Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FBPP2 Probe Info |
 |
6.21 | LDD0318 | [1] | |
|
CY-1 Probe Info |
 |
100.00 | LDD0243 | [2] | |
|
CY4 Probe Info |
 |
100.00 | LDD0244 | [2] | |
|
N1 Probe Info |
 |
100.00 | LDD0242 | [2] | |
|
YN-1 Probe Info |
 |
3.82 | LDD0444 | [3] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [3] | |
|
ONAyne Probe Info |
 |
K245(0.80) | LDD0274 | [4] | |
|
STPyne Probe Info |
 |
K245(9.54); K255(8.36); K256(7.97); K477(8.61) | LDD0277 | [4] | |
|
AZ-9 Probe Info |
 |
E462(10.00) | LDD2209 | [5] | |
|
DBIA Probe Info |
 |
C133(1.76) | LDD3365 | [6] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [7] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C027 Probe Info |
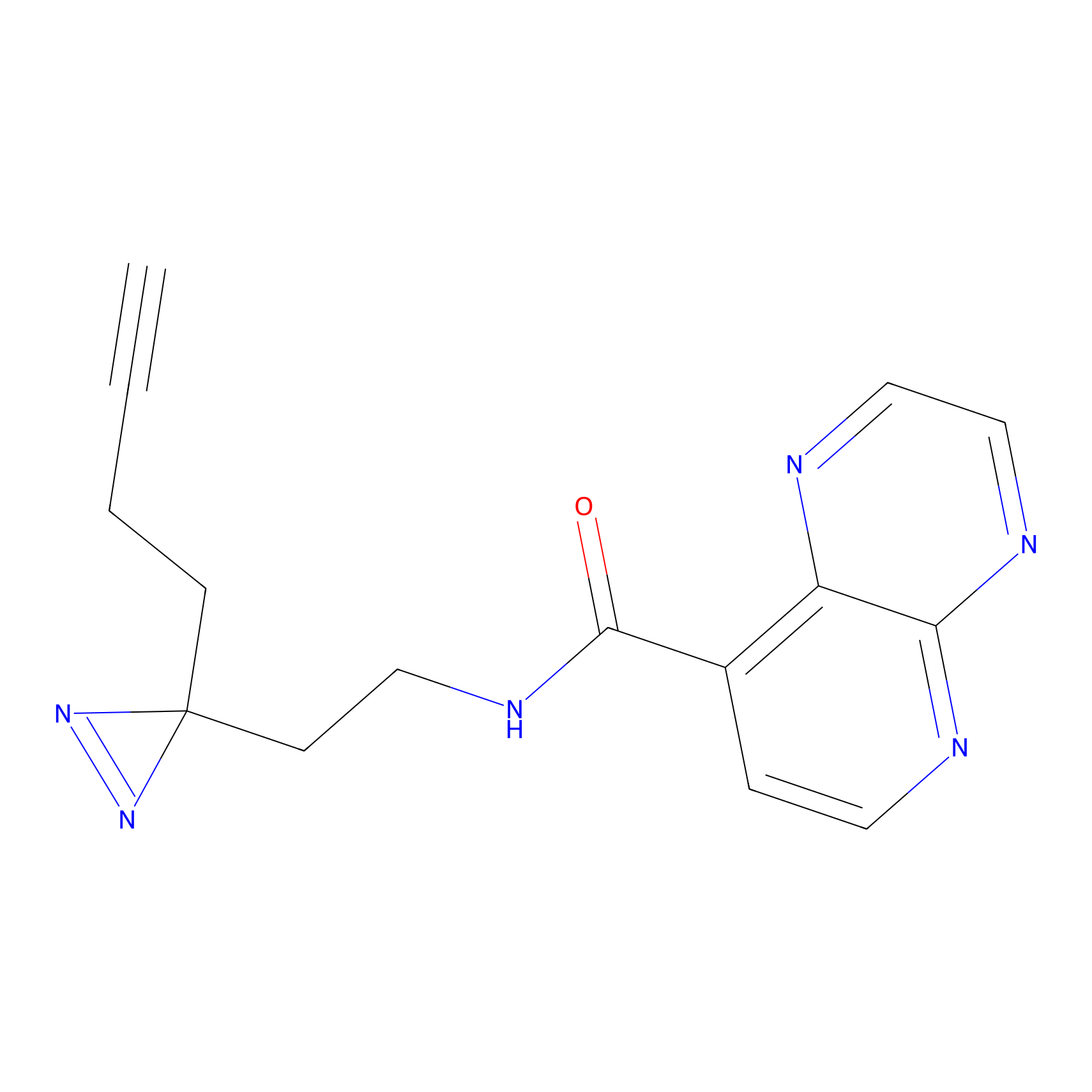 |
6.73 | LDD1733 | [8] | |
|
C063 Probe Info |
 |
10.48 | LDD1760 | [8] | |
|
C158 Probe Info |
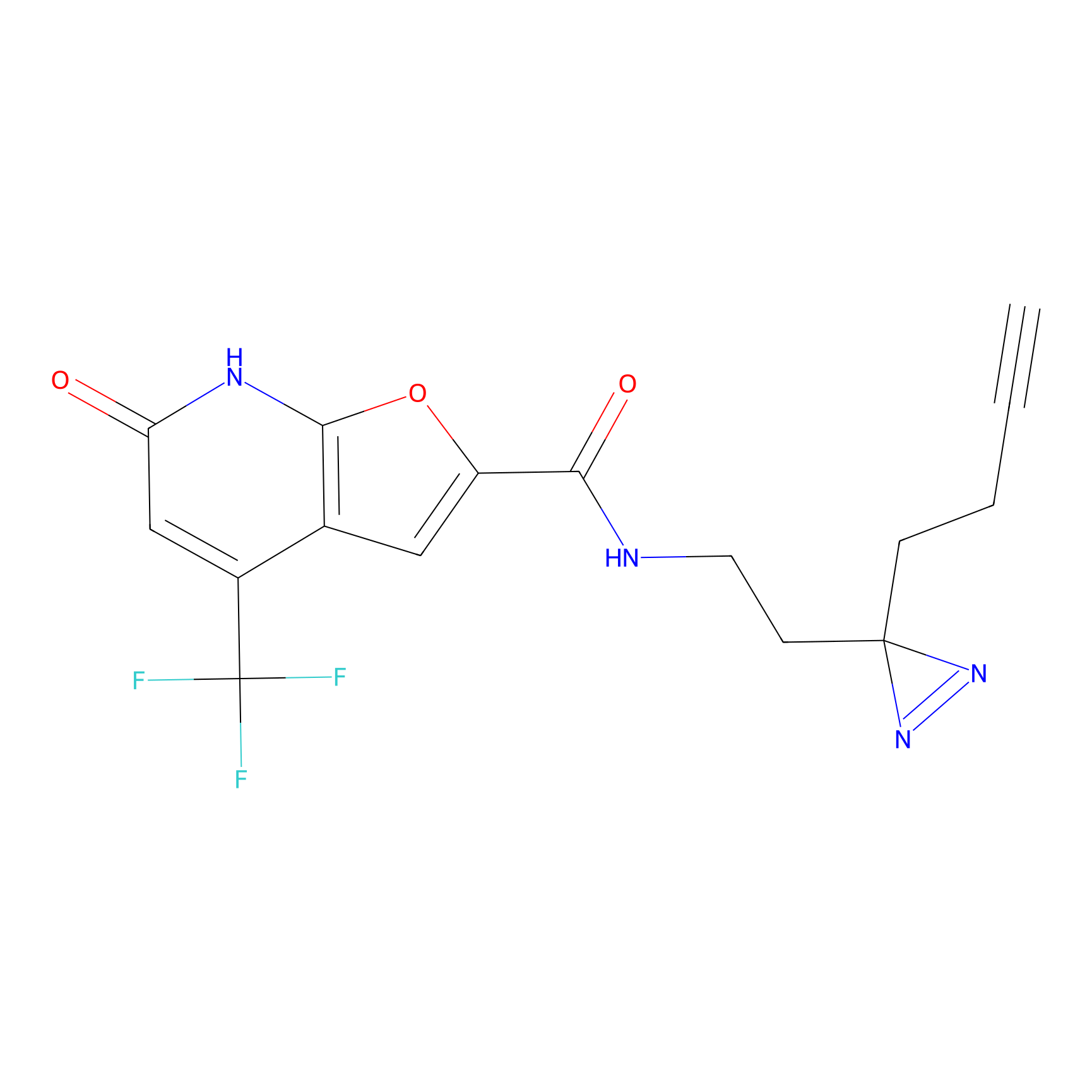 |
11.55 | LDD1838 | [8] | |
|
C159 Probe Info |
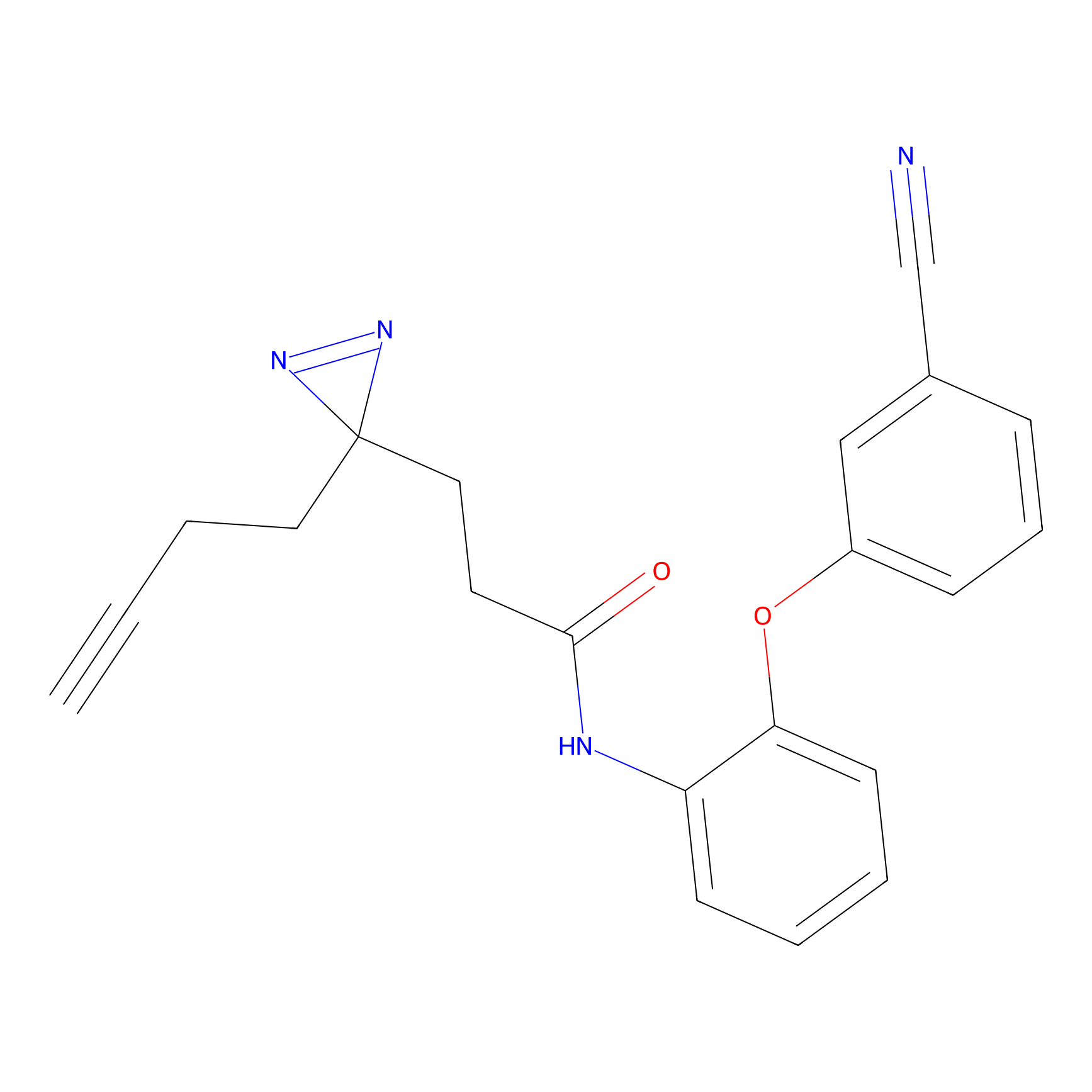 |
8.63 | LDD1839 | [8] | |
|
C160 Probe Info |
