Details of the Target
General Information of Target
| Target ID | LDTP12042 | |||||
|---|---|---|---|---|---|---|
| Target Name | Proton-activated chloride channel (PACC1) | |||||
| Gene Name | PACC1 | |||||
| Gene ID | 55248 | |||||
| Synonyms |
C1orf75; TMEM206; Proton-activated chloride channel; PAC; hPAC; Acid-sensitive outwardly-rectifying anion channel; ASOR; Proton-activated outwardly rectifying anion channel; PAORAC; Transmembrane protein 206; hTMEM206
|
|||||
| 3D Structure | ||||||
| Sequence |
MTSRDQPRPKGPPKSTSPCPGISNSESSPTLNYQGILNRLKQFPRFSPHFAAELESIYYS
LHKIQQDVAEHHKQIGNVLQIVESCSQLQGFQSEEVSPAEPASPGTPQQVKDKTLQESSF EDIMATRSSDWLRRPLGEDNQPETQLFWDKEPWFWHDTLTEQLWRIFAGVHDEKAKPRDR QQAPGLGQESKAPGSCDPGTDPCPEDASTPRPPEASSSPPEGSQDRNTSWGVVQEPPGRA SRFLQSISWDPEDFEDAWKRPDALPGQSKRLAVPCKLEKMRILAHGELVLATAISSFTRH VFTCGRRGIKVWSLTGQVAEDRFPESHLPIQTPGAFLRTCLLSSNSRSLLTGGYNLASVS VWDLAAPSLHVKEQLPCAGLNCQALDANLDANLAFASFTSGVVRIWDLRDQSVVRDLKGY