Details of the Target
General Information of Target
| Target ID | LDTP04111 | |||||
|---|---|---|---|---|---|---|
| Target Name | Small ribosomal subunit protein eS10 (RPS10) | |||||
| Gene Name | RPS10 | |||||
| Gene ID | 6204 | |||||
| Synonyms |
Small ribosomal subunit protein eS10; 40S ribosomal protein S10 |
|||||
| 3D Structure | ||||||
| Sequence |
MLMPKKNRIAIYELLFKEGVMVAKKDVHMPKHPELADKNVPNLHVMKAMQSLKSRGYVKE
QFAWRHFYWYLTNEGIQYLRDYLHLPPEIVPATLRRSRPETGRPRPKGLEGERPARLTRG EADRDTYRRSAVPPGADKKAEAGAGSATEFQFRGGFGRGRGQPPQ |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
Eukaryotic ribosomal protein eS10 family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function | Component of the 40S ribosomal subunit. The ribosome is a large ribonucleoprotein complex responsible for the synthesis of proteins in the cell. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C-Sul Probe Info |
 |
18.58 | LDD0066 | [1] | |
|
TH216 Probe Info |
 |
Y127(20.00) | LDD0259 | [2] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [3] | |
|
AZ-9 Probe Info |
 |
E149(0.84); E141(0.93); E110(0.98) | LDD2208 | [4] | |
|
ONAyne Probe Info |
 |
K53(0.00); K107(0.00) | LDD0273 | [5] | |
|
Probe 1 Probe Info |
 |
Y127(53.17) | LDD3495 | [6] | |
|
HPAP Probe Info |
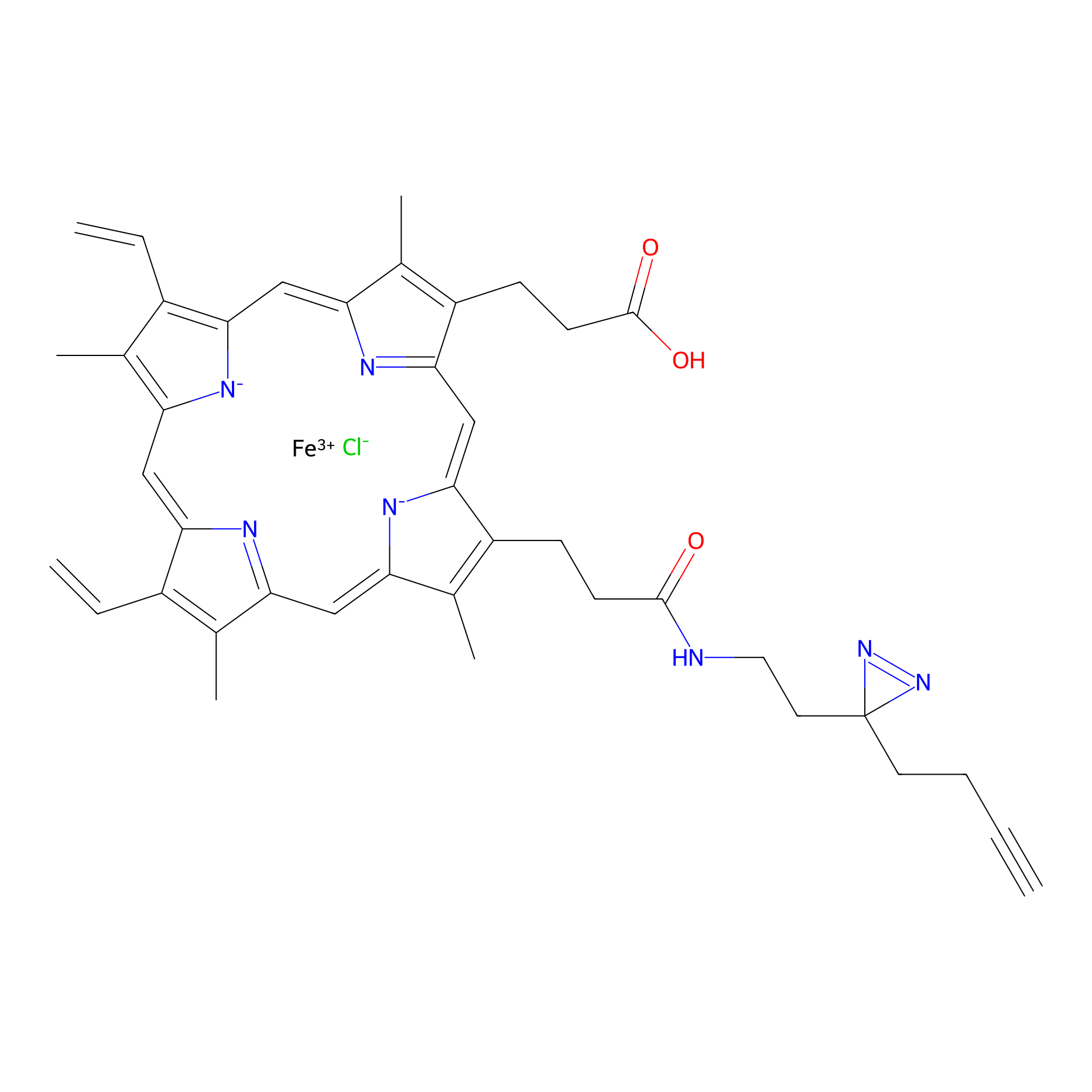 |
3.43 | LDD0063 | [7] | |
|
HHS-465 Probe Info |
 |
Y127(10.00) | LDD2237 | [8] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [9] | |