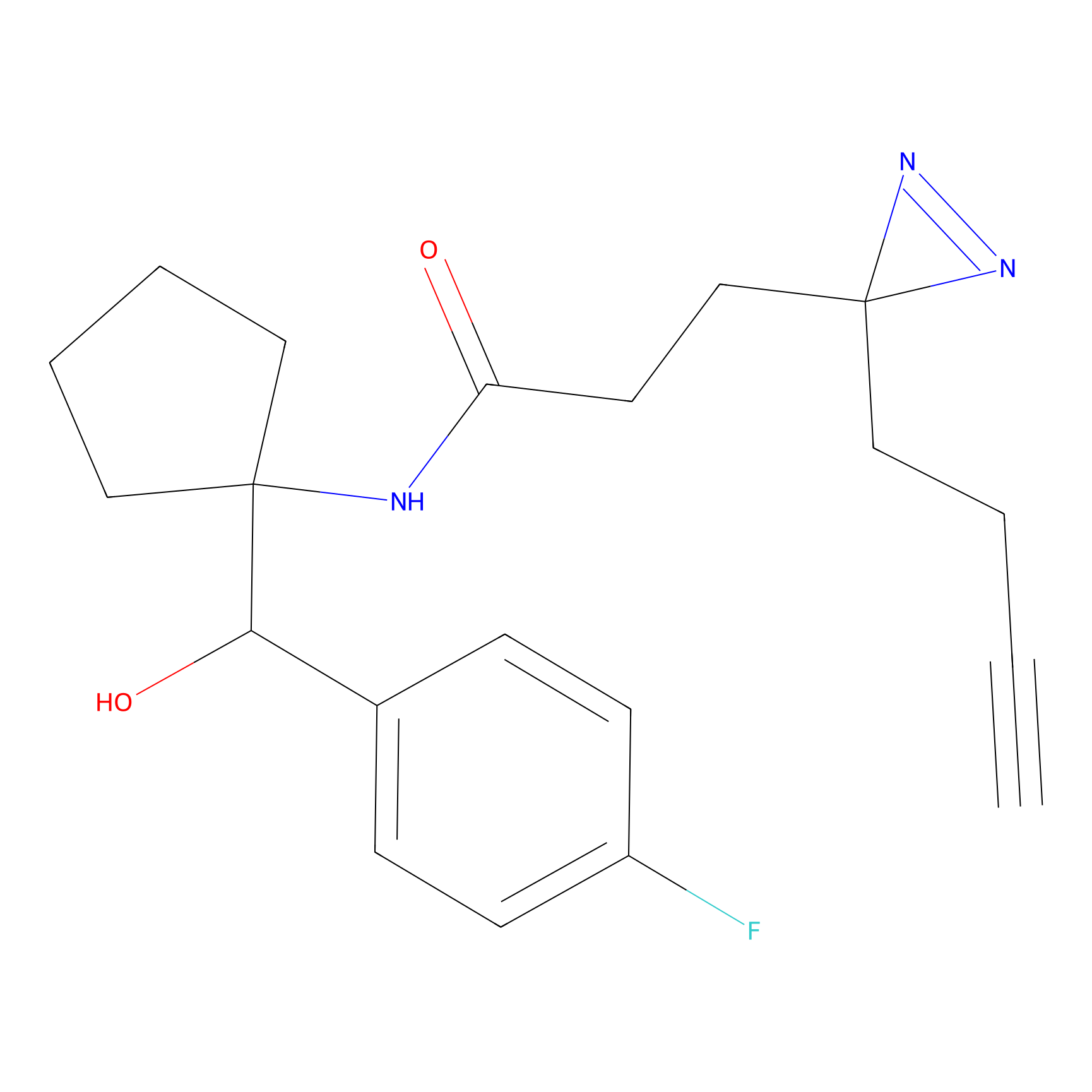 |
7.36 | LDD1840 | [8] | |
|
C161 Probe Info |
 |
15.14 | LDD1841 | [8] | |
|
C212 Probe Info |
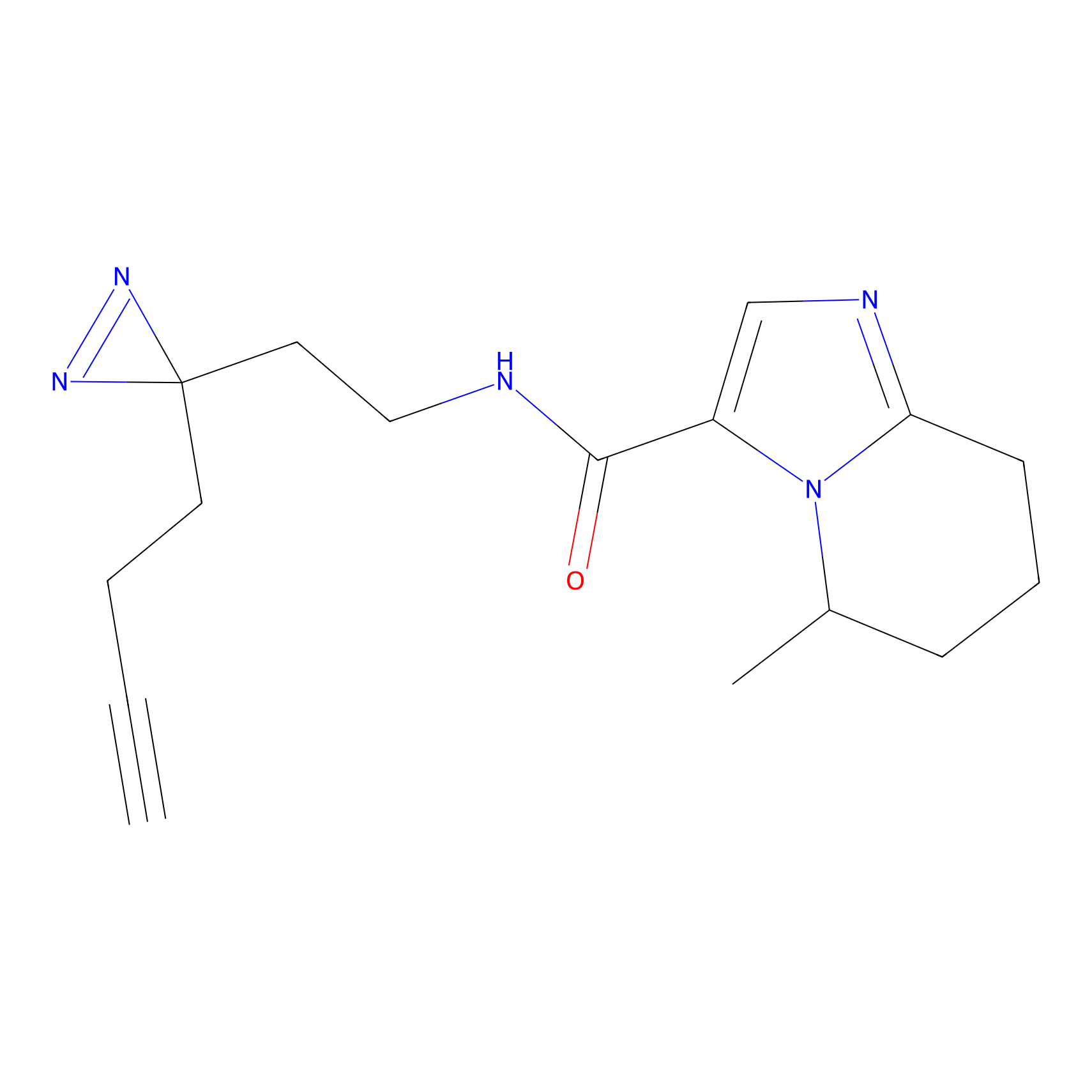 |
5.94 | LDD1886 | [8] | |
|
C246 Probe Info |
 |
13.93 | LDD1919 | [8] | |
|
C310 Probe Info |
 |
11.63 | LDD1977 | [8] | |
|
FFF probe13 Probe Info |
 |
5.71 | LDD0476 | [9] | |
|
FFF probe3 Probe Info |
 |
7.90 | LDD0465 | [9] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0136 | [10] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0138 | [10] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [11] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0214 | AC1 | HEK-293T | C133(0.95) | LDD1507 | [12] |
| LDCM0215 | AC10 | HEK-293T | C133(0.91) | LDD1508 | [12] |
| LDCM0259 | AC14 | HEK-293T | C133(1.10) | LDD1512 | [12] |
| LDCM0276 | AC17 | HEK-293T | C133(0.92) | LDD1515 | [12] |
| LDCM0277 | AC18 | HEK-293T | C133(0.86) | LDD1516 | [12] |
| LDCM0279 | AC2 | HEK-293T | C133(1.09) | LDD1518 | [12] |
| LDCM0281 | AC21 | HEK-293T | C133(0.83) | LDD1520 | [12] |