PDGVKSIVVKGYNIWTGGPDACLRCWDQRTIMKPLEYQFKSQIMSLSHSPQEDWVLLGMA NGQQWLQSTSGSQRHMVGQKDSVILSVKFSPFGQWWASVGMDDFLGVYSMPAGTKVFEVP EMSPVTCCDVSSNNRLVVTGSGEHASVYQITY |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
Proton-activated chloride channel family
|
|||||
| Subcellular location |
Cell membrane
|
|||||
| Function |
Chloride channel gated by pH that facilitates the entry of chloride ions into cells upon exposure to extracellular acidic pH. Involved in acidosis-induced cell death by mediating chloride influx and subsequent cell swelling.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| CAL51 | Deletion: p.S277VfsTer22 | . | |||
| MCC26 | SNV: p.A320E | . | |||
| MOLT4 | Deletion: p.S277VfsTer22 | IA-alkyne Probe Info | |||
| NCIH2286 | SNV: p.P144A | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FBPP2 Probe Info |
 |
5.36 | LDD0054 | [1] | |
|
DBIA Probe Info |
 |
C223(1.13) | LDD1508 | [2] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [3] | |
|
AOyne Probe Info |
 |
14.80 | LDD0443 | [4] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C170 Probe Info |
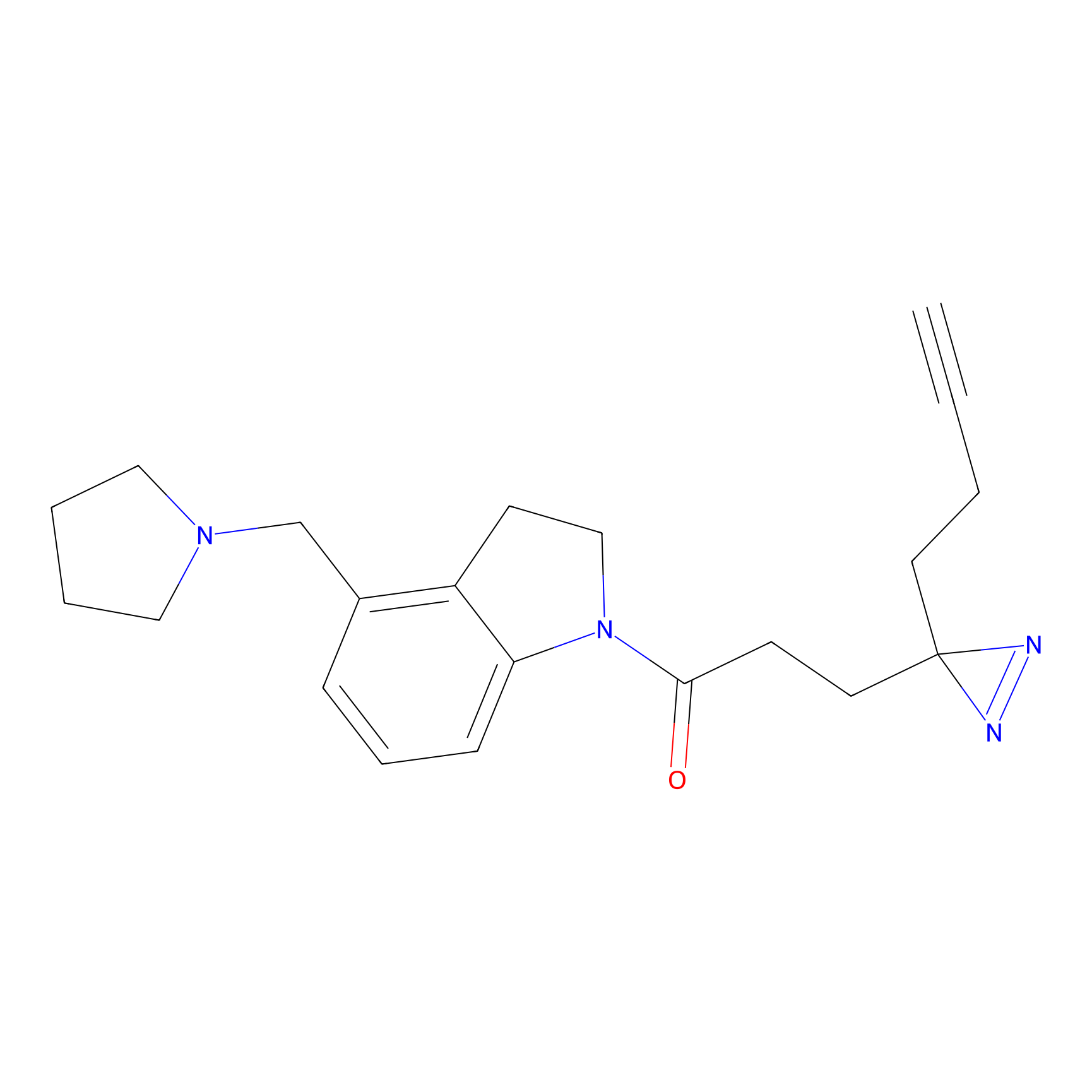 |
26.17 | LDD1850 | [5] | |
|
C171 Probe Info |
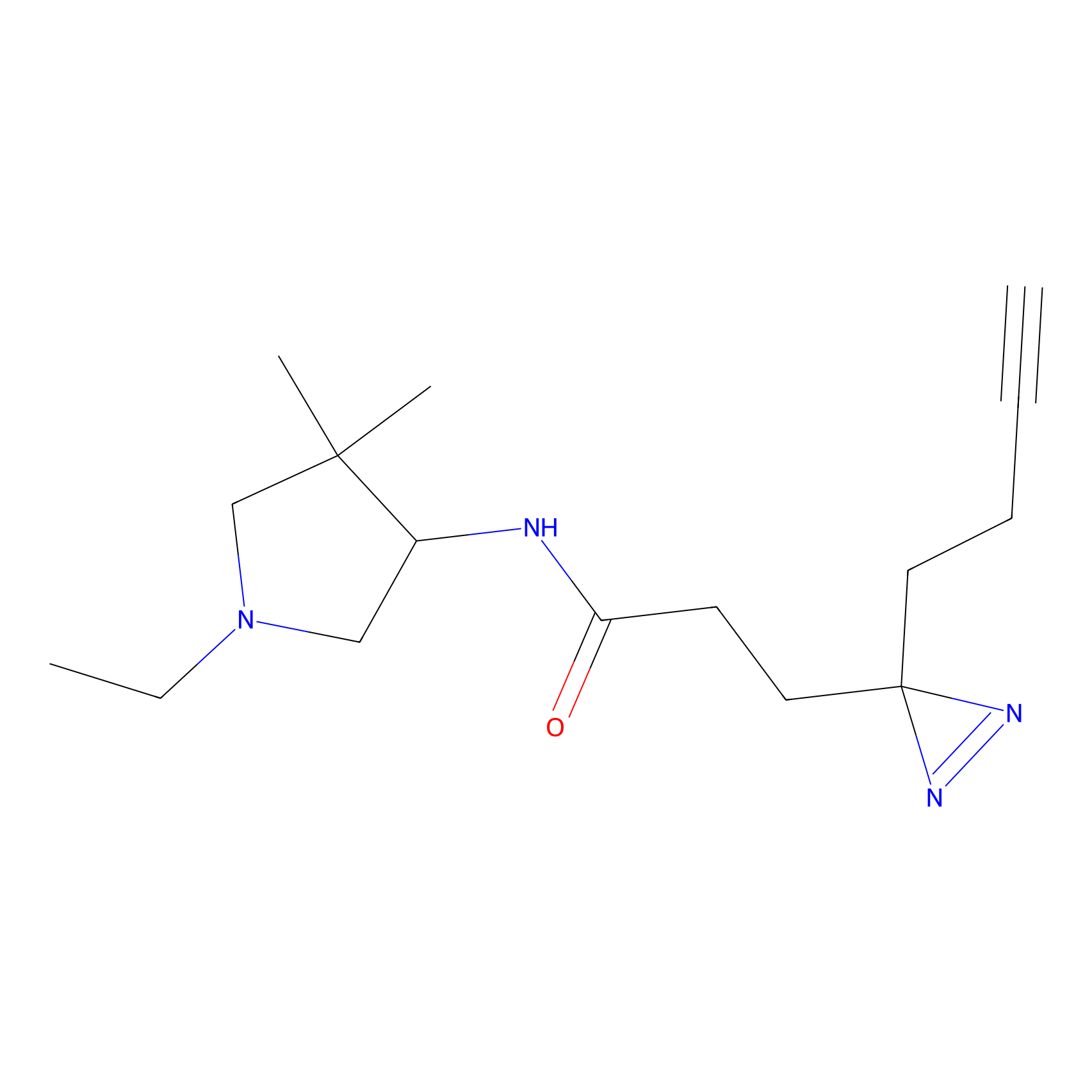 |
14.83 | LDD1851 | [5] | |
|
C281 Probe Info |
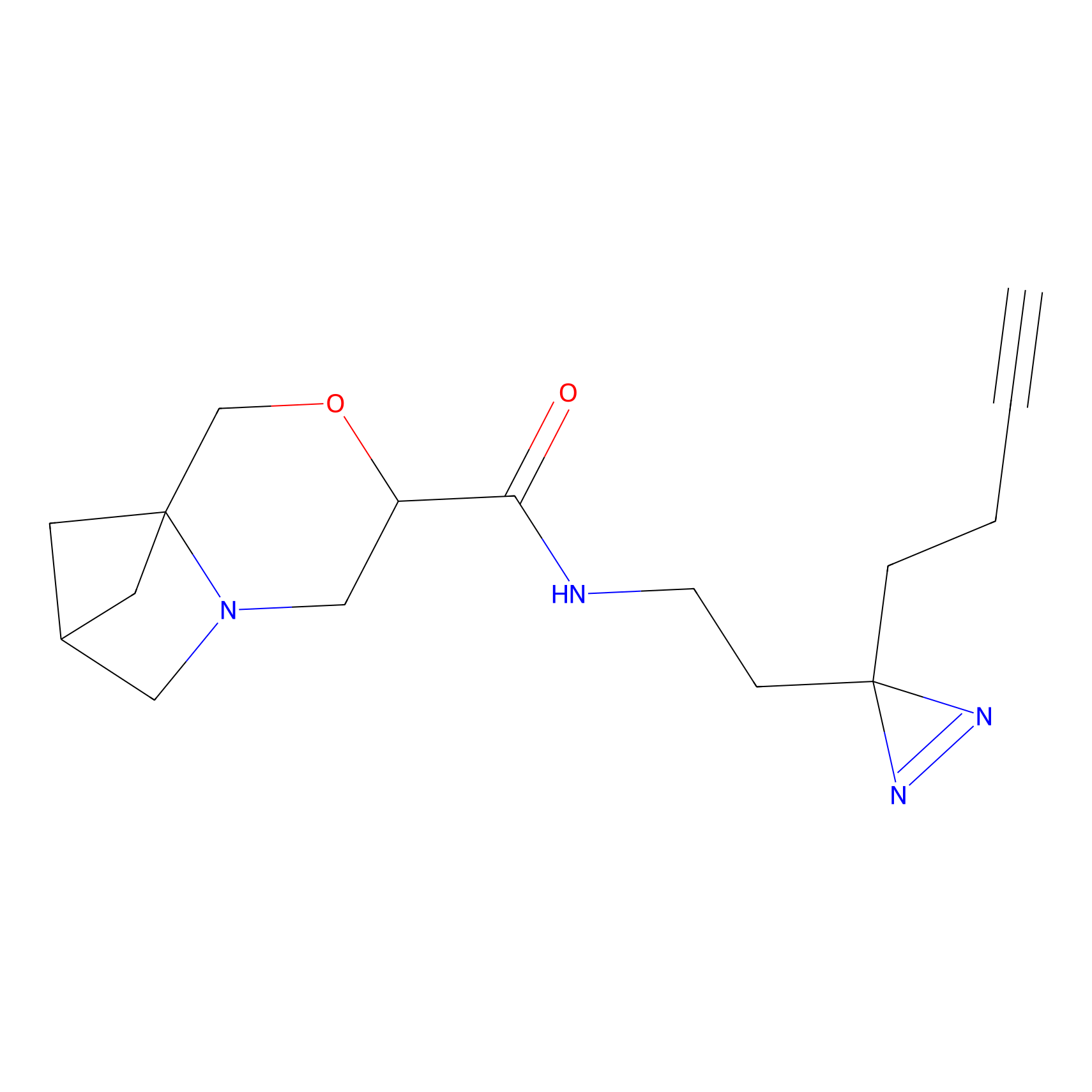 |
23.75 | LDD1951 | [5] | |
|
C282 Probe Info |
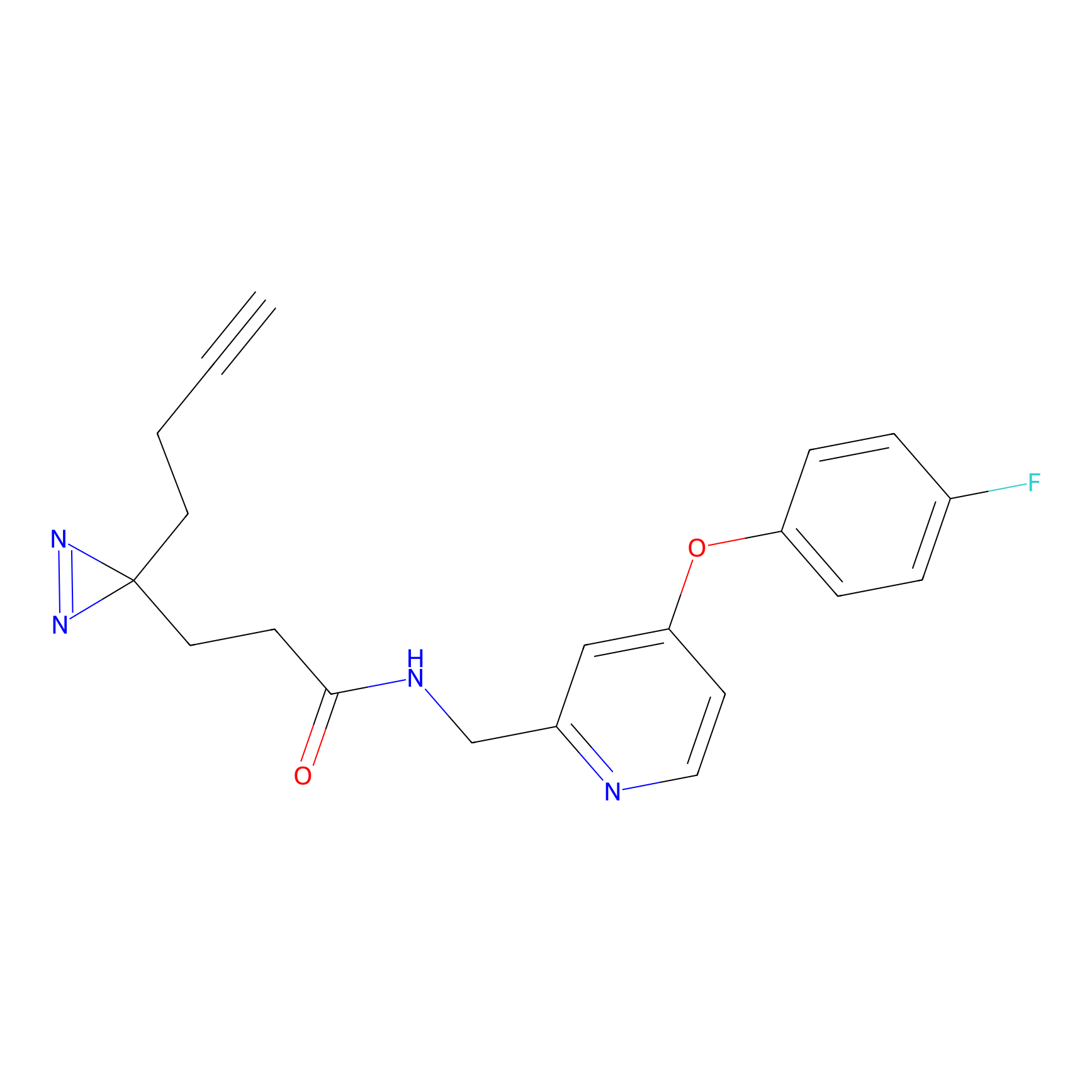 |
19.03 | LDD1952 | [5] | |
|
C310 Probe Info |
 |
20.68 | LDD1977 | [5] | |
|
FFF probe11 Probe Info |
 |
13.30 | LDD0471 | [6] | |
|
FFF probe12 Probe Info |
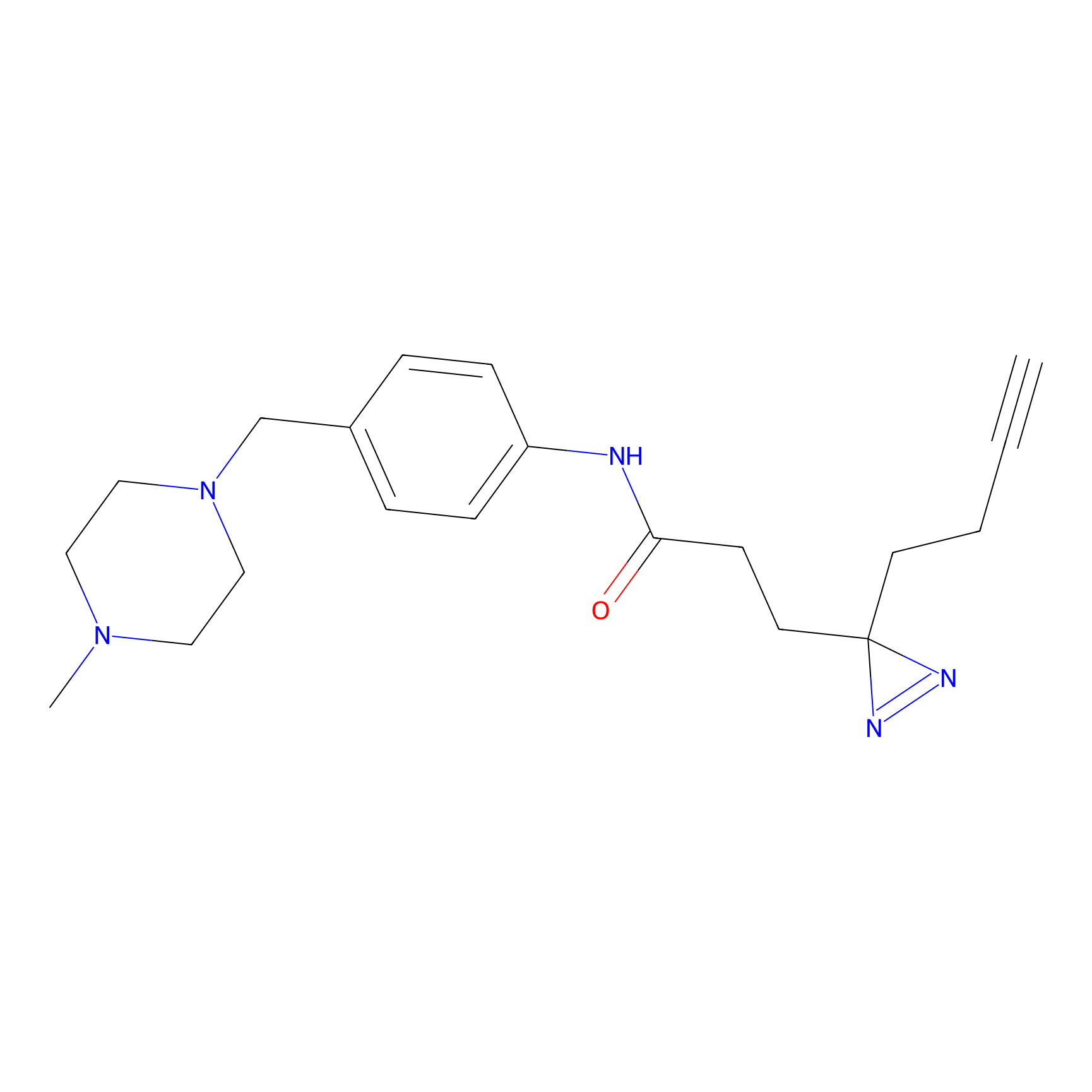 |
17.88 | LDD0473 | [6] | |
|
FFF probe15 Probe Info |
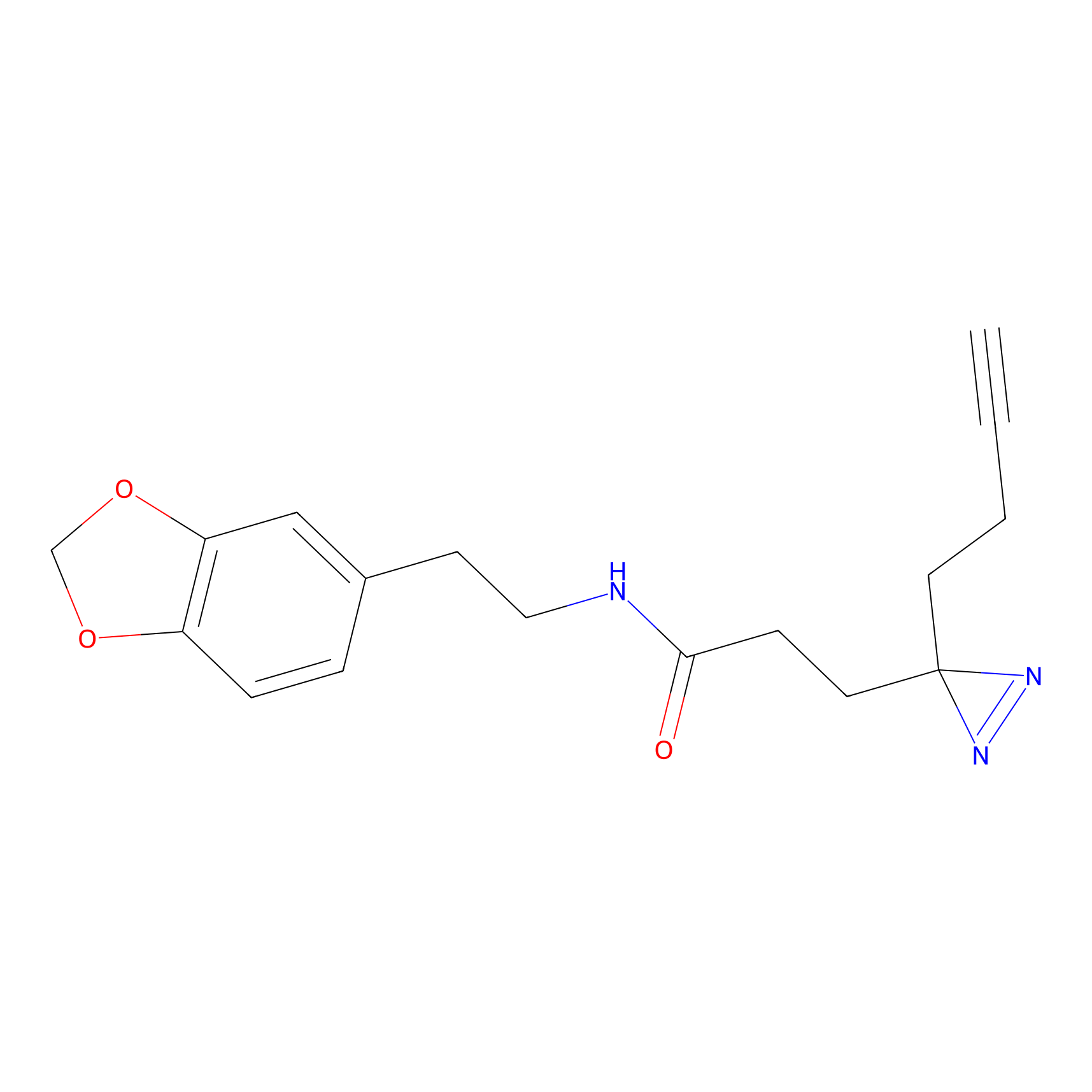 |
8.84 | LDD0478 | [6] | |
|
FFF probe2 Probe Info |
 |
16.41 | LDD0463 | [6] | |
|
FFF probe4 Probe Info |
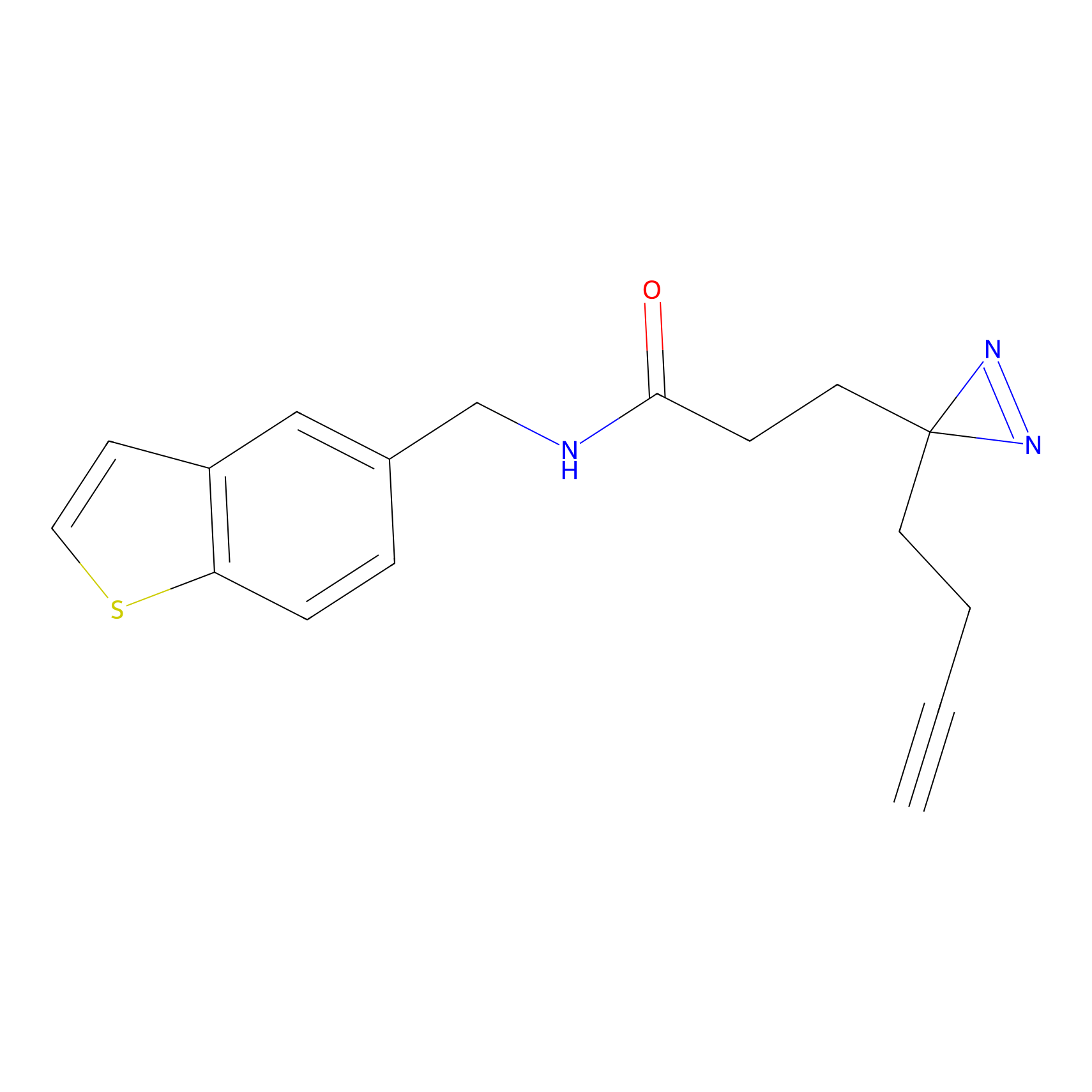 |
6.32 | LDD0466 | [6] | |
|
FFF probe6 Probe Info |
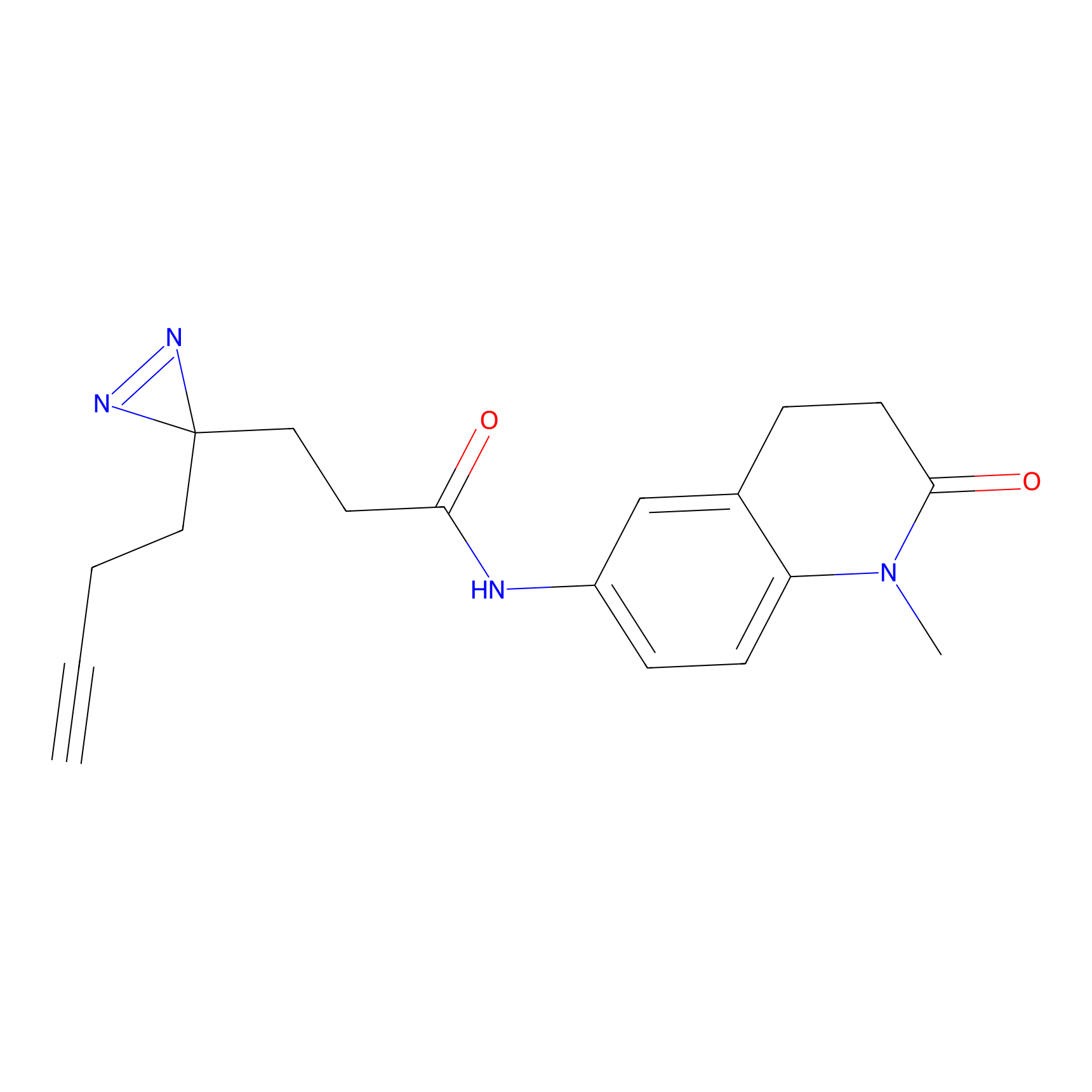 |
