Details of the Target
General Information of Target
| Target ID | LDTP10148 | |||||
|---|---|---|---|---|---|---|
| Target Name | Store-operated calcium entry-associated regulatory factor (SARAF) | |||||
| Gene Name | SARAF | |||||
| Gene ID | 51669 | |||||
| Synonyms |
TMEM66; XTP3; Store-operated calcium entry-associated regulatory factor; SARAF; SOCE-associated regulatory factor; HBV X-transactivated gene 3 protein; HBV XAg-transactivated protein 3; Protein FOAP-7; Transmembrane protein 66
|
|||||
| 3D Structure | ||||||
| Sequence |
MPWPFSESIKKRACRYLLQRYLGHFLQEKLSLEQLSLDLYQGTGSLAQVPLDKWCLNEIL
ESADAPLEVTEGFIQSISLSVPWGSLLQDNCALEVRGLEMVFRPRPRPATGSEPMYWSSF MTSSMQLAKECLSQKLTDEQGEGSQPFEGLEKFAETIETVLRRVKVTFIDTVLRIEHVPE NSKTGTALEIRIERTVYCDETADESSGINVHQPTAFAHKLLQLSGVSLFWDEFSASAKSS PVCSTAPVETEPKLSPSWNPKIIYEPHPQLTRNLPEIAPSDPVQIGRLIGRLELSLTLKQ NEVLPGAKLDVDGQIDSIHLLLSPRQVHLLLDMLAAIAGPENSSKIGLANKDRKNRPMQQ EDEYRIQMELNRYYLRKDSLSVGVSSEQSFYETETARTPSSREEEVFFSMADMDMSHSLS SLPPLGDPPNMDLELSLTSTYTNTPAGSPLSATVLQPTWGEFLDHHKEQPVRGSTFPSNL