| LDCM0282 | AC22 | HEK-293T | C133(1.00) | LDD1521 | [12] |
| LDCM0285 | AC25 | HEK-293T | C133(0.96) | LDD1524 | [12] |
| LDCM0286 | AC26 | HEK-293T | C133(0.89) | LDD1525 | [12] |
| LDCM0289 | AC29 | HEK-293T | C133(0.76) | LDD1528 | [12] |
| LDCM0291 | AC30 | HEK-293T | C133(1.25) | LDD1530 | [12] |
| LDCM0294 | AC33 | HEK-293T | C133(0.92) | LDD1533 | [12] |
| LDCM0295 | AC34 | HEK-293T | C133(0.90) | LDD1534 | [12] |
| LDCM0298 | AC37 | HEK-293T | C133(0.95) | LDD1537 | [12] |
| LDCM0299 | AC38 | HEK-293T | C133(1.13) | LDD1538 | [12] |
| LDCM0303 | AC41 | HEK-293T | C133(0.96) | LDD1542 | [12] |
| LDCM0304 | AC42 | HEK-293T | C133(0.83) | LDD1543 | [12] |
| LDCM0307 | AC45 | HEK-293T | C133(1.19) | LDD1546 | [12] |
| LDCM0308 | AC46 | HEK-293T | C133(1.07) | LDD1547 | [12] |
| LDCM0311 | AC49 | HEK-293T | C133(0.98) | LDD1550 | [12] |
| LDCM0312 | AC5 | HEK-293T | C133(1.01) | LDD1551 | [12] |
| LDCM0313 | AC50 | HEK-293T | C133(0.87) | LDD1552 | [12] |
| LDCM0316 | AC53 | HEK-293T | C133(0.85) | LDD1555 | [12] |