8.66 | LDD0467 | [6] | |
|
FFF probe9 Probe Info |
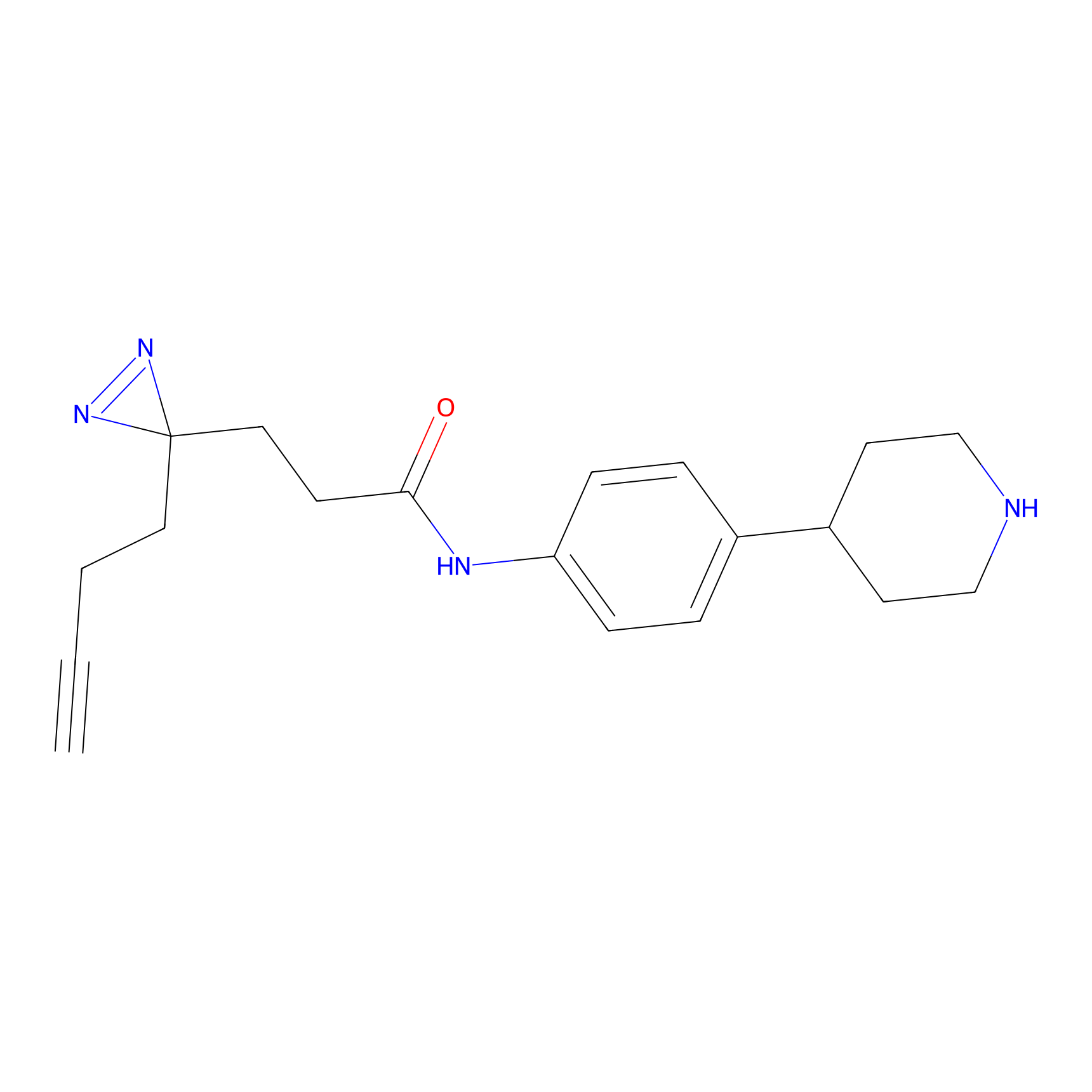 |
17.89 | LDD0470 | [6] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0137 | [7] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0215 | AC10 | HEK-293T | C223(1.13) | LDD1508 | [2] |
| LDCM0226 | AC11 | HEK-293T | C128(0.93) | LDD1509 | [2] |
| LDCM0259 | AC14 | HEK-293T | C223(0.87) | LDD1512 | [2] |
| LDCM0277 | AC18 | HEK-293T | C223(1.16) | LDD1516 | [2] |
| LDCM0278 | AC19 | HEK-293T | C128(1.10) | LDD1517 | [2] |
| LDCM0279 | AC2 | HEK-293T | C223(1.18) | LDD1518 | [2] |
| LDCM0282 | AC22 | HEK-293T | C223(0.90) | LDD1521 | [2] |
| LDCM0286 | AC26 | HEK-293T | C223(1.06) | LDD1525 | [2] |
| LDCM0287 | AC27 | HEK-293T | C128(0.93) | LDD1526 | [2] |
| LDCM0290 | AC3 | HEK-293T | C128(0.99) | LDD1529 | [2] |
| LDCM0291 | AC30 | HEK-293T | C223(0.92) | LDD1530 | [2] |
| LDCM0295 | AC34 | HEK-293T | C223(1.05) | LDD1534 | [2] |