VHPTPLQKTSLPSRSVSVDESRPELIFRLAVGTFSISVLHIDPLSPPETSQNLNPLTPMA VAFFTCIEKIDPARFSTEDFKSFRAVFAEACSHDHLRFIGTGIKVSYEQRQRSASRYFST DMSIGQMEFLECLFPTDFHSVPPHYTELLTFHSKEETGSHSPVCLQLHYKHSENRGPQGN QARLSSVPHKAELQIKLNPVCCELDISIVDRLNSLLQPQKLATVEMMASHMYTSYNKHIS LHKAFTEVFLDDSHSPANCRISVQVATPALNLSVRFPIPDLRSDQERGPWFKKSLQKEIL YLAFTDLEFKTEFIGGSTPEQIKLELTFRELIGSFQEEKGDPSIKFFHVSSGVDGDTTSS DDFDWPRIVLKINPPAMHSILERIAAEEEEENDGHYQEEEEGGAHSLKDVCDLRRPAPSP FSSRRVMFENEQMVMPGDPVEMTEFQDKAISNSHYVLELTLPNIYVTLPNKSFYEKLYNR IFNDLLLWEPTAPSPVETFENISYGIGLSVASQLINTFNKDSFSAFKSAVHYDEESGSEE ETLQYFSTVDPNYRSRRKKKLDSQNKNSQSFLSVLLNINHGLIAVFTDVKQDNGDLLENK HGEFWLEFNSGSLFCVTKYEGFDDKHYICLHSSSFSLYHKGIVNGVILPTETRLPSSTRP HWLEPTIYSSEEDGLSKTSSDGVGGDSLNMLSVAVKILSDKSESNTKEFLIAVGLKGATL QHRMLPSGLSWHEQILYFLNIADEPVLGYNPPTSFTTFHVHLWSCALDYRPLYLPIRSLL TVETFSVSSSVALDKSSSTLRIILDEAALHLSDKCNTVTINLSRDYVRVMDMGLLELTIT AVKSDSDGEQTEPRFELHCSSDVVHIRTCSDSCAALMNLIQYIASYGDLQTPNKADMKPG AFQRRSKVDSSGRSSSRGPVLPEADQQMLRDLMSDAMEEIDMQQGTSSVKPQANGVLDEK SQIQEPCCSDLFLFPDESGNVSQESGPTYASFSHHFISDAMTGVPTENDDFCILFAPKAA