| LDCM0317 | AC54 | HEK-293T | C133(0.93) | LDD1556 | [12] |
| LDCM0320 | AC57 | HEK-293T | C133(0.92) | LDD1559 | [12] |
| LDCM0321 | AC58 | HEK-293T | C133(0.98) | LDD1560 | [12] |
| LDCM0323 | AC6 | HEK-293T | C133(0.83) | LDD1562 | [12] |
| LDCM0325 | AC61 | HEK-293T | C133(0.97) | LDD1564 | [12] |
| LDCM0326 | AC62 | HEK-293T | C133(1.16) | LDD1565 | [12] |
| LDCM0248 | AKOS034007472 | HEK-293T | C133(0.93) | LDD1511 | [12] |
| LDCM0356 | AKOS034007680 | HEK-293T | C133(1.04) | LDD1570 | [12] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [7] |
| LDCM0368 | CL10 | HEK-293T | C133(1.17) | LDD1572 | [12] |
| LDCM0369 | CL100 | HEK-293T | C133(0.88) | LDD1573 | [12] |
| LDCM0373 | CL104 | HEK-293T | C133(0.95) | LDD1577 | [12] |
| LDCM0377 | CL108 | HEK-293T | C133(0.84) | LDD1581 | [12] |
| LDCM0382 | CL112 | HEK-293T | C133(0.93) | LDD1586 | [12] |
| LDCM0386 | CL116 | HEK-293T | C133(1.08) | LDD1590 | [12] |
| LDCM0391 | CL120 | HEK-293T | C133(0.91) | LDD1595 | [12] |
| LDCM0395 | CL124 | HEK-293T | C133(0.85) | LDD1599 | [12] |