| LDCM0296 | AC35 | HEK-293T | C128(1.04) | LDD1535 | [2] |
| LDCM0299 | AC38 | HEK-293T | C223(0.92) | LDD1538 | [2] |
| LDCM0304 | AC42 | HEK-293T | C223(1.22) | LDD1543 | [2] |
| LDCM0305 | AC43 | HEK-293T | C128(0.95) | LDD1544 | [2] |
| LDCM0308 | AC46 | HEK-293T | C223(0.87) | LDD1547 | [2] |
| LDCM0313 | AC50 | HEK-293T | C223(1.22) | LDD1552 | [2] |
| LDCM0314 | AC51 | HEK-293T | C128(1.01) | LDD1553 | [2] |
| LDCM0317 | AC54 | HEK-293T | C223(0.95) | LDD1556 | [2] |
| LDCM0321 | AC58 | HEK-293T | C223(1.29) | LDD1560 | [2] |
| LDCM0322 | AC59 | HEK-293T | C128(0.96) | LDD1561 | [2] |
| LDCM0323 | AC6 | HEK-293T | C223(1.03) | LDD1562 | [2] |
| LDCM0326 | AC62 | HEK-293T | C223(1.05) | LDD1565 | [2] |
| LDCM0368 | CL10 | HEK-293T | C223(0.96) | LDD1572 | [2] |
| LDCM0371 | CL102 | HEK-293T | C223(1.13); C128(1.17) | LDD1575 | [2] |
| LDCM0375 | CL106 | HEK-293T | C223(0.94); C128(1.19) | LDD1579 | [2] |
