Details of the Target
General Information of Target
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
P2 Probe Info |
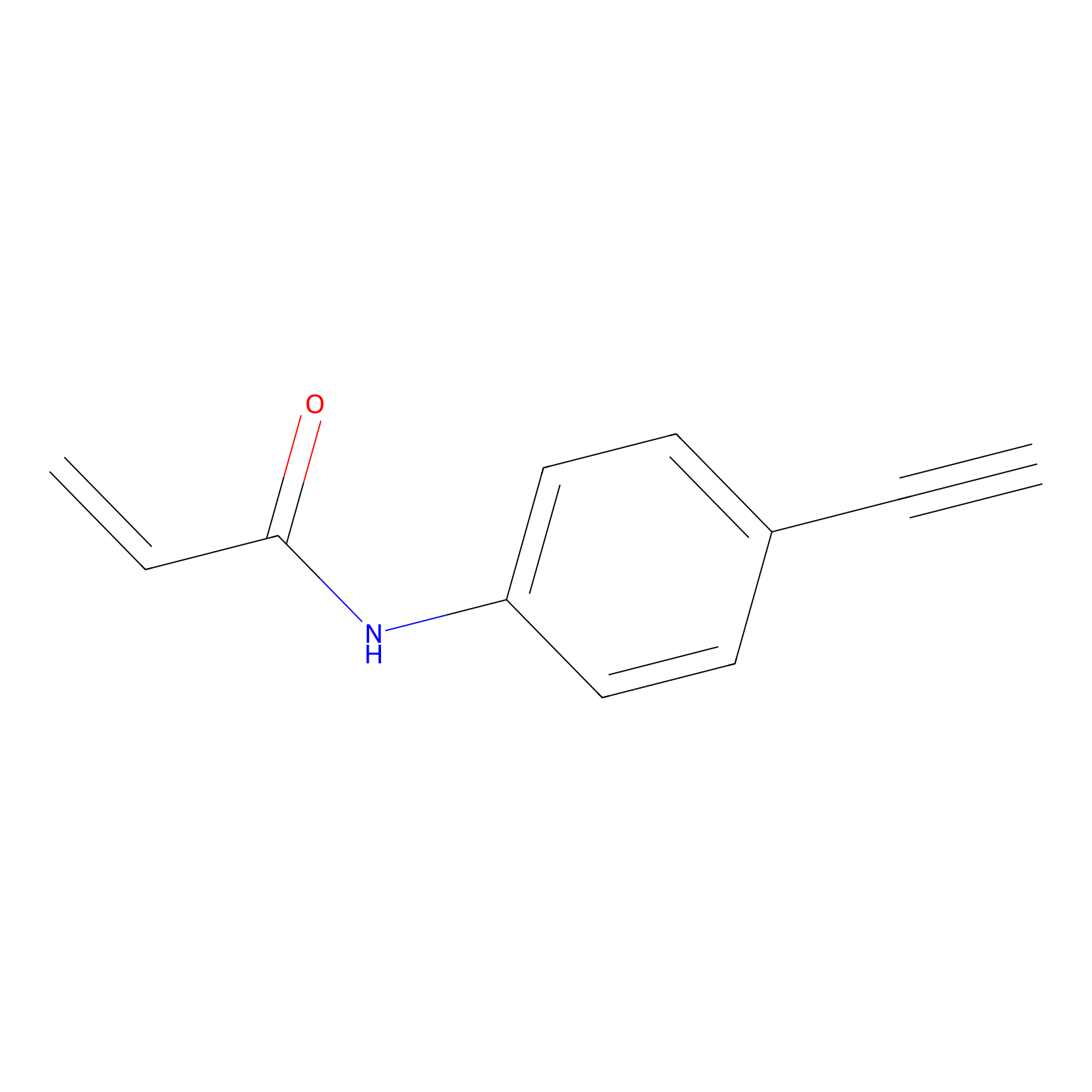 |