|
AMP probe Probe Info |
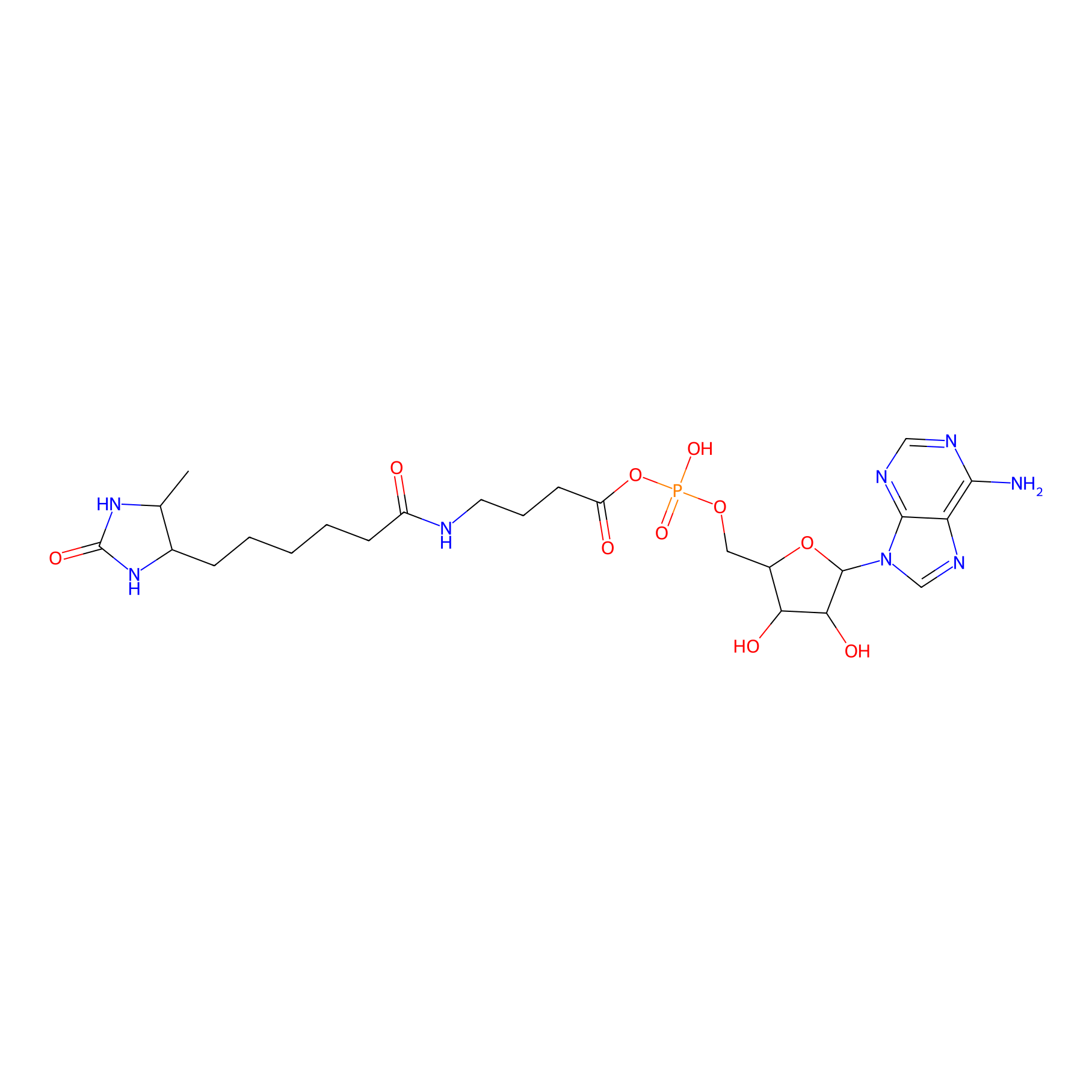 |
N.A. | LDD0200 | [10] | |
|
ATP probe Probe Info |
 |
K139(0.00); K38(0.00); K59(0.00); K31(0.00) | LDD0199 | [10] | |
|
m-APA Probe Info |
 |
N.A. | LDD2231 | [9] | |
|
ATP probe Probe Info |
 |
K59(0.00); K47(0.00); K139(0.00) | LDD0035 | [11] | |
|
1d-yne Probe Info |
 |
N.A. | LDD0356 | [12] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [13] | |
|
OSF Probe Info |