| LDCM0380 | CL110 | HEK-293T | C223(1.10); C128(1.12) | LDD1584 | [2] |
| LDCM0384 | CL114 | HEK-293T | C223(1.13); C128(1.13) | LDD1588 | [2] |
| LDCM0388 | CL118 | HEK-293T | C223(1.10); C128(1.16) | LDD1592 | [2] |
| LDCM0393 | CL122 | HEK-293T | C223(1.07); C128(1.03) | LDD1597 | [2] |
| LDCM0397 | CL126 | HEK-293T | C223(1.23); C128(1.14) | LDD1601 | [2] |
| LDCM0401 | CL14 | HEK-293T | C223(1.08); C128(1.20) | LDD1605 | [2] |
| LDCM0405 | CL18 | HEK-293T | C223(1.09) | LDD1609 | [2] |
| LDCM0406 | CL19 | HEK-293T | C128(0.93) | LDD1610 | [2] |
| LDCM0407 | CL2 | HEK-293T | C223(1.02); C128(1.06) | LDD1611 | [2] |
| LDCM0410 | CL22 | HEK-293T | C223(1.01) | LDD1614 | [2] |
| LDCM0414 | CL26 | HEK-293T | C223(0.83); C128(1.03) | LDD1618 | [2] |
| LDCM0419 | CL30 | HEK-293T | C223(1.00) | LDD1623 | [2] |
| LDCM0420 | CL31 | HEK-293T | C128(0.90) | LDD1624 | [2] |
| LDCM0423 | CL34 | HEK-293T | C223(0.77) | LDD1627 | [2] |
