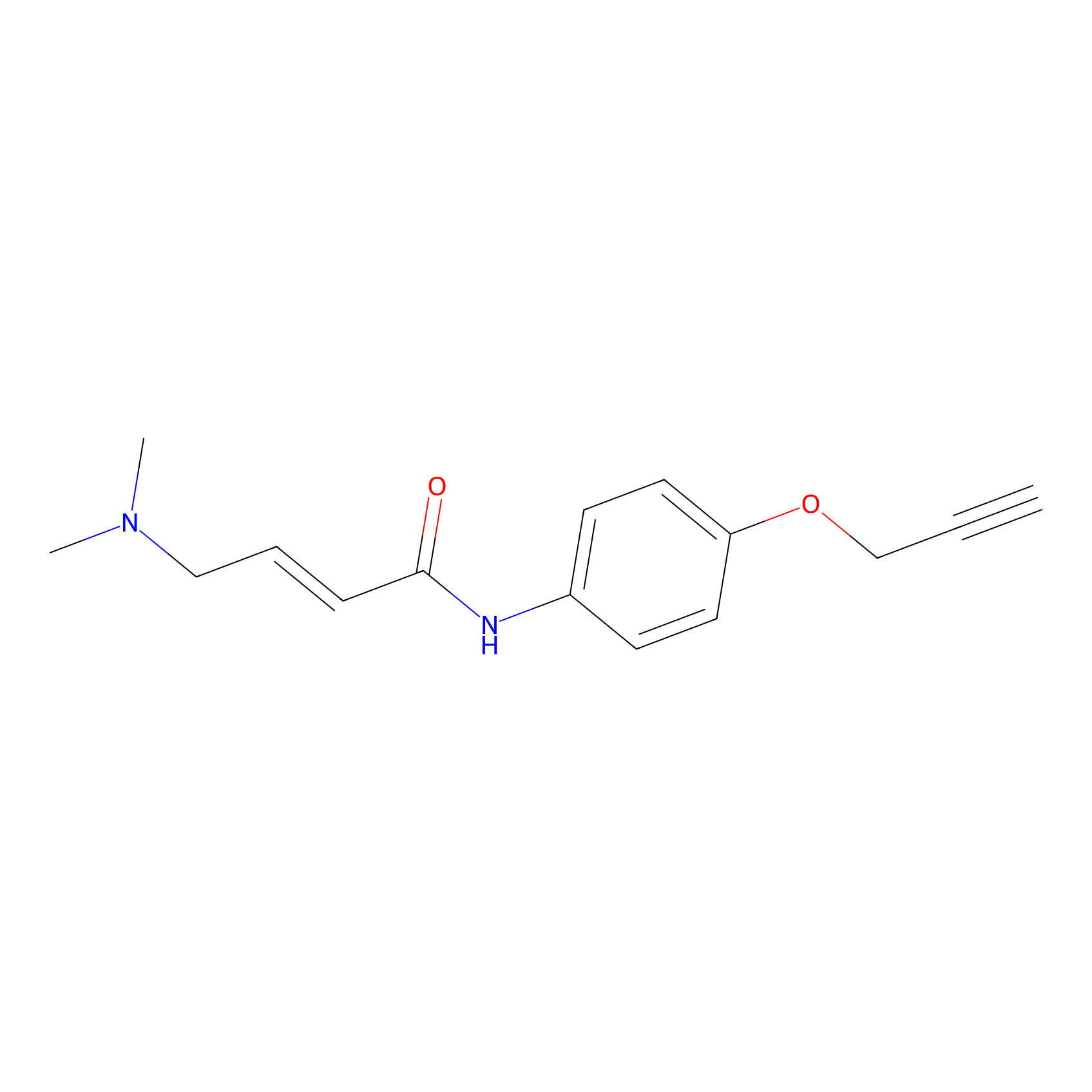 |
7.72 | LDD0454 | [1] | |
|
11RK72 Probe Info |
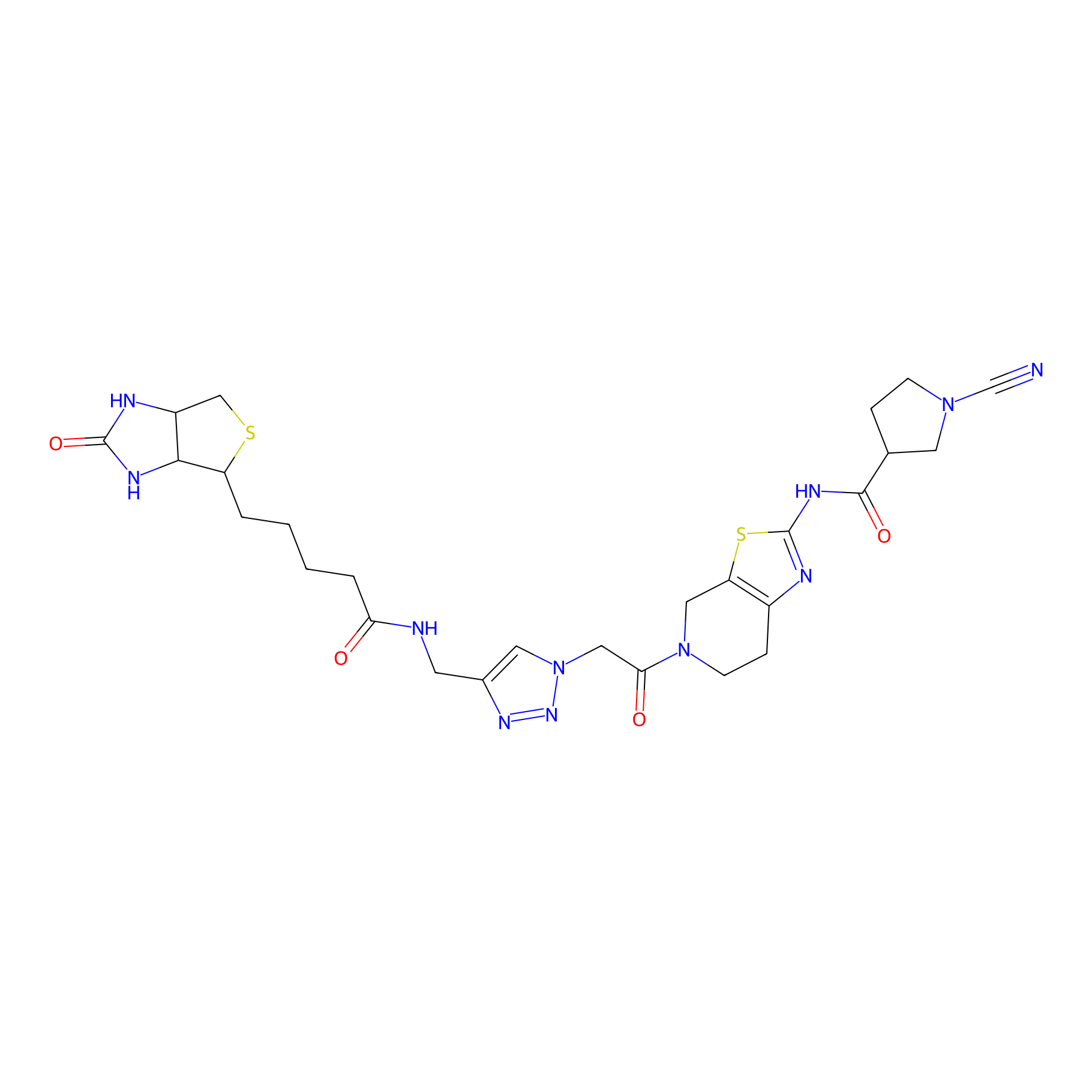 |
3.13 | LDD0327 | [2] | |
|
11RK73 Probe Info |
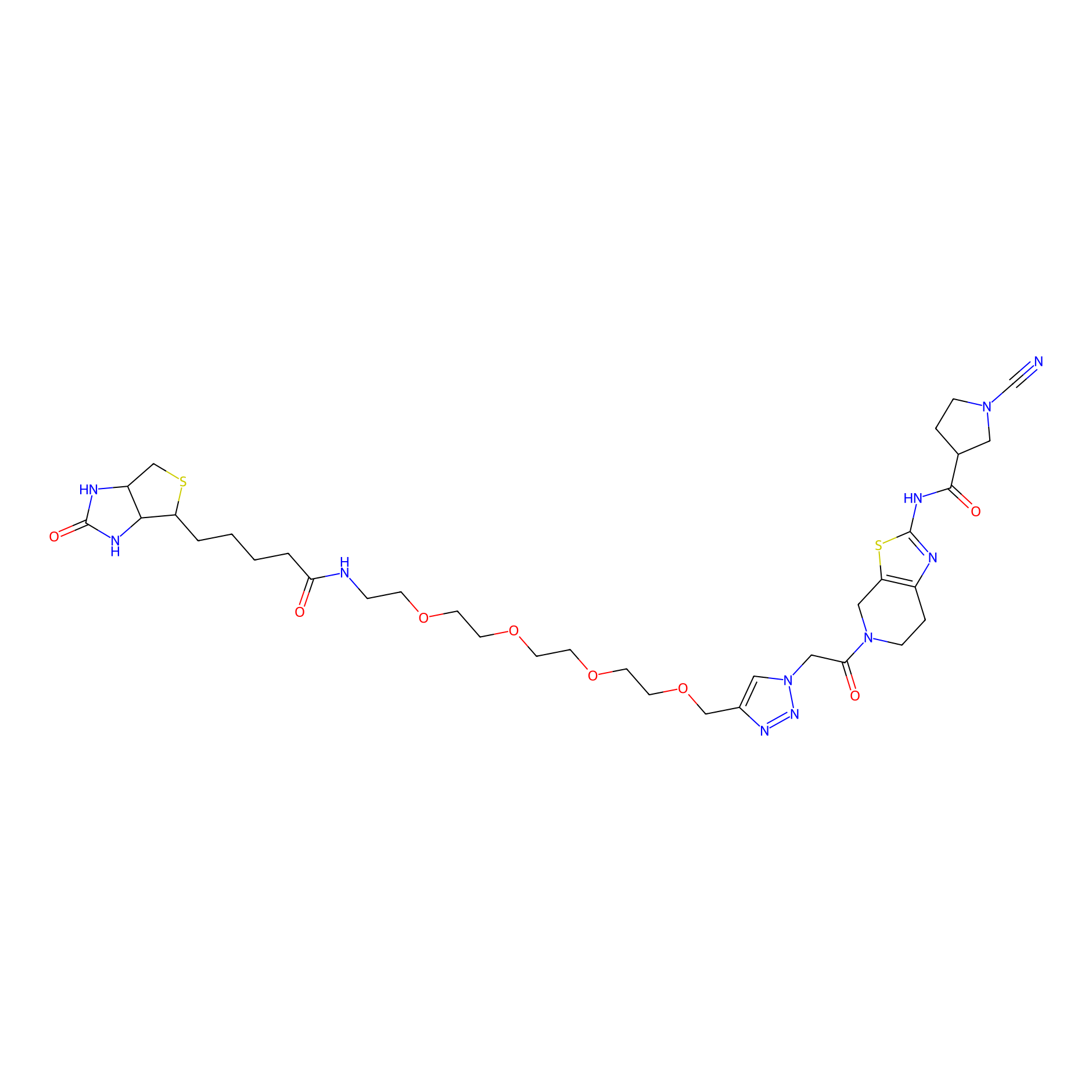 |
3.44 | LDD0328 | [2] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [3] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [3] | |
|
Nap-2 Probe Info |
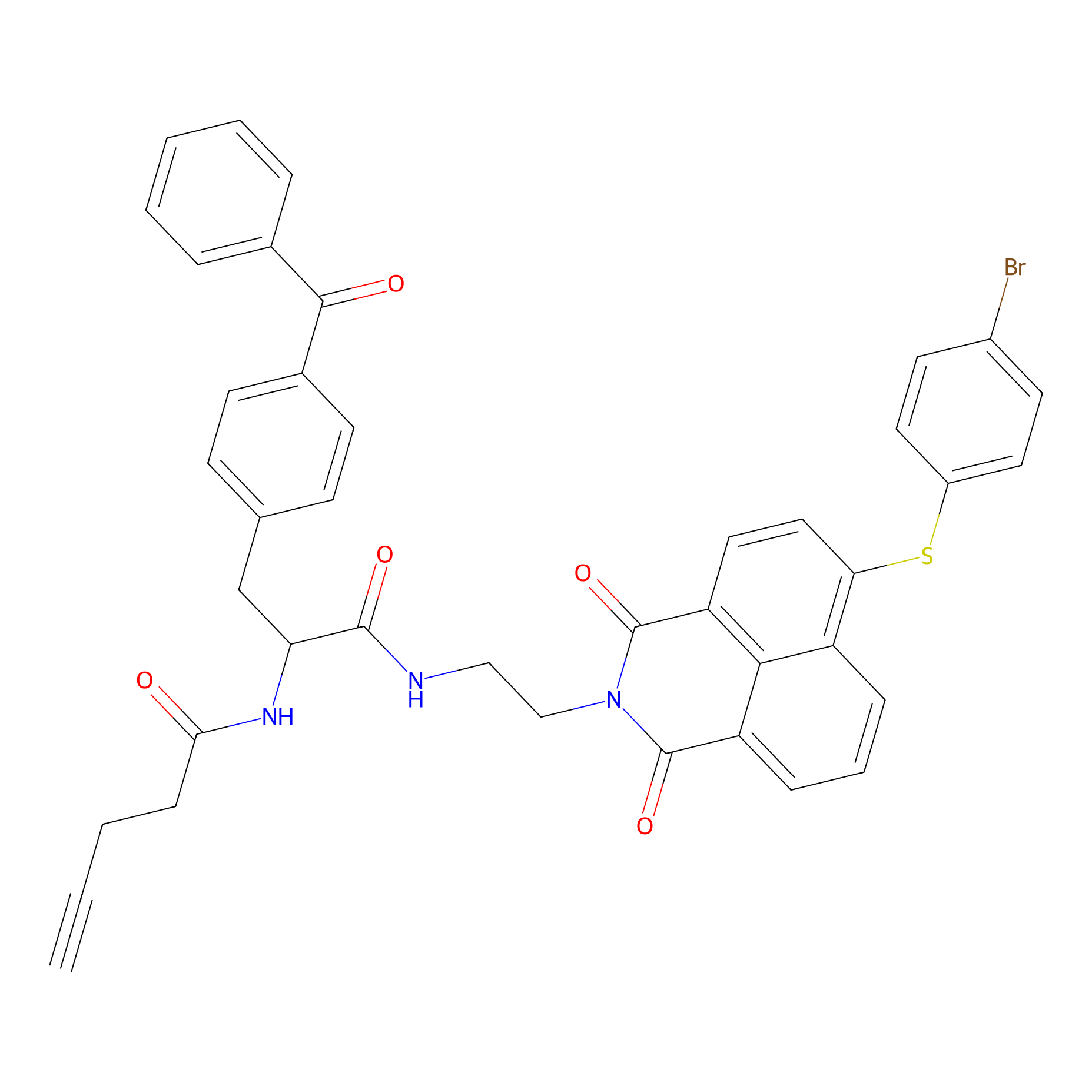 |
N.A. | LDD0435 | [4] | |
|
ONAyne Probe Info |
 |
K1916(0.39) | LDD0274 | [5] | |
|
STPyne Probe Info |
 |
K1028(5.88); K1056(5.00); K1073(2.19); K1075(9.07) | LDD0277 | [5] | |
|
BTD Probe Info |
 |
C2656(1.51) | LDD1700 | [6] | |
|
AHL-Pu-1 Probe Info |
 |
C899(6.51); C798(2.44); C467(7.57); C2776(3.41) | LDD0168 | [7] | |