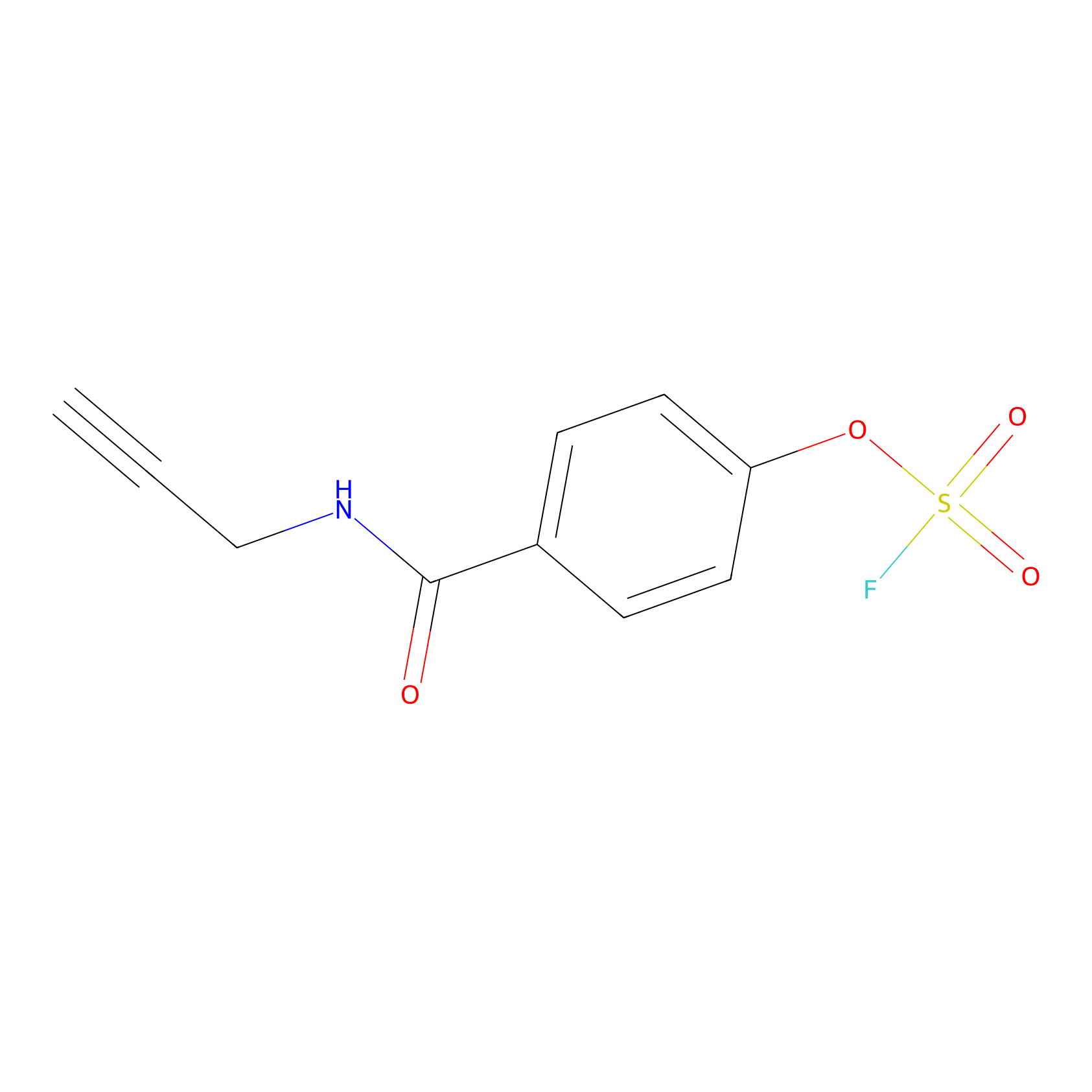 |
N.A. | LDD0029 | [14] | |
|
SF Probe Info |
 |
Y82(0.00); Y12(0.00); K139(0.00) | LDD0028 | [14] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [13] | |
|
Ox-W18 Probe Info |
 |
W69(0.00); W64(0.00) | LDD2175 | [15] | |
|
1c-yne Probe Info |
 |
K139(0.00); K38(0.00); K17(0.00) | LDD0228 | [12] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [16] | |
|
W1 Probe Info |
 |
Y68(0.00); W69(0.00); H66(0.00) | LDD0236 | [17] | |