1.51 | LDD0449 | [1] | |
|
P8 Probe Info |
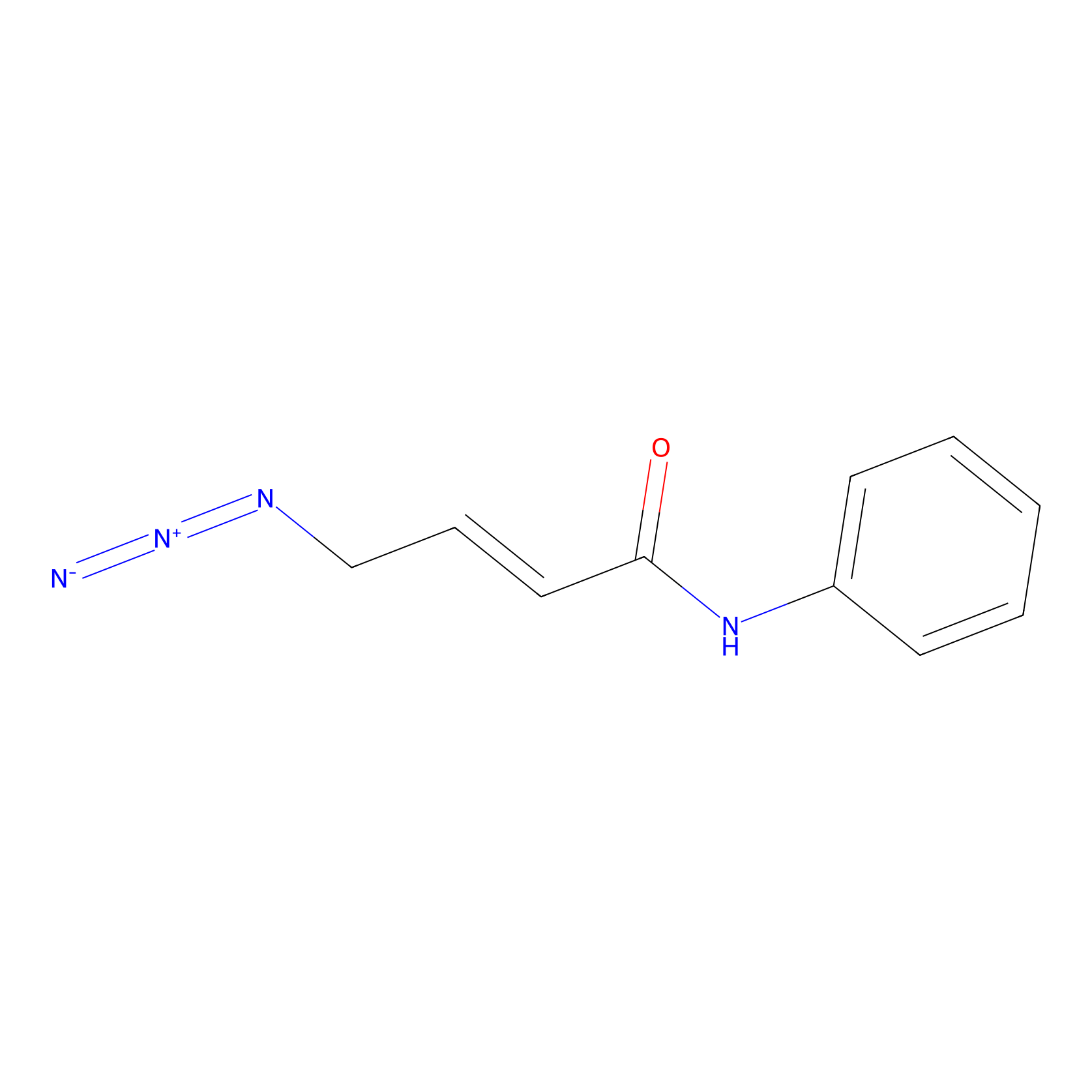 |
1.64 | LDD0451 | [1] | |
|
DAyne Probe Info |