|
DBIA Probe Info |
 |
C1499(6.38) | LDD0206 | [8] | |
|
W1 Probe Info |
 |
C174(1.02) | LDD0239 | [9] | |
|
5E-2FA Probe Info |
 |
H2742(0.00); H2195(0.00) | LDD2235 | [10] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [11] | |
|
m-APA Probe Info |
 |
H2742(0.00); H2195(0.00) | LDD2231 | [10] | |
|
N1 Probe Info |
 |
E1211(0.00); E1213(0.00) | LDD0245 | [12] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [13] | |
|
IA-alkyne Probe Info |
 |
C1069(0.00); C110(0.00) | LDD0032 | [14] | |
|
WYneN Probe Info |
 |
C3(0.00); C2776(0.00); C1805(0.00); C482(0.00) | LDD0021 | [15] | |
|
WYneO Probe Info |
 |
C1805(0.00); C135(0.00) | LDD0022 | [15] | |
|
ENE Probe Info |
 |
C3(0.00); C1052(0.00); C1280(0.00); C1589(0.00) | LDD0006 | [15] | |
|
IPM Probe Info |
 |
C174(0.00); C740(0.00); C1805(0.00); C1069(0.00) | LDD0005 | [15] | |
|
NPM Probe Info |
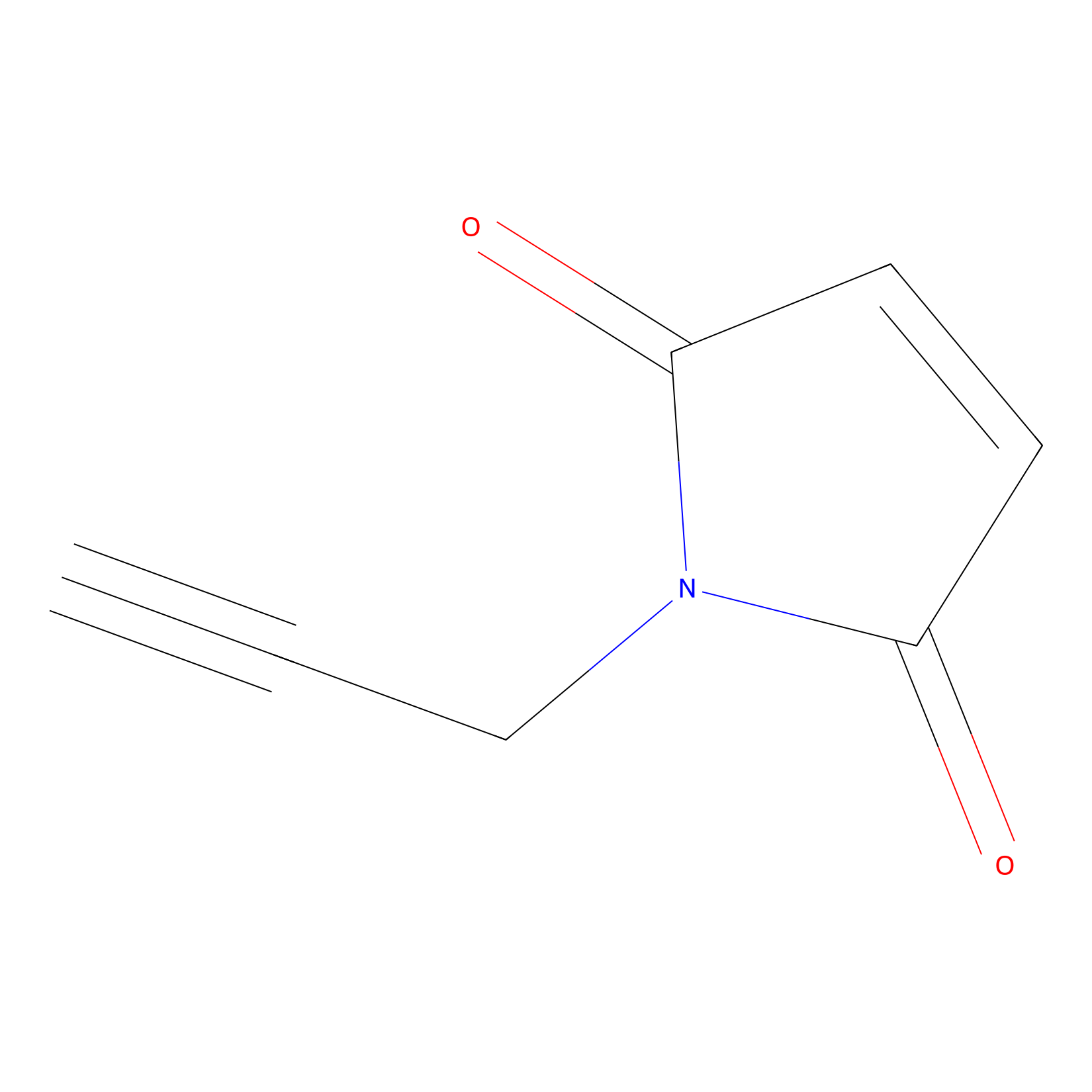 |
N.A. | LDD0016 | [15] | |
|
PF-06672131 Probe Info |
 |
C174(0.00); C2776(0.00) | LDD0017 | [16] | |
|
PPMS Probe Info |
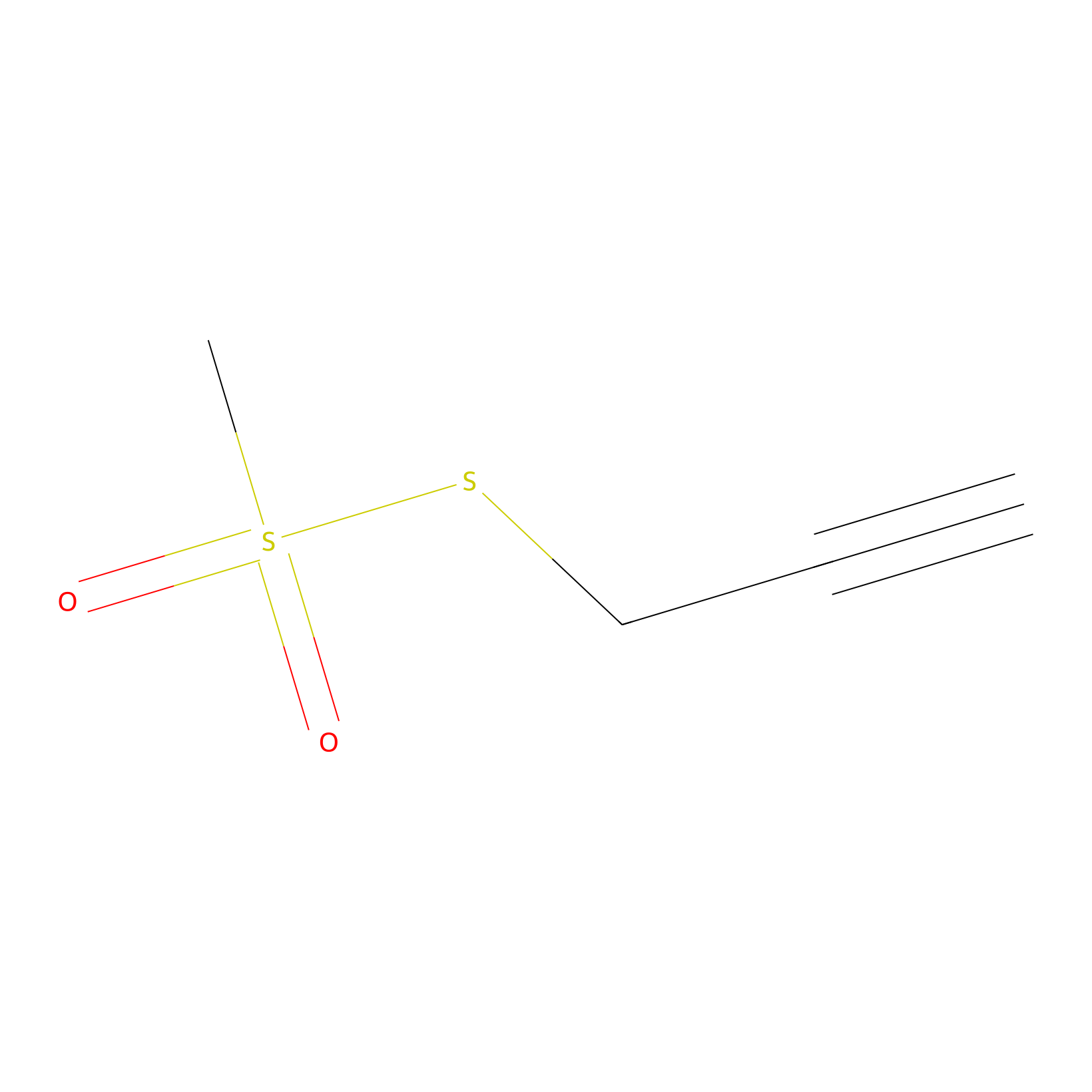 |
N.A. | LDD0008 | [15] | |
|
SF Probe Info |
 |
K1692(0.00); Y2316(0.00) | LDD0028 | [17] | |
|
TFBX Probe Info |
 |
C1280(0.00); C954(0.00); C1052(0.00); C1100(0.00) | LDD0148 | [18] | |
|
VSF Probe Info |
 |
C57(0.00); C1589(0.00); C1957(0.00); C1805(0.00) | LDD0007 | [15] | |
|
Phosphinate-6 Probe Info |
 |
C1805(0.00); C1589(0.00); C1148(0.00); C1052(0.00) | LDD0018 | [19] | |
