Details of the Target
General Information of Target
| Target ID | LDTP05533 | |||||
|---|---|---|---|---|---|---|
| Target Name | Kinesin light chain 1 (KLC1) | |||||
| Gene Name | KLC1 | |||||
| Gene ID | 3831 | |||||
| Synonyms |
KLC; KNS2; Kinesin light chain 1; KLC 1 |
|||||
| 3D Structure | ||||||
| Sequence |
MYDNMSTMVYIKEDKLEKLTQDEIISKTKQVIQGLEALKNEHNSILQSLLETLKCLKKDD
ESNLVEEKSNMIRKSLEMLELGLSEAQVMMALSNHLNAVESEKQKLRAQVRRLCQENQWL RDELANTQQKLQKSEQSVAQLEEEKKHLEFMNQLKKYDDDISPSEDKDTDSTKEPLDDLF PNDEDDPGQGIQQQHSSAAAAAQQGGYEIPARLRTLHNLVIQYASQGRYEVAVPLCKQAL EDLEKTSGHDHPDVATMLNILALVYRDQNKYKDAANLLNDALAIREKTLGKDHPAVAATL NNLAVLYGKRGKYKEAEPLCKRALEIREKVLGKDHPDVAKQLNNLALLCQNQGKYEEVEY YYQRALEIYQTKLGPDDPNVAKTKNNLASCYLKQGKFKQAETLYKEILTRAHEREFGSVD DENKPIWMHAEEREECKGKQKDGTSFGEYGGWYKACKVDSPTVTTTLKNLGALYRRQGKF EAAETLEEAAMRSRKQGLDNVHKQRVAEVLNDPENMEKRRSRESLNVDVVKYESGPDGGE EVSMSVEWNGGVSGRASFCGKRQQQQWPGRRHR |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
Kinesin light chain family
|
|||||
| Subcellular location |
Cell projection, growth cone
|
|||||
| Function |
Kinesin is a microtubule-associated force-producing protein that may play a role in organelle transport. The light chain may function in coupling of cargo to the heavy chain or in the modulation of its ATPase activity.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
CY-1 Probe Info |
 |
100.00 | LDD0243 | [2] | |
|
TH216 Probe Info |
 |
Y223(10.61) | LDD0259 | [3] | |
|
ONAyne Probe Info |
 |
K503(0.43) | LDD0274 | [4] | |
|
STPyne Probe Info |
 |