|
HHS-482 Probe Info |
 |
Y12(1.01) | LDD2239 | [8] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C055 Probe Info |
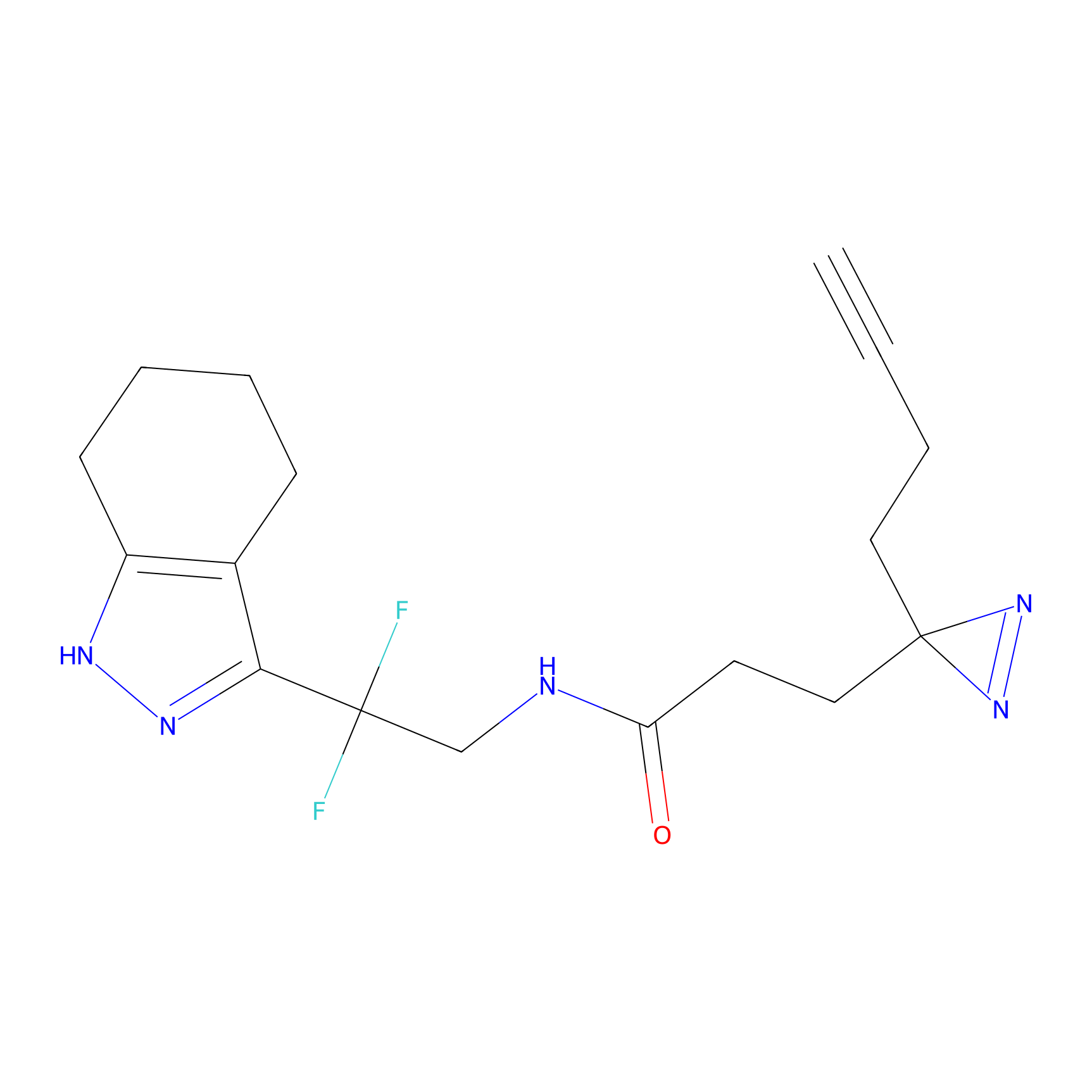 |
11.88 | LDD1752 | [18] | |
|
C087 Probe Info |
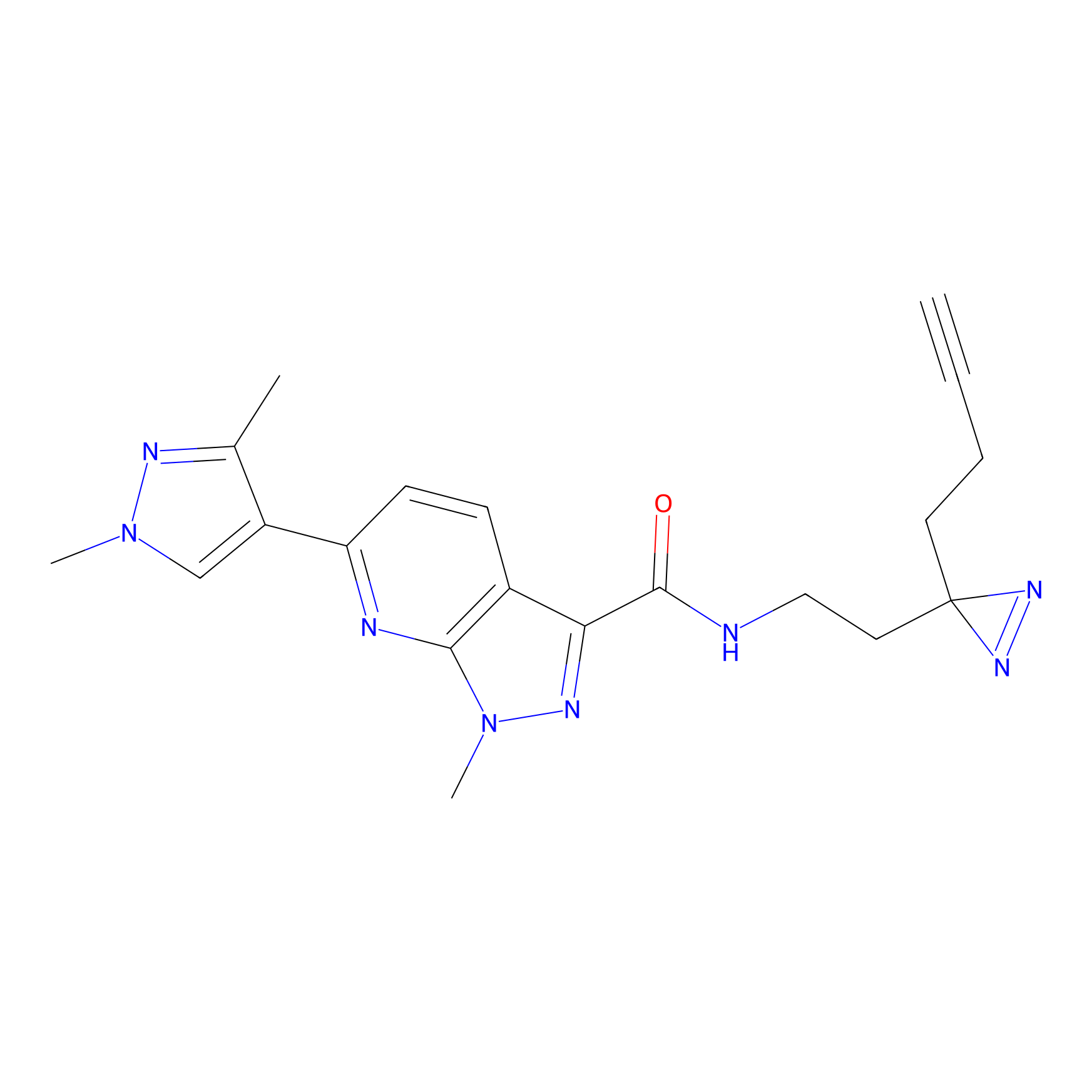 |
7.46 | LDD1779 | [18] | |
|
C244 Probe Info |
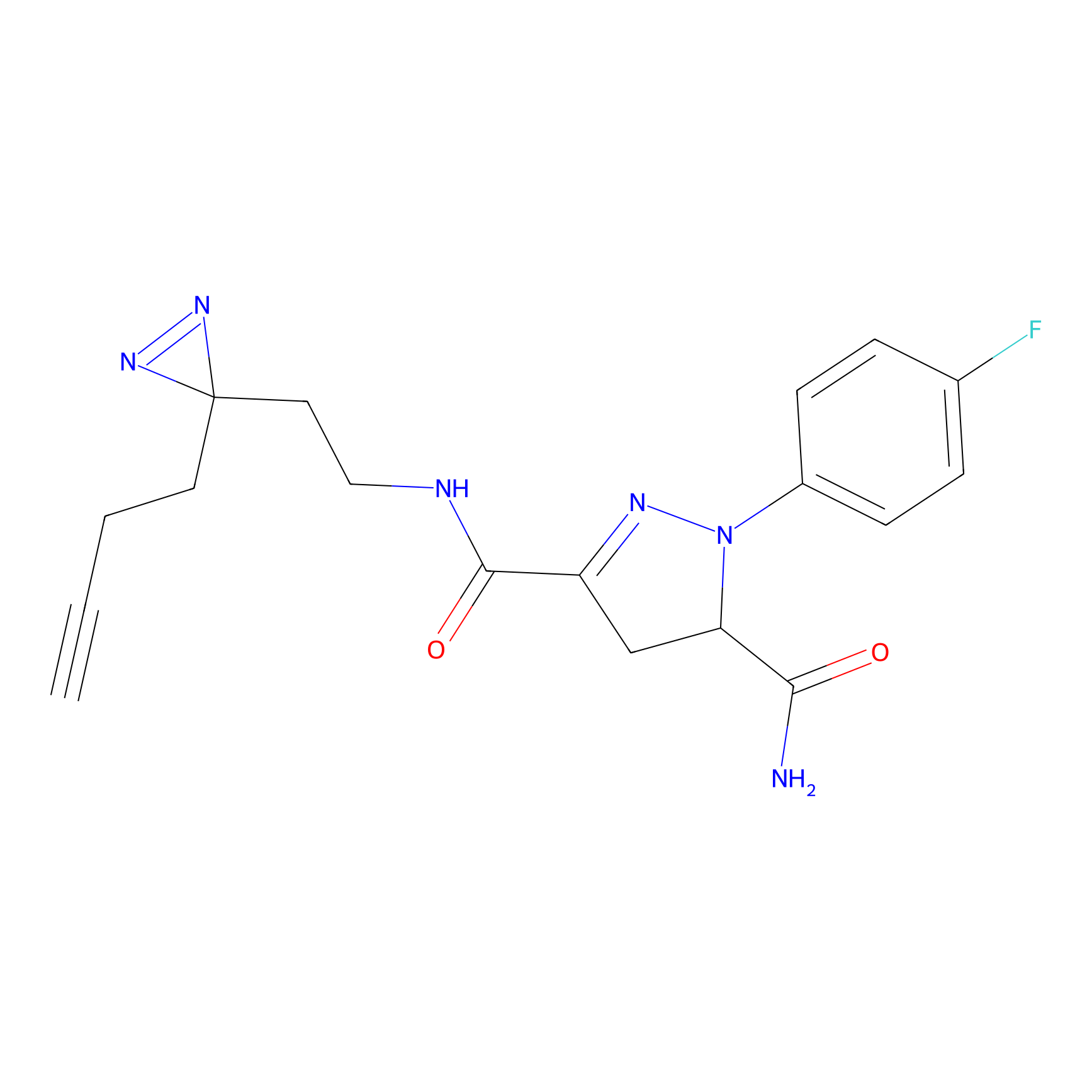 |
14.22 | LDD1917 | [18] | |
|
C313 Probe Info |
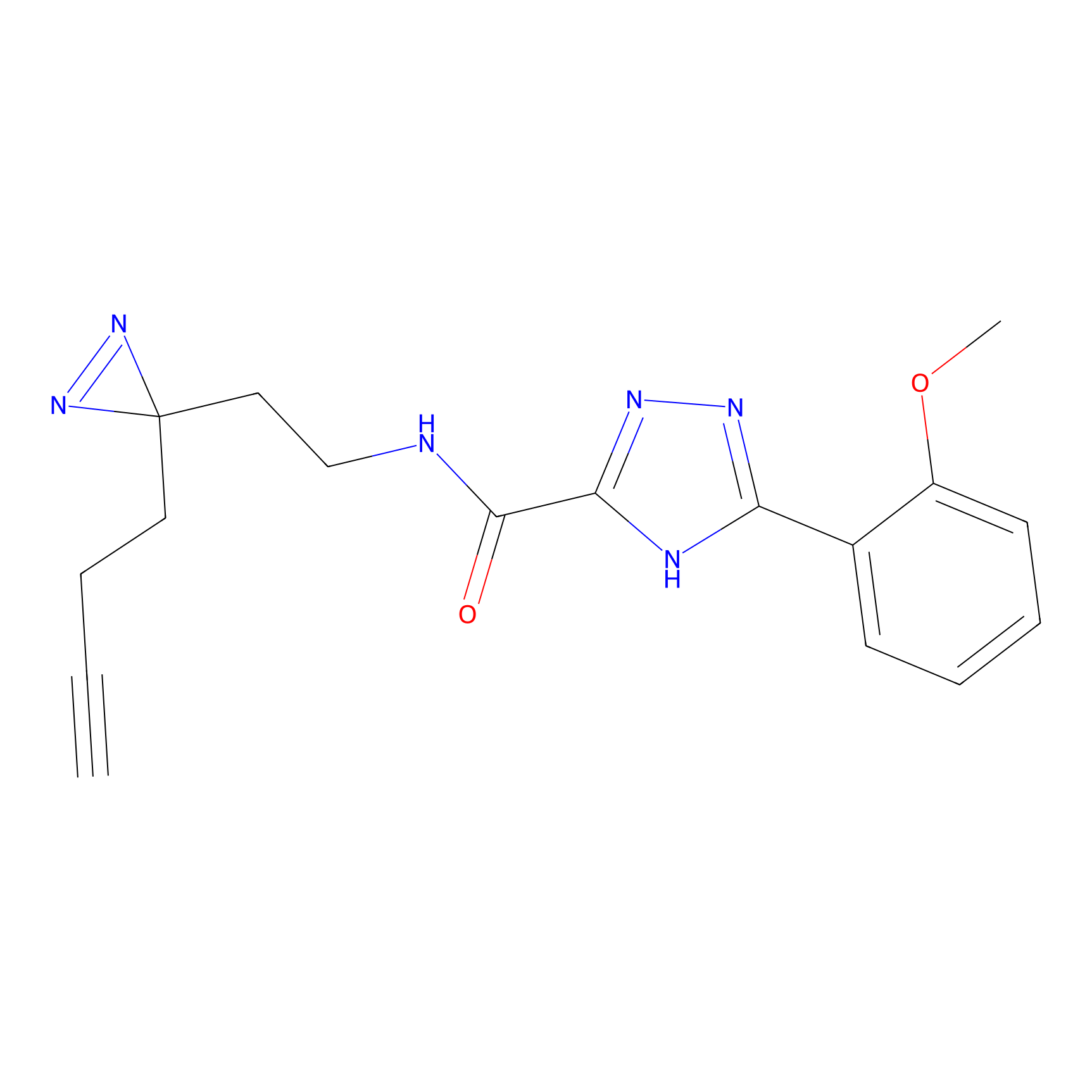 |
12.73 | LDD1980 | [18] | |
|