| LDCM0432 | CL42 | HEK-293T | C223(1.07) | LDD1636 | [2] |
| LDCM0433 | CL43 | HEK-293T | C128(0.85) | LDD1637 | [2] |
| LDCM0436 | CL46 | HEK-293T | C223(0.84) | LDD1640 | [2] |
| LDCM0441 | CL50 | HEK-293T | C223(0.99); C128(1.18) | LDD1645 | [2] |
| LDCM0445 | CL54 | HEK-293T | C223(0.96) | LDD1648 | [2] |
| LDCM0446 | CL55 | HEK-293T | C128(0.92) | LDD1649 | [2] |
| LDCM0449 | CL58 | HEK-293T | C223(0.92) | LDD1652 | [2] |
| LDCM0451 | CL6 | HEK-293T | C223(1.22) | LDD1654 | [2] |
| LDCM0454 | CL62 | HEK-293T | C223(0.92); C128(1.03) | LDD1657 | [2] |
| LDCM0458 | CL66 | HEK-293T | C223(1.19) | LDD1661 | [2] |
| LDCM0459 | CL67 | HEK-293T | C128(0.90) | LDD1662 | [2] |
| LDCM0462 | CL7 | HEK-293T | C128(0.86) | LDD1665 | [2] |
| LDCM0463 | CL70 | HEK-293T | C223(0.86) | LDD1666 | [2] |
| LDCM0467 | CL74 | HEK-293T | C223(0.98); C128(1.12) | LDD1670 | [2] |
| LDCM0471 | CL78 | HEK-293T | C223(1.10) | LDD1674 | [2] |
| LDCM0472 | CL79 | HEK-293T | C128(0.88) | LDD1675 | [2] |