MQEKEEEPVIKIMVDDAIVIRDNYFSLPVNKTDTSKAPLHFPIPVIRYVVKEVSLVWHLY GGKDFGIVPPTSPAKSYISPHSSPSHTPTRHGRNTVCGGKGRNHDFLMEIQLSKVKFQHE VYPPCKPDCDSSLSEHPVSRQVFIVQDLEIRDRLATSQMNKFLYLYCSKEMPRKAHSNML TVKALHVCPESGRSPQECCLRVSLMPLRLNIDQDALFFLKDFFTSLSAEVELQMTPDPEV KKSPGADVTCSLPRHLSTSKEPNLVISFSGPKQPSQNDSANSVEVVNGMEEKNFSAEEAS FRDQPVFFREFRFTSEVPIRLDYHGKHVSMDQGTLAGILIGLAQLNCSELKLKRLSYRHG LLGVDKLFSYAITEWLNDIKKNQLPGILGGVGPMHSLVQLVQGLKDLVWLPIEQYRKDGR IVRGFQRGAASFGTSTAMAALELTNRMVQTIQAAAETAYDMVSPGTLSIEPKKTKRFPHH RLAHQPVDLREGVAKAYSVVKEGITDTAQTIYETAAREHESRGVTGAVGEVLRQIPPAVV KPLIVATEATSNVLGGMRNQIRPDVRQDESQKWRHGDD |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
SARAF family
|
|||||
| Subcellular location |
Endoplasmic reticulum membrane
|
|||||
| Function |
Negative regulator of store-operated Ca(2+) entry (SOCE) involved in protecting cells from Ca(2+) overfilling. In response to cytosolic Ca(2+) elevation after endoplasmic reticulum Ca(2+) refilling, promotes a slow inactivation of STIM (STIM1 or STIM2)-dependent SOCE activity: possibly act by facilitating the