C317 Probe Info |
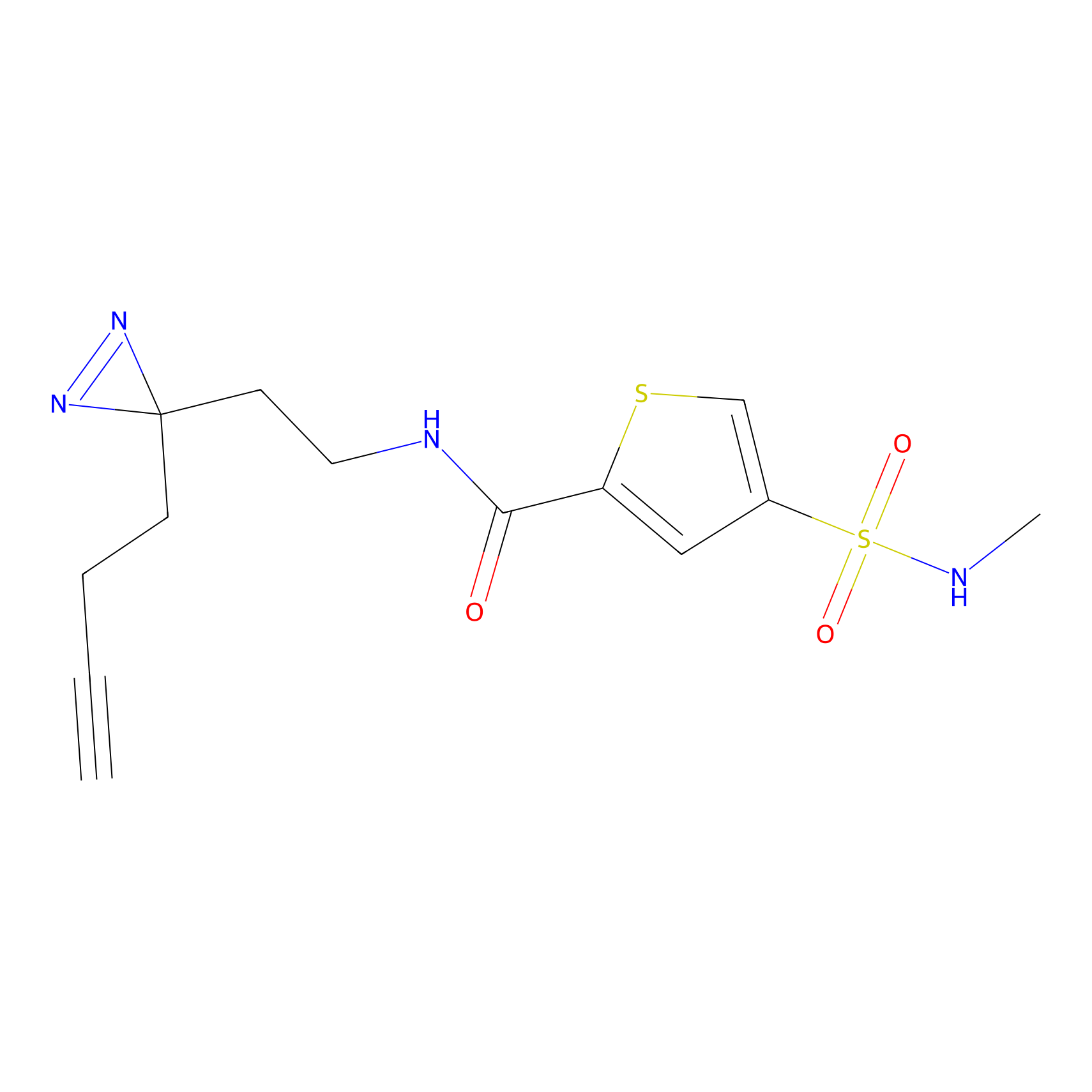 |
6.06 | LDD1983 | [18] | |
|
C354 Probe Info |
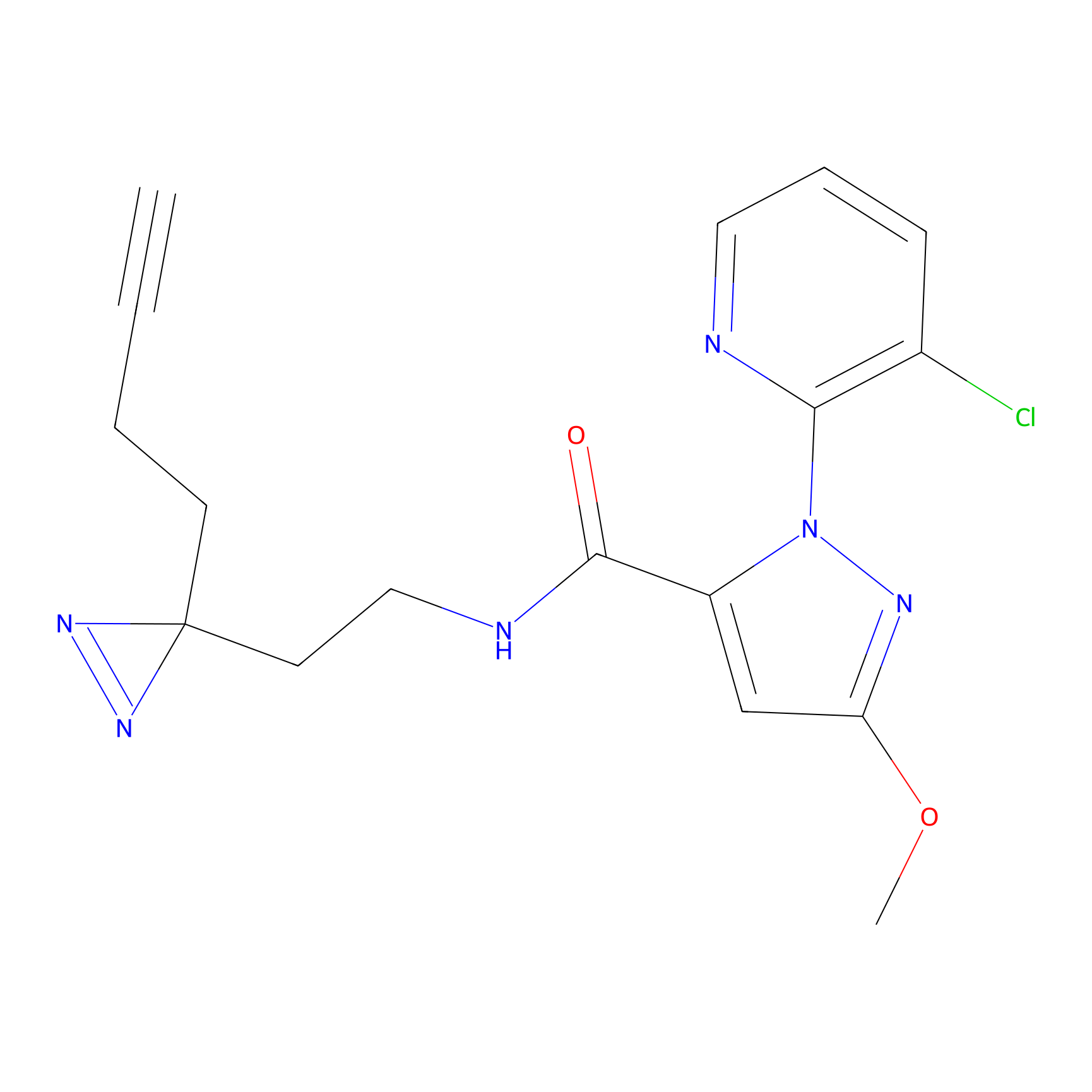 |
8.57 | LDD2015 | [18] | |
|
C391 Probe Info |
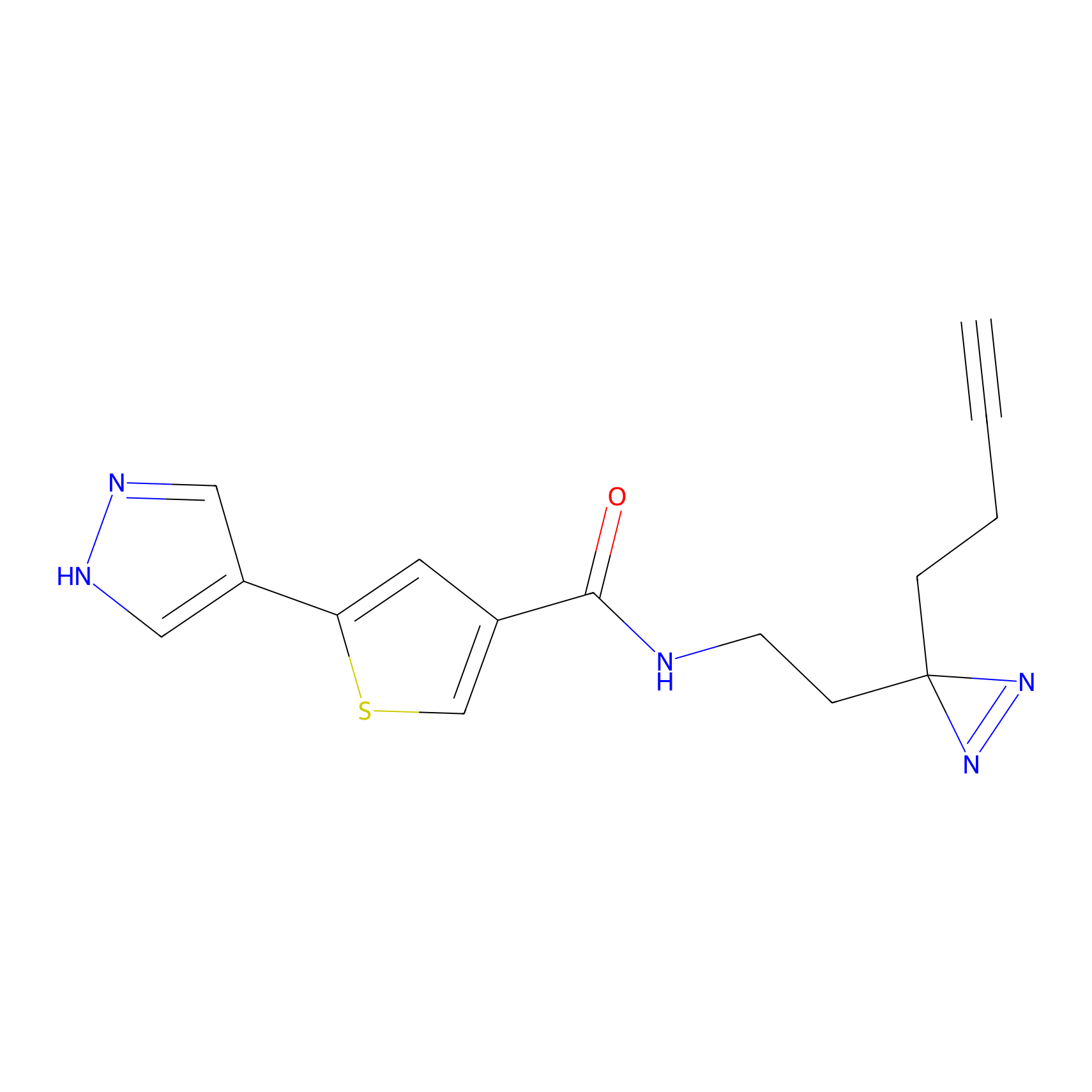 |
14.42 | LDD2050 | [18] | |
|
C403 Probe Info |
 |
17.03 | LDD2061 | [18] | |
|
FFF probe13 Probe Info |
 |
5.94 | LDD0475 | [19] | |
|
FFF probe3 Probe Info |
 |
5.99 | LDD0464 | [19] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0138 | [20] | |