| LDCM0476 | CL82 | HEK-293T | C223(0.85) | LDD1679 | [2] |
| LDCM0480 | CL86 | HEK-293T | C223(1.11); C128(1.27) | LDD1683 | [2] |
| LDCM0485 | CL90 | HEK-293T | C223(1.24) | LDD1688 | [2] |
| LDCM0486 | CL91 | HEK-293T | C128(0.88) | LDD1689 | [2] |
| LDCM0489 | CL94 | HEK-293T | C223(0.79) | LDD1692 | [2] |
| LDCM0493 | CL98 | HEK-293T | C223(1.06); C128(1.03) | LDD1696 | [2] |
| LDCM0427 | Fragment51 | HEK-293T | C223(0.90); C128(1.04) | LDD1631 | [2] |
| LDCM0008 | Tranylcypromine | SH-SY5Y | 5.36 | LDD0054 | [1] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| BOS complex subunit TMEM147 (TMEM147) | TMEM147 family | Q9BVK8 | |||
| Proteolipid protein 2 (PLP2) | . | Q04941 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Cortexin-3 (CTXN3) | Cortexin family | Q4LDR2 | |||
| Plasmolipin (PLLP) | MAL family | Q9Y342 | |||
| Vesicle-trafficking protein SEC22a (SEC22A) | Synaptobrevin family | Q96IW7 | |||
References