| LDCM0399 | CL128 | HEK-293T | C133(0.92) | LDD1603 | [12] |
| LDCM0403 | CL16 | HEK-293T | C133(0.97) | LDD1607 | [12] |
| LDCM0404 | CL17 | HEK-293T | C133(1.10) | LDD1608 | [12] |
| LDCM0405 | CL18 | HEK-293T | C133(0.91) | LDD1609 | [12] |
| LDCM0409 | CL21 | HEK-293T | C133(0.91) | LDD1613 | [12] |
| LDCM0410 | CL22 | HEK-293T | C133(1.15) | LDD1614 | [12] |
| LDCM0416 | CL28 | HEK-293T | C133(0.90) | LDD1620 | [12] |
| LDCM0417 | CL29 | HEK-293T | C133(1.02) | LDD1621 | [12] |
| LDCM0419 | CL30 | HEK-293T | C133(0.77) | LDD1623 | [12] |
| LDCM0422 | CL33 | HEK-293T | C133(0.94) | LDD1626 | [12] |
| LDCM0423 | CL34 | HEK-293T | C133(1.41) | LDD1627 | [12] |
| LDCM0429 | CL4 | HEK-293T | C133(0.99) | LDD1633 | [12] |
| LDCM0430 | CL40 | HEK-293T | C133(0.83) | LDD1634 | [12] |
| LDCM0431 | CL41 | HEK-293T | C133(1.04) | LDD1635 | [12] |
| LDCM0432 | CL42 | HEK-293T | C133(0.93) | LDD1636 | [12] |
| LDCM0435 | CL45 | HEK-293T | C133(0.79) | LDD1639 | [12] |
| LDCM0436 | CL46 | HEK-293T | C133(1.13) | LDD1640 | [12] |
| LDCM0440 | CL5 | HEK-293T | C133(0.99) | LDD1644 | [12] |