|
Diazir Probe Info |
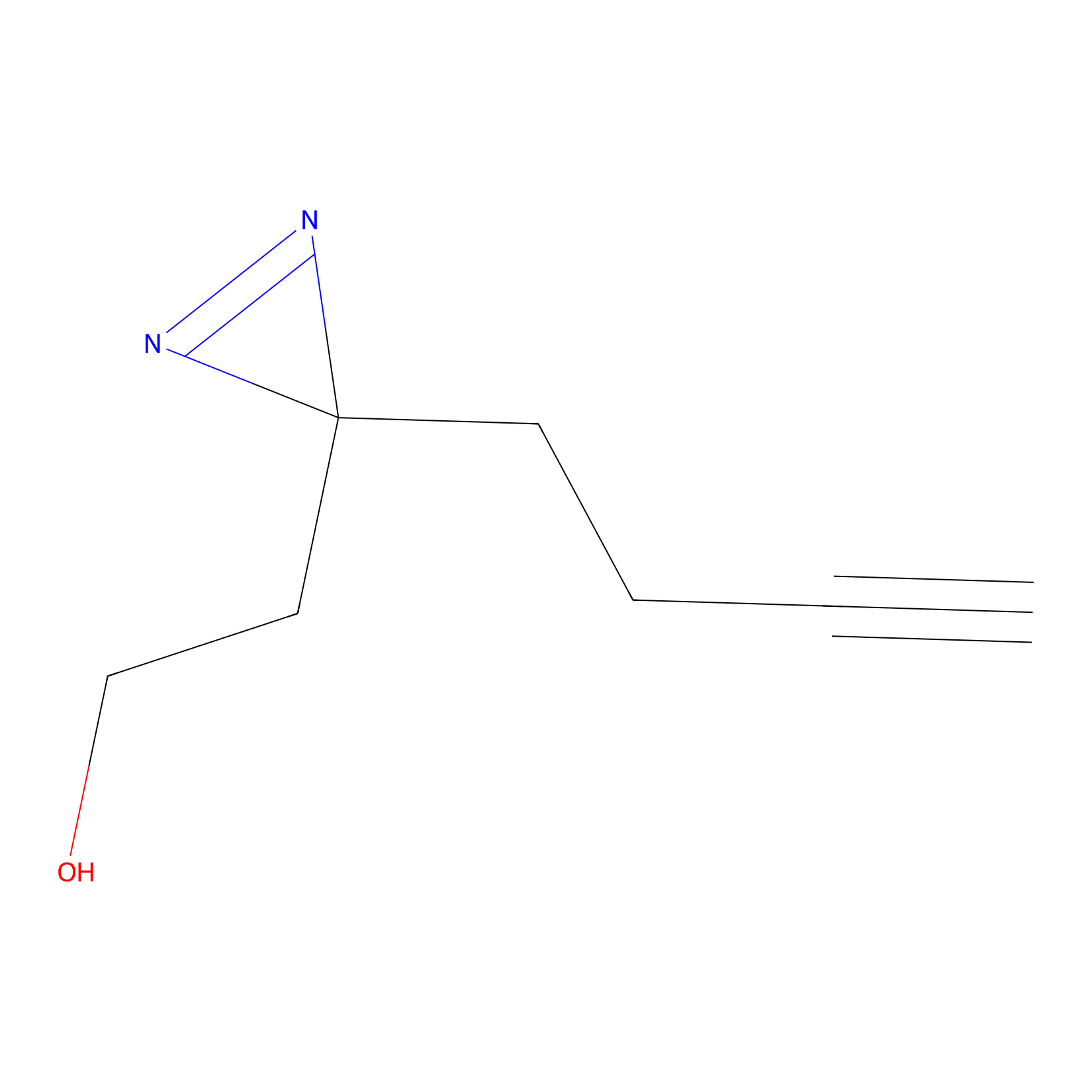 |
N.A. | LDD0011 | [13] | |
|
BD-F Probe Info |
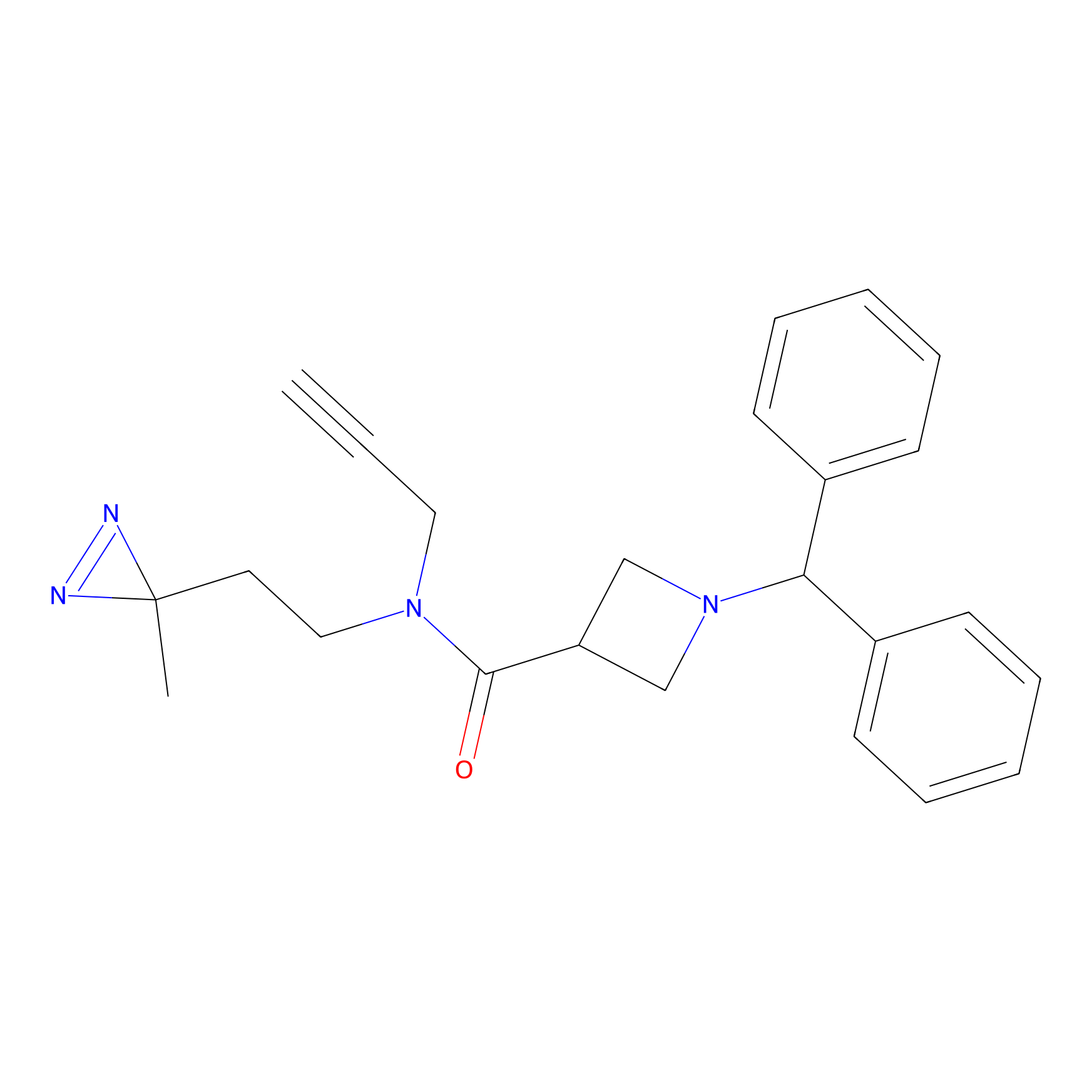 |
N.A. | LDD0024 | [21] | |
|
LD-F Probe Info |
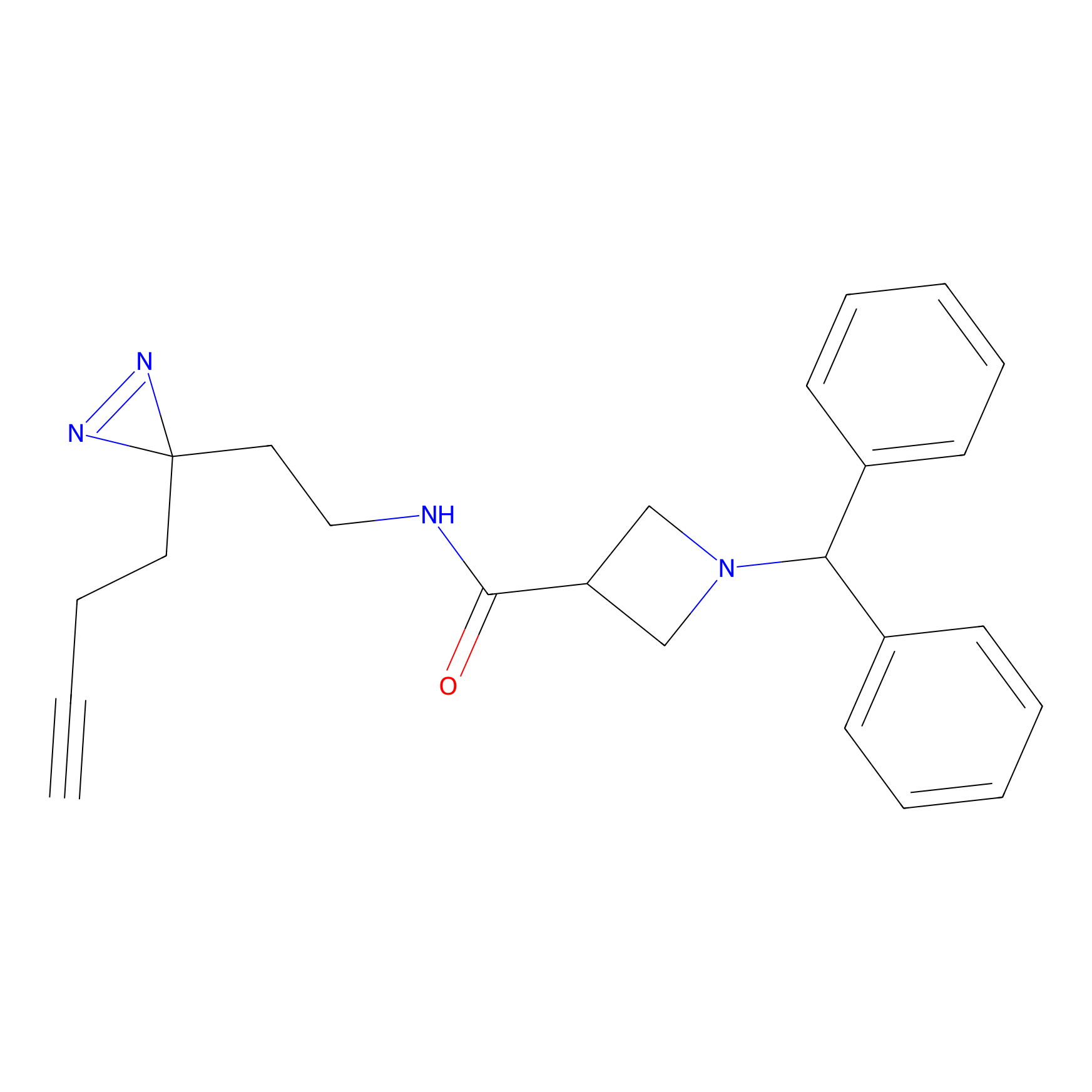 |
L83(0.00); H84(0.00); D81(0.00) | LDD0015 | [21] | |
|
DA-2 Probe Info |
 |
N.A. | LDD0072 | [22] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0068 | [23] | |
Competitor(s) Related to This Target
References