K27(10.00); K272(6.35); K372(0.92); K398(3.85) | LDD0277 | [4] | |
|
Probe 1 Probe Info |
 |
Y355(3.71); Y360(48.52); Y369(12.64) | LDD3495 | [5] | |
|
DBIA Probe Info |
 |
C621(1.35) | LDD3310 | [6] | |
|
BTD Probe Info |
 |
C320(1.10); C559(1.28); C390(0.75); C349(1.23) | LDD2099 | [7] | |
|
Sulforaphane-probe2 Probe Info |
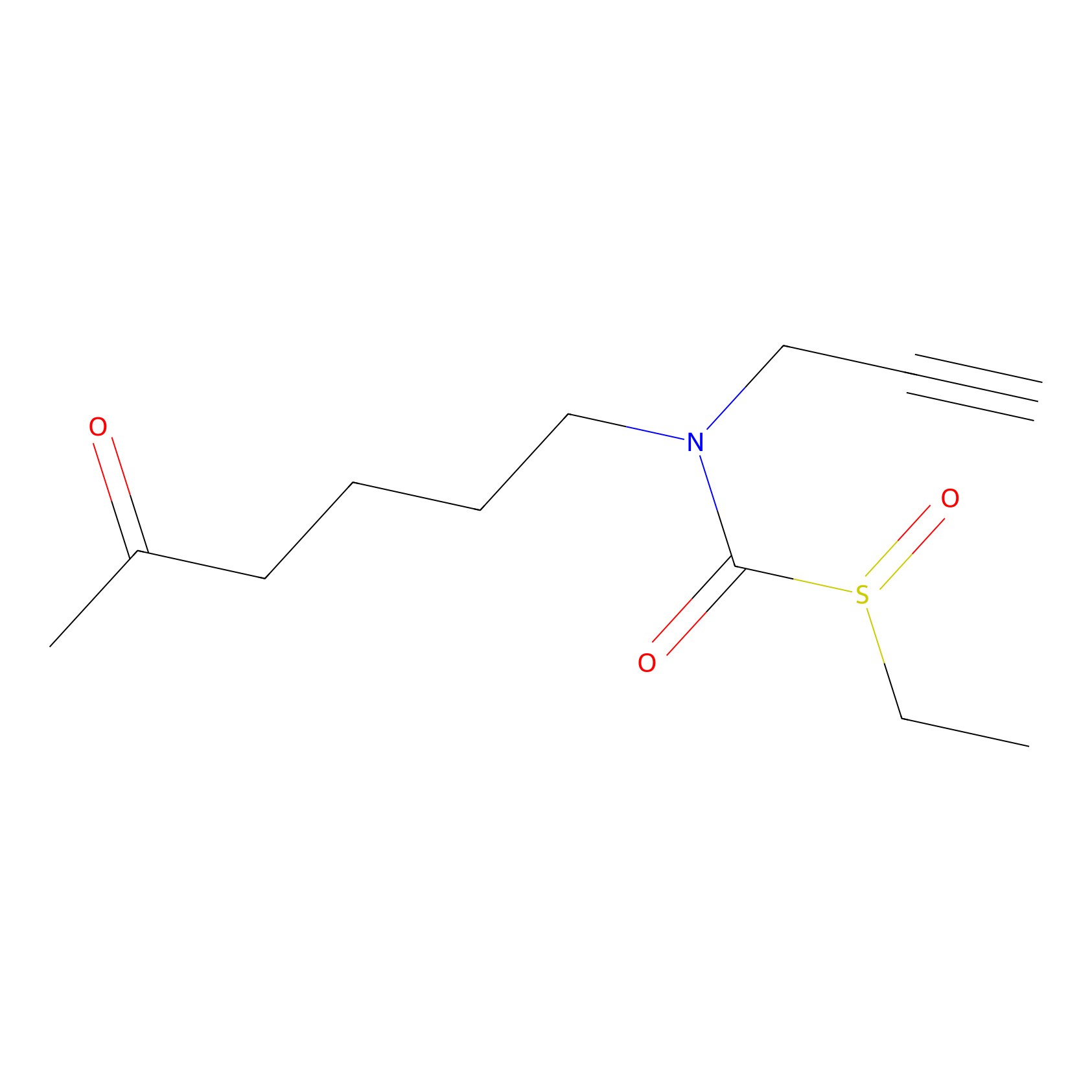 |
1.63 | LDD0160 | [8] | |
|
AHL-Pu-1 Probe Info |
 |
C114(2.60) | LDD0169 | [9] | |
|
HHS-482 Probe Info |
 |
Y223(1.44); Y307(0.65) | LDD0285 | [10] | |
|
HHS-475 Probe Info |
 |
Y265(0.68); Y449(0.91); Y223(0.97); Y307(1.08) | LDD0264 | [11] | |
|
HHS-465 Probe Info |
 |
Y223(10.00); Y307(7.29); Y449(10.00) | LDD2237 | [12] | |
|
5E-2FA Probe Info |
 |
H147(0.00); H217(0.00) | LDD2235 | [13] | |
|
ATP probe Probe Info |
 |
K309(0.00); K393(0.00); K272(0.00); K457(0.00) | LDD0199 | [14] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C114(0.00); C349(0.00); C390(0.00); C236(0.00) | LDD0038 | [15] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0032 | [16] | |
|
Lodoacetamide azide Probe Info |
 |
C320(0.00); C114(0.00); C390(0.00); C349(0.00) | LDD0037 | [15] | |
|
NAIA_4 Probe Info |
 |
C114(0.00); C456(0.00); C320(0.00); C390(0.00) | LDD2226 | [17] | |
|
TFBX Probe Info |
 |
C456(0.00); C236(0.00) | LDD0027 | [18] | |
|
Compound 10 Probe Info |
 |
N.A. | LDD2216 | [19] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [19] | |
|
IPM Probe Info |
 |
C559(0.00); C114(0.00) | LDD0005 | [20] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [20] | |