|
1c-yne Probe Info |
 |
K2523(0.00); K245(0.00) | LDD0228 | [20] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [21] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [21] | |
|
Methacrolein Probe Info |
 |
C1805(0.00); C682(0.00); C174(0.00) | LDD0218 | [21] | |
|
AOyne Probe Info |
 |
8.10 | LDD0443 | [22] | |
|
NAIA_5 Probe Info |
 |
C467(0.00); C1052(0.00); C2442(0.00); C110(0.00) | LDD2223 | [23] | |
|
HHS-465 Probe Info |
 |
K2317(0.00); Y2720(0.00); Y95(0.00) | LDD2240 | [24] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FFF probe11 Probe Info |
 |
20.00 | LDD0471 | [25] | |
|
FFF probe12 Probe Info |
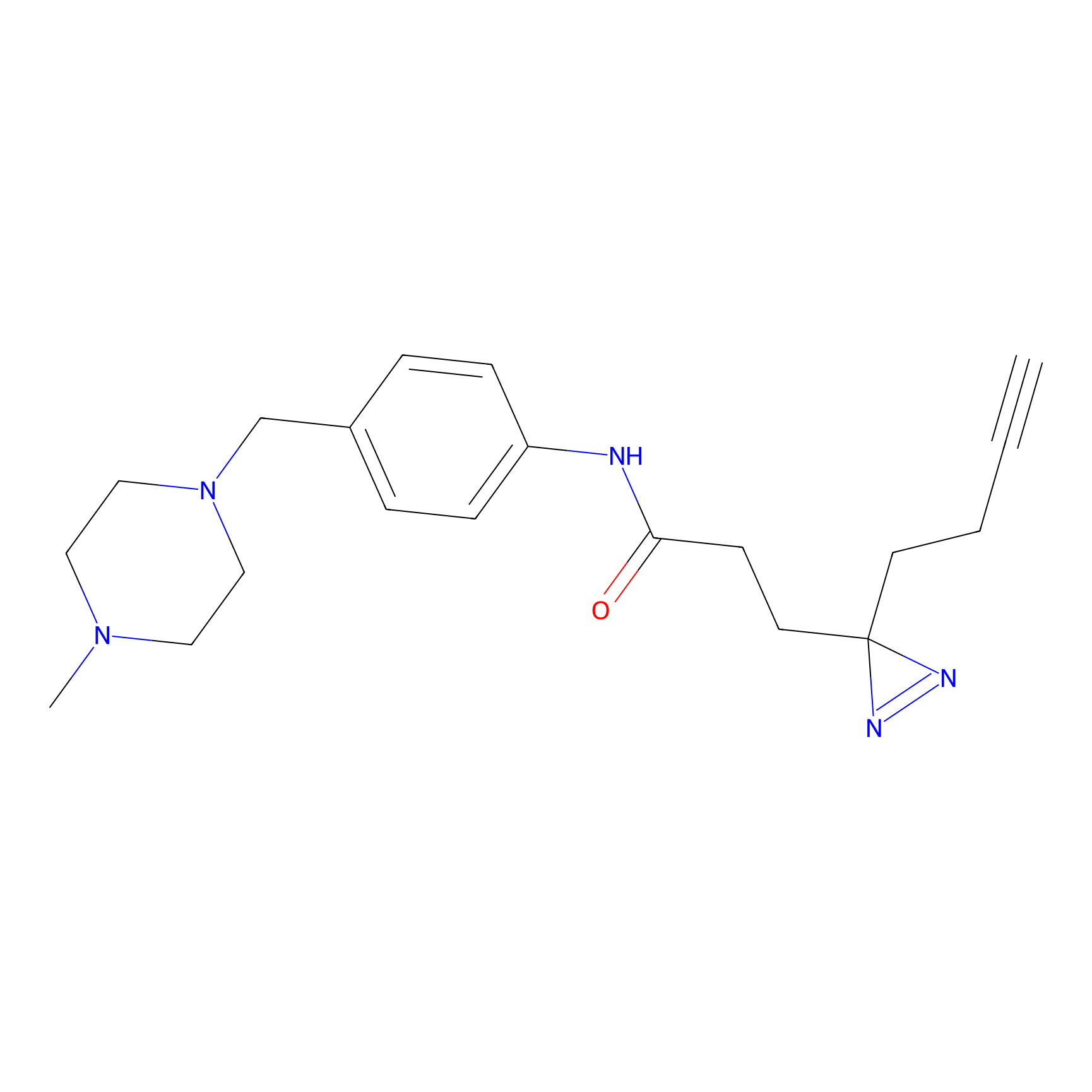 |
20.00 | LDD0474 | [25] | |
|