| LDCM0443 | CL52 | HEK-293T | C133(0.95) | LDD1646 | [12] |
| LDCM0444 | CL53 | HEK-293T | C133(1.01) | LDD1647 | [12] |
| LDCM0445 | CL54 | HEK-293T | C133(0.95) | LDD1648 | [12] |
| LDCM0448 | CL57 | HEK-293T | C133(1.01) | LDD1651 | [12] |
| LDCM0449 | CL58 | HEK-293T | C133(1.14) | LDD1652 | [12] |
| LDCM0451 | CL6 | HEK-293T | C133(1.13) | LDD1654 | [12] |
| LDCM0456 | CL64 | HEK-293T | C133(0.79) | LDD1659 | [12] |
| LDCM0457 | CL65 | HEK-293T | C133(1.01) | LDD1660 | [12] |
| LDCM0458 | CL66 | HEK-293T | C133(0.83) | LDD1661 | [12] |
| LDCM0461 | CL69 | HEK-293T | C133(0.87) | LDD1664 | [12] |
| LDCM0463 | CL70 | HEK-293T | C133(1.77) | LDD1666 | [12] |
| LDCM0469 | CL76 | HEK-293T | C133(0.80) | LDD1672 | [12] |
| LDCM0470 | CL77 | HEK-293T | C133(0.93) | LDD1673 | [12] |
| LDCM0471 | CL78 | HEK-293T | C133(0.81) | LDD1674 | [12] |
| LDCM0475 | CL81 | HEK-293T | C133(0.82) | LDD1678 | [12] |
| LDCM0476 | CL82 | HEK-293T | C133(1.03) | LDD1679 | [12] |
| LDCM0482 | CL88 | HEK-293T | C133(1.09) | LDD1685 | [12] |
| LDCM0483 | CL89 | HEK-293T | C133(1.01) | LDD1686 | [12] |
| LDCM0484 | CL9 | HEK-293T | C133(1.14) | LDD1687 | [12] |
| LDCM0485 | CL90 | HEK-293T | C133(1.17) | LDD1688 | [12] |
| LDCM0488 | CL93 | HEK-293T | C133(1.05) | LDD1691 | [12] |
| LDCM0489 | CL94 | HEK-293T | C133(1.27) | LDD1692 | [12] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [7] |
| LDCM0022 | KB02 | HEK-293T | C133(1.08) | LDD1492 | [12] |
| LDCM0023 | KB03 | HEK-293T | C133(1.10) | LDD1497 | [12] |
| LDCM0024 | KB05 | OVCAR-3 | C133(1.76) | LDD3365 | [6] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [7] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Solute carrier family 2, facilitated glucose transporter member 1 (SLC2A1) | Sugar transporter (TC 2.A.1.1) family | P11166 | |||
The Drug(s) Related To This Target
Approved
Preclinical
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Wzb-117 | Small molecular drug | DCL8J5 | |||
Investigative
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Resveratrol | Small molecular drug | DB02709 | |||
References