|
PF-06672131 Probe Info |
 |
N.A. | LDD0017 | [21] | |
|
Phosphinate-6 Probe Info |
 |
C114(0.00); C559(0.00); C320(0.00); C456(0.00) | LDD0018 | [22] | |
|
1c-yne Probe Info |
 |
K479(0.00); K309(0.00); K74(0.00) | LDD0228 | [23] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [24] | |
|
W1 Probe Info |
 |
N218(0.00); H217(0.00) | LDD0236 | [25] | |
|
NAIA_5 Probe Info |
 |
C456(0.00); C55(0.00); C320(0.00); C390(0.00) | LDD2223 | [17] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C158 Probe Info |
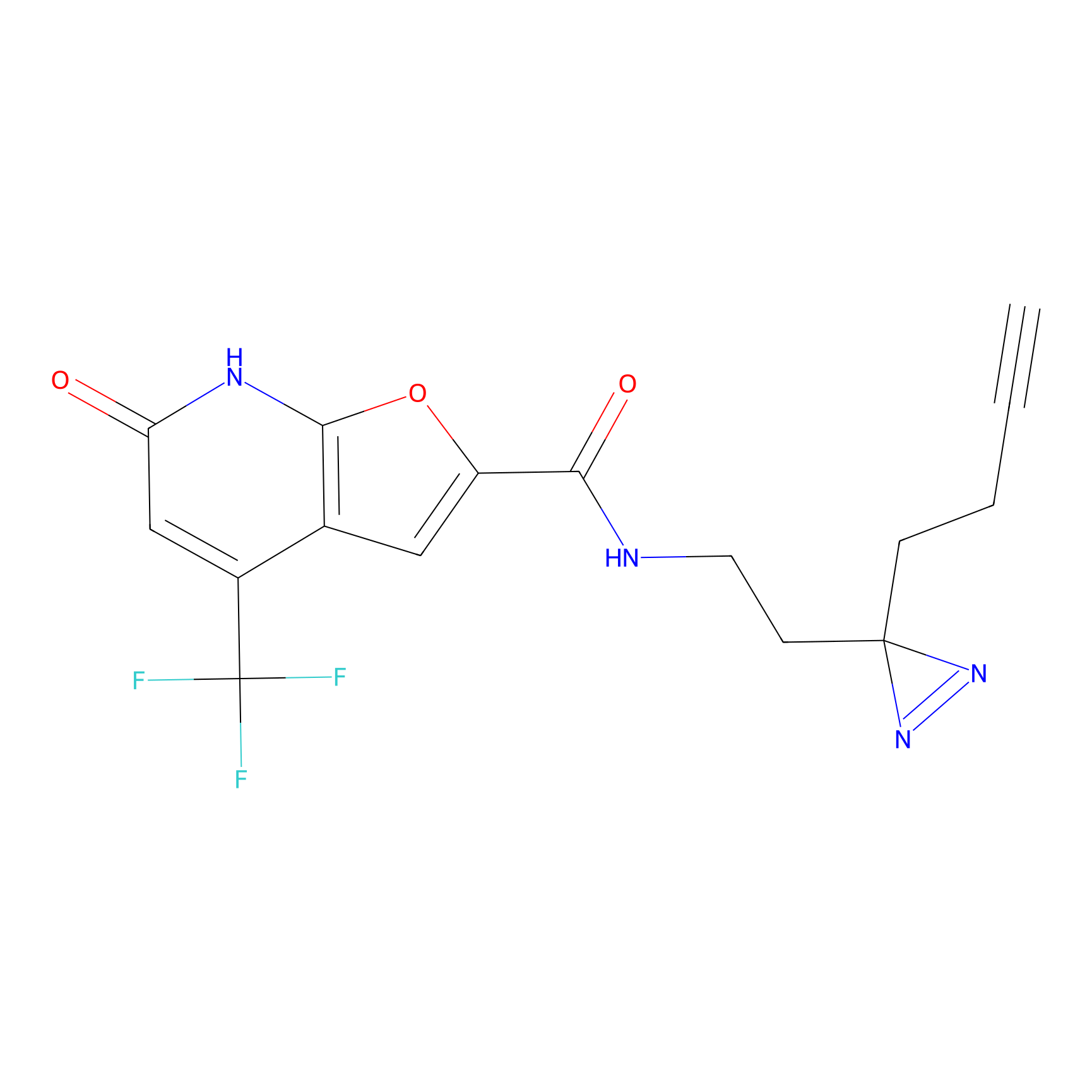 |
9.58 | LDD1838 | [26] | |
|
C187 Probe Info |
 |
16.68 | LDD1865 | [26] | |
|
C191 Probe Info |
 |
9.51 | LDD1868 | [26] | |
|
C275 Probe Info |
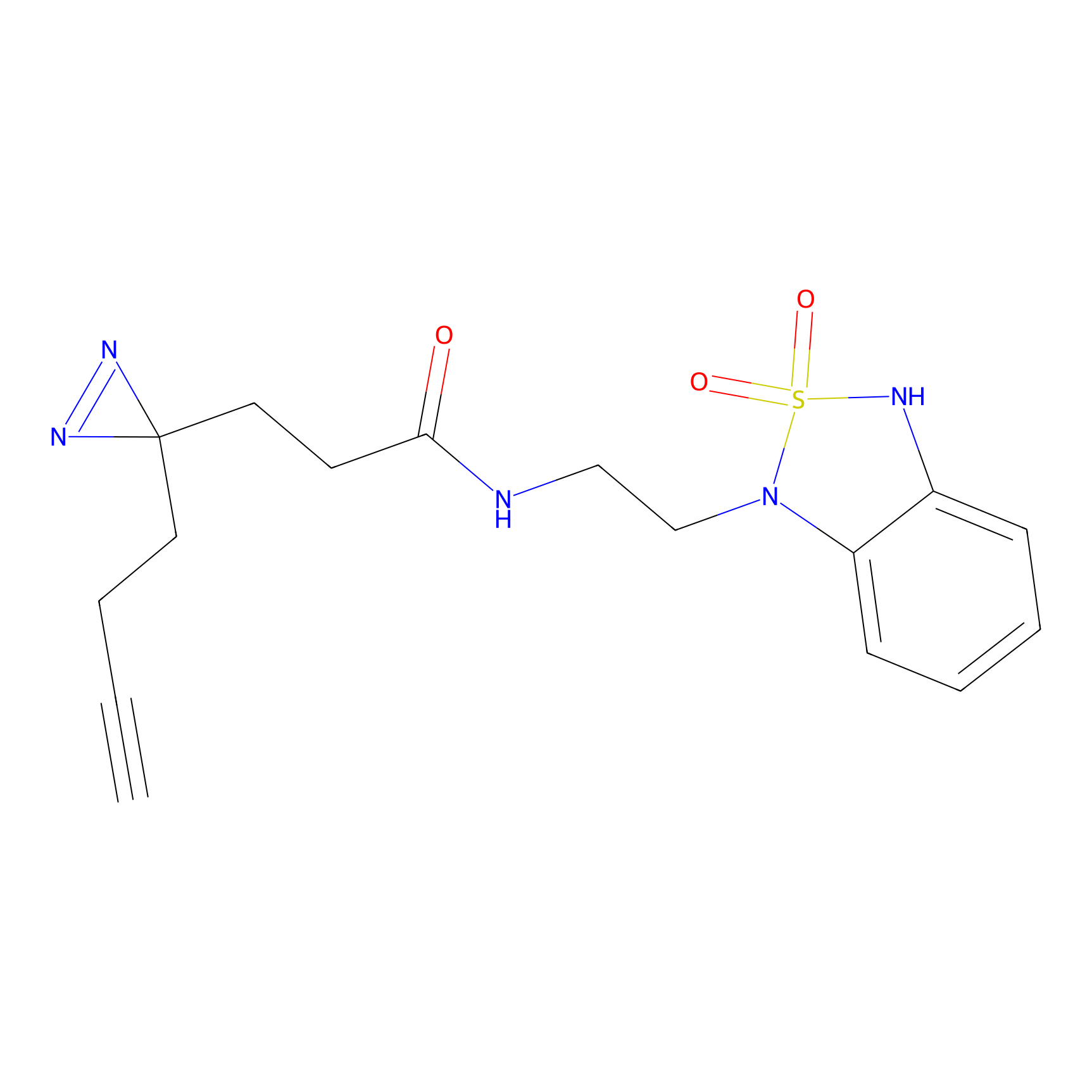 |
7.01 | LDD1945 | [26] | |
|
FFF probe11 Probe Info |
 |
6.67 | LDD0472 | [27] | |
|
FFF probe13 Probe Info |
 |
20.00 | LDD0475 | [27] | |
|
FFF probe14 Probe Info |
 |
16.49 | LDD0477 | [27] | |
|
FFF probe2 Probe Info |
 |
20.00 | LDD0463 | [27] | |
|
FFF probe3 Probe Info |
 |
9.11 | LDD0464 | [27] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C320(1.48); C236(1.30); C559(1.22); C114(1.14) | LDD2117 | [7] |
| LDCM0025 | 4SU-RNA | DM93 | C436(2.26) | LDD0170 | [9] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C114(2.60) | LDD0169 | [9] |
| LDCM0156 | Aniline | NCI-H1299 | 11.53 | LDD0403 | [1] |
| LDCM0632 | CL-Sc | Hep-G2 | C114(20.00); C236(2.90); C114(1.17); C390(1.06) | LDD2227 | [17] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C114(0.92); C559(0.81) | LDD1702 | [7] |