FFF probe13 Probe Info |
 |
20.00 | LDD0475 | [25] | |
|
FFF probe2 Probe Info |
 |
20.00 | LDD0479 | [25] | |
|
FFF probe6 Probe Info |
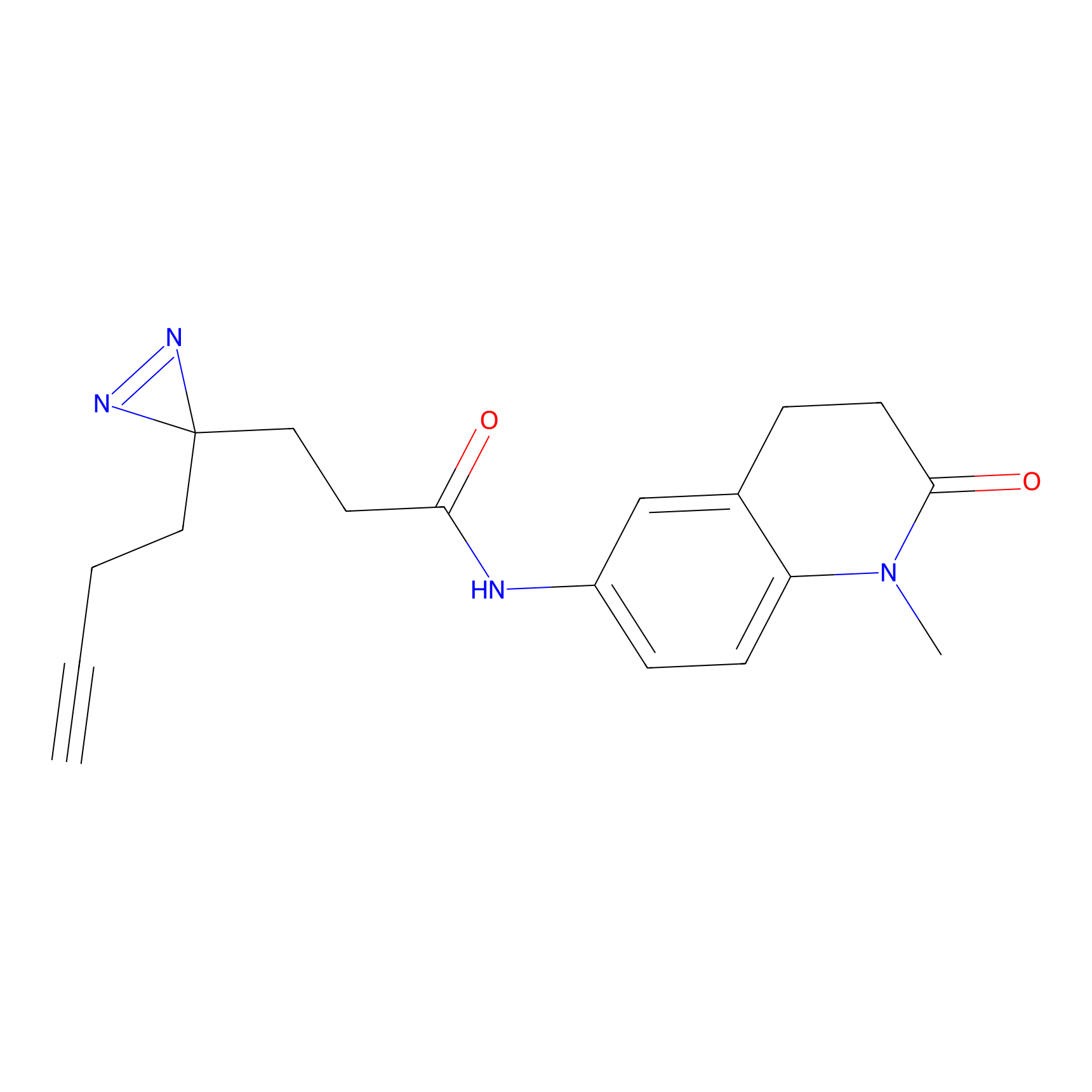 |
20.00 | LDD0482 | [25] | |
|
FFF probe9 Probe Info |
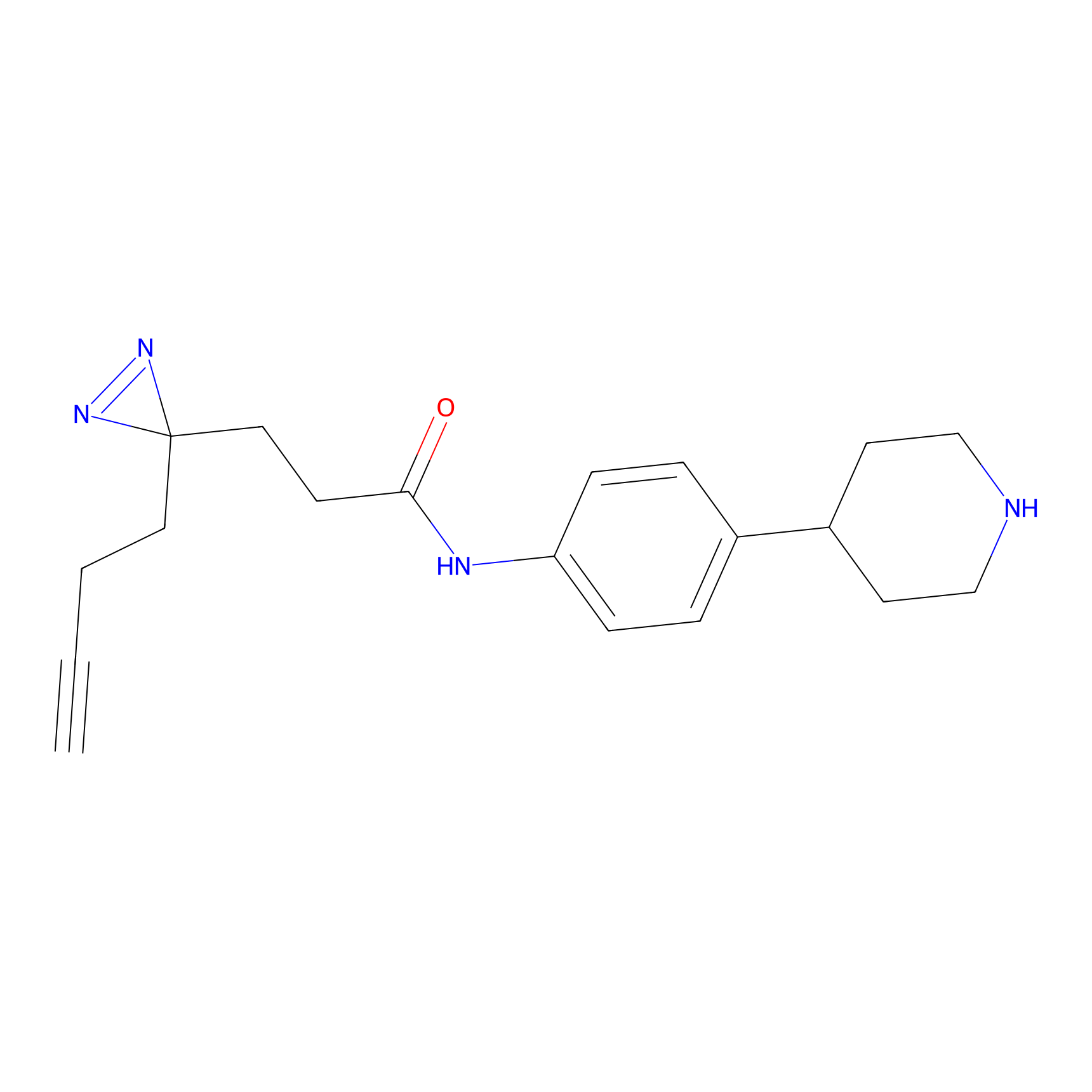 |
20.00 | LDD0470 | [25] | |
|
JN0003 Probe Info |
 |
20.00 | LDD0484 | [25] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0136 | [26] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0138 | [26] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | HEK-293T | C899(6.51); C798(2.44); C467(7.57); C2776(3.41) | LDD0168 | [7] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C899(2.60); C798(2.38); C768(2.25); C740(2.11) | LDD0169 | [7] |