deoligomerization of STIM to efficiently turn off ORAI when the endoplasmic reticulum lumen is filled with the appropriate Ca(2+) levels, and thus preventing the overload of the cell with excessive Ca(2+) ions.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
TG42 Probe Info |
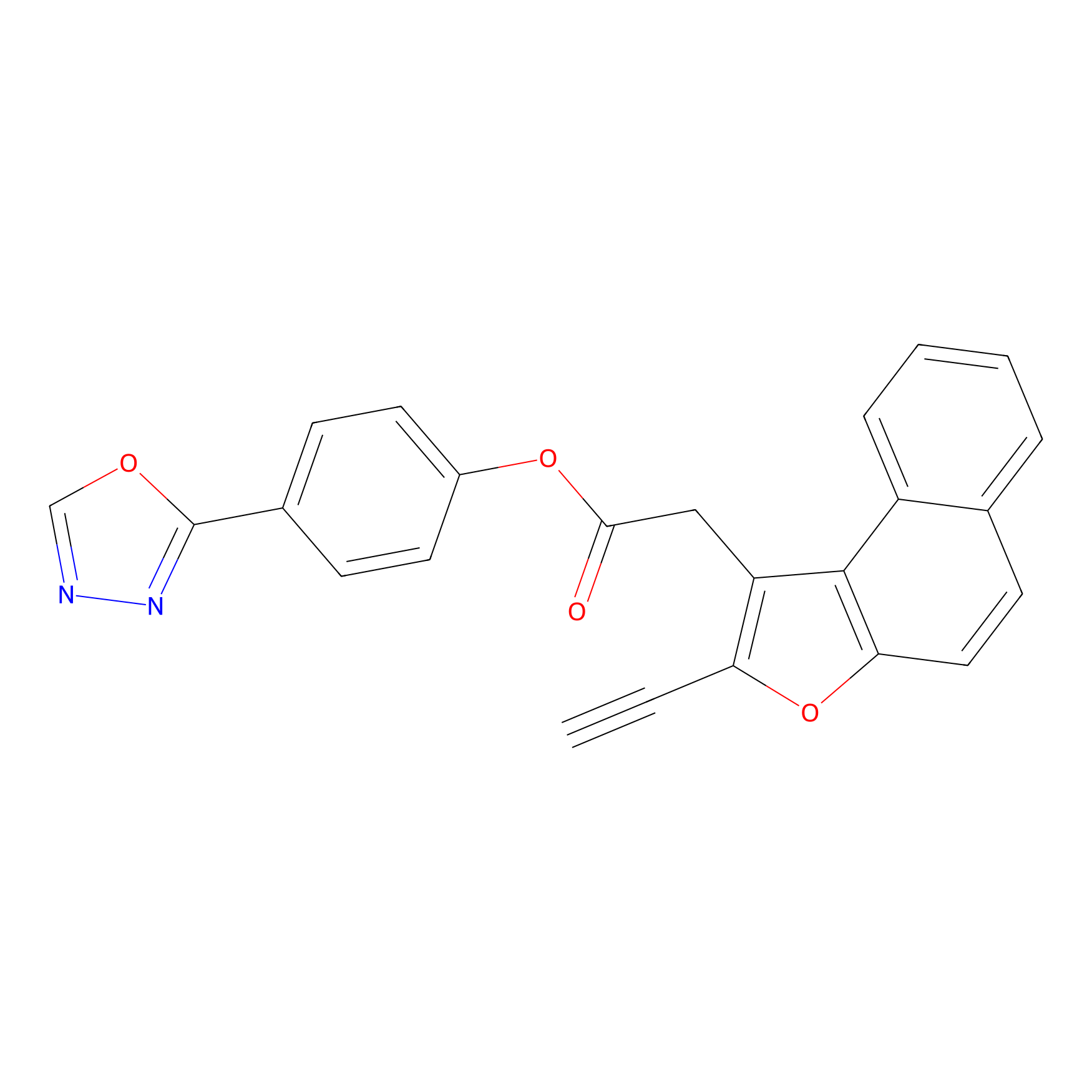 |
63.60 | LDD0326 | [1] | |
|
Curcusone 37 Probe Info |
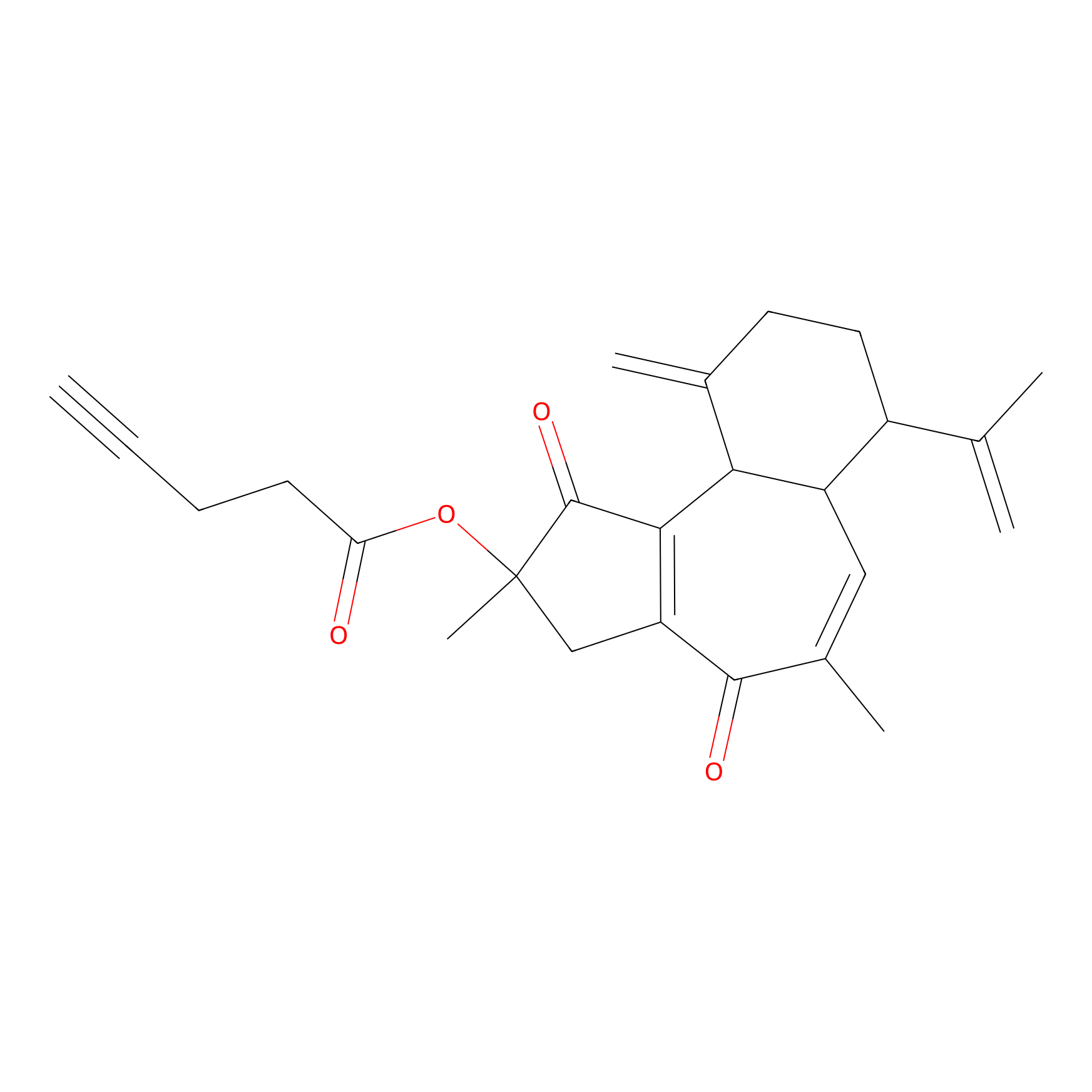 |
2.66 | LDD0188 | [2] | |
|
Johansson_61 Probe Info |
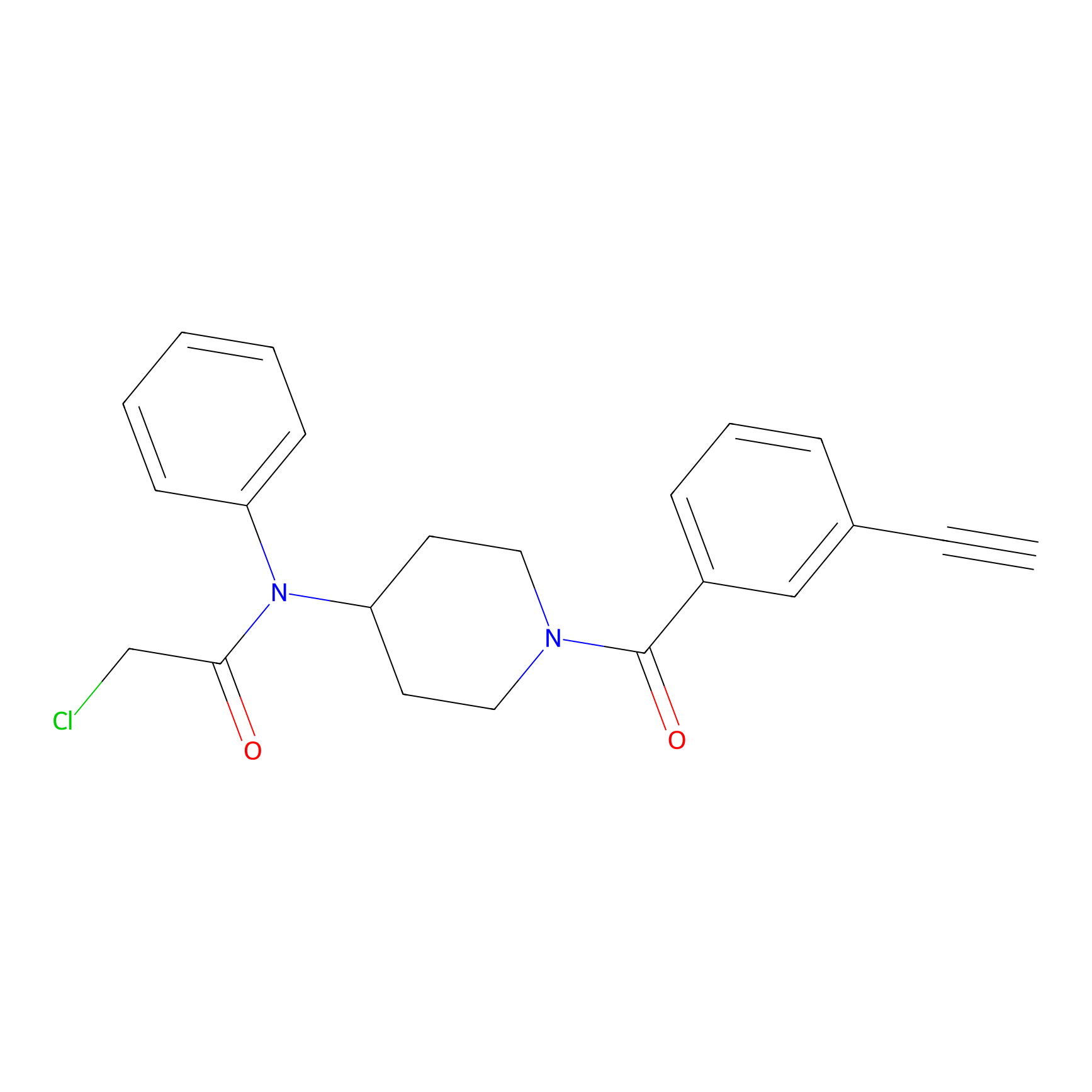 |
_(5.15) | LDD1487 | [3] | |
|
AHL-Pu-1 Probe Info |
 |
C320(2.51) | LDD0171 | [4] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0162 | [5] | |
|
IPM Probe Info |
 |
N.A. | LDD0025 | [6] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [7] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2224 | [7] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [6] | |
|
DA-P3 Probe Info |
 |
N.A. | LDD0178 | [8] | |
|
Compound 10 Probe Info |
 |
N.A. | LDD2216 | [9] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [9] | |