| LDCM0625 | F8 | Ramos | C236(1.64) | LDD2187 | [28] |
| LDCM0572 | Fragment10 | Ramos | C236(1.07) | LDD2189 | [28] |
| LDCM0573 | Fragment11 | Ramos | C236(3.34) | LDD2190 | [28] |
| LDCM0574 | Fragment12 | Ramos | C236(0.67) | LDD2191 | [28] |
| LDCM0575 | Fragment13 | Ramos | C236(0.96) | LDD2192 | [28] |
| LDCM0576 | Fragment14 | Ramos | C236(0.99) | LDD2193 | [28] |
| LDCM0579 | Fragment20 | Ramos | C236(0.44) | LDD2194 | [28] |
| LDCM0580 | Fragment21 | Ramos | C236(1.01) | LDD2195 | [28] |
| LDCM0582 | Fragment23 | Ramos | C236(0.88) | LDD2196 | [28] |
| LDCM0578 | Fragment27 | Ramos | C236(0.94) | LDD2197 | [28] |
| LDCM0586 | Fragment28 | Ramos | C236(0.48) | LDD2198 | [28] |
| LDCM0588 | Fragment30 | Ramos | C236(1.01) | LDD2199 | [28] |
| LDCM0589 | Fragment31 | Ramos | C236(1.10) | LDD2200 | [28] |
| LDCM0590 | Fragment32 | Ramos | C236(0.96) | LDD2201 | [28] |
| LDCM0468 | Fragment33 | Ramos | C236(0.79) | LDD2202 | [28] |
| LDCM0596 | Fragment38 | Ramos | C236(0.63) | LDD2203 | [28] |
| LDCM0566 | Fragment4 | Ramos | C236(0.60) | LDD2184 | [28] |
| LDCM0610 | Fragment52 | Ramos | C236(0.86) | LDD2204 | [28] |
| LDCM0614 | Fragment56 | Ramos | C236(1.45) | LDD2205 | [28] |
| LDCM0569 | Fragment7 | Ramos | C236(0.58) | LDD2186 | [28] |
| LDCM0571 | Fragment9 | Ramos | C236(1.85) | LDD2188 | [28] |
| LDCM0149 | GA | MCF-7 | C55(2.60) | LDD0379 | [29] |
| LDCM0116 | HHS-0101 | DM93 | Y265(0.68); Y449(0.91); Y223(0.97); Y307(1.08) | LDD0264 | [11] |