| LDCM0104 | BDHI 13 | Jurkat | C1499(6.38) | LDD0206 | [8] |
| LDCM0108 | Chloroacetamide | HeLa | C682(0.00); C1805(0.00) | LDD0222 | [21] |
| LDCM0632 | CL-Sc | Hep-G2 | C1280(20.00); C1499(20.00); C135(20.00); C1069(7.26) | LDD2227 | [23] |
| LDCM0634 | CY-0357 | Hep-G2 | C1052(0.98); C3(0.88); C57(0.84); C1957(0.78) | LDD2228 | [23] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C367(2.24); C2776(0.98) | LDD1702 | [6] |
| LDCM0022 | KB02 | 22RV1 | C1280(2.59); C1499(1.40); C1052(1.35); C174(3.55) | LDD2243 | [27] |
| LDCM0023 | KB03 | Jurkat | C174(47.43) | LDD0209 | [8] |
| LDCM0024 | KB05 | G361 | C1280(1.37) | LDD3311 | [27] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C1499(0.81) | LDD2093 | [6] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C1499(0.61) | LDD2098 | [6] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C1499(1.03) | LDD2099 | [6] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C2656(0.56) | LDD2109 | [6] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C1499(0.92); C2776(0.99); C1100(1.22) | LDD2125 | [6] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C1499(0.70); C2776(0.92); C1280(1.02) | LDD2127 | [6] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C2776(1.02) | LDD2128 | [6] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C2656(1.30); C1499(0.77) | LDD2129 | [6] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C1499(0.72) | LDD2135 | [6] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C1280(1.51) | LDD2137 | [6] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C2656(1.51) | LDD1700 | [6] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C1499(0.94); C2776(0.75) | LDD2140 | [6] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C2776(0.83) | LDD2143 | [6] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C2656(1.07) | LDD2146 | [6] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C1499(1.12) | LDD2150 | [6] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C57(0.70); C57(0.07) | LDD2206 | [28] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C57(0.79) | LDD2207 | [28] |
| LDCM0112 | W16 | Hep-G2 | C174(1.02) | LDD0239 | [9] |
The Interaction Atlas With This Target
References