|
PF-06672131 Probe Info |
 |
N.A. | LDD0152 | [10] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [11] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C091 Probe Info |
 |
12.04 | LDD1782 | [12] | |
|
C092 Probe Info |
 |
23.59 | LDD1783 | [12] | |
|
C094 Probe Info |
 |
24.93 | LDD1785 | [12] | |
|
C112 Probe Info |
 |
36.50 | LDD1799 | [12] | |
|
C231 Probe Info |
 |
15.35 | LDD1904 | [12] | |
|
C235 Probe Info |
 |
19.43 | LDD1908 | [12] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | HEK-293T | C320(2.61) | LDD0172 | [4] |
| LDCM0026 | 4SU-RNA+native RNA | DM93 | C320(2.51) | LDD0171 | [4] |
| LDCM0033 | Curcusone1d | MCF-7 | 2.66 | LDD0188 | [2] |
| LDCM0615 | Fragment63-R | Jurkat | _(5.15) | LDD1487 | [3] |
| LDCM0617 | Fragment63-S | Jurkat | _(20.00) | LDD1490 | [3] |
| LDCM0022 | KB02 | T cell | C320(5.33) | LDD1703 | [13] |
| LDCM0024 | KB05 | T cell | C320(18.73) | LDD1709 | [13] |
| LDCM0170 | Structure8 | Ramos | 7.10 | LDD0433 | [14] |
The Interaction Atlas With This Target
References