| LDCM0117 | HHS-0201 | DM93 | Y223(0.80); Y265(0.82); Y307(0.82); Y313(1.49) | LDD0265 | [11] |
| LDCM0118 | HHS-0301 | DM93 | Y313(0.06); Y265(0.60); Y449(0.71); Y223(0.71) | LDD0266 | [11] |
| LDCM0119 | HHS-0401 | DM93 | Y265(0.72); Y223(0.74); Y307(0.91); Y449(0.92) | LDD0267 | [11] |
| LDCM0120 | HHS-0701 | DM93 | Y307(0.85); Y223(0.92); Y265(0.99); Y449(1.03) | LDD0268 | [11] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [24] |
| LDCM0123 | JWB131 | DM93 | Y223(1.44); Y307(0.65) | LDD0285 | [10] |
| LDCM0124 | JWB142 | DM93 | Y223(0.28); Y307(0.70) | LDD0286 | [10] |
| LDCM0125 | JWB146 | DM93 | Y223(2.28); Y307(0.80) | LDD0287 | [10] |
| LDCM0126 | JWB150 | DM93 | Y223(3.79); Y307(2.18) | LDD0288 | [10] |
| LDCM0127 | JWB152 | DM93 | Y223(2.15); Y307(1.29) | LDD0289 | [10] |
| LDCM0128 | JWB198 | DM93 | Y223(1.52); Y307(0.39) | LDD0290 | [10] |
| LDCM0129 | JWB202 | DM93 | Y223(0.47); Y307(0.50) | LDD0291 | [10] |
| LDCM0130 | JWB211 | DM93 | Y223(1.04); Y307(0.78) | LDD0292 | [10] |
| LDCM0022 | KB02 | T cell-activated | C559(5.18) | LDD1704 | [30] |
| LDCM0023 | KB03 | Jurkat | C390(4.09); C456(7.91) | LDD0315 | [16] |
| LDCM0024 | KB05 | COLO792 | C621(1.35) | LDD3310 | [6] |
| LDCM0509 | N-(4-bromo-3,5-dimethylphenyl)-2-nitroacetamide | MDA-MB-231 | C236(1.02) | LDD2102 | [7] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [24] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C320(1.10); C559(1.28); C390(0.75); C349(1.23) | LDD2099 | [7] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C320(0.81) | LDD2105 | [7] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C320(0.97); C559(1.09) | LDD2107 | [7] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C349(0.56) | LDD2109 | [7] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C559(0.31) | LDD2114 | [7] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C320(2.57) | LDD2119 | [7] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C320(0.84) | LDD2123 | [7] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C236(0.97); C114(1.44); C349(0.89) | LDD2125 | [7] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C320(1.39); C236(1.32); C559(0.78) | LDD2127 | [7] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C390(1.13) | LDD2136 | [7] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C236(1.14); C559(1.19); C390(1.20) | LDD2137 | [7] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C320(1.20); C236(0.98) | LDD2140 | [7] |
| LDCM0553 | Nucleophilic fragment 6b | MDA-MB-231 | C390(1.42) | LDD2147 | [7] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C456(0.67) | LDD2206 | [31] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C456(0.99) | LDD2207 | [31] |
| LDCM0003 | Sulforaphane | MDA-MB-231 | 1.63 | LDD0160 | [8] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| BCL2/adenovirus E1B 19 kDa protein-interacting protein 2 (BNIP2) | . | Q12982 | |||
| Caytaxin (ATCAY) | . | Q86WG3 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Amyloid-beta precursor protein (APP) | APP family | P05067 | |||
Other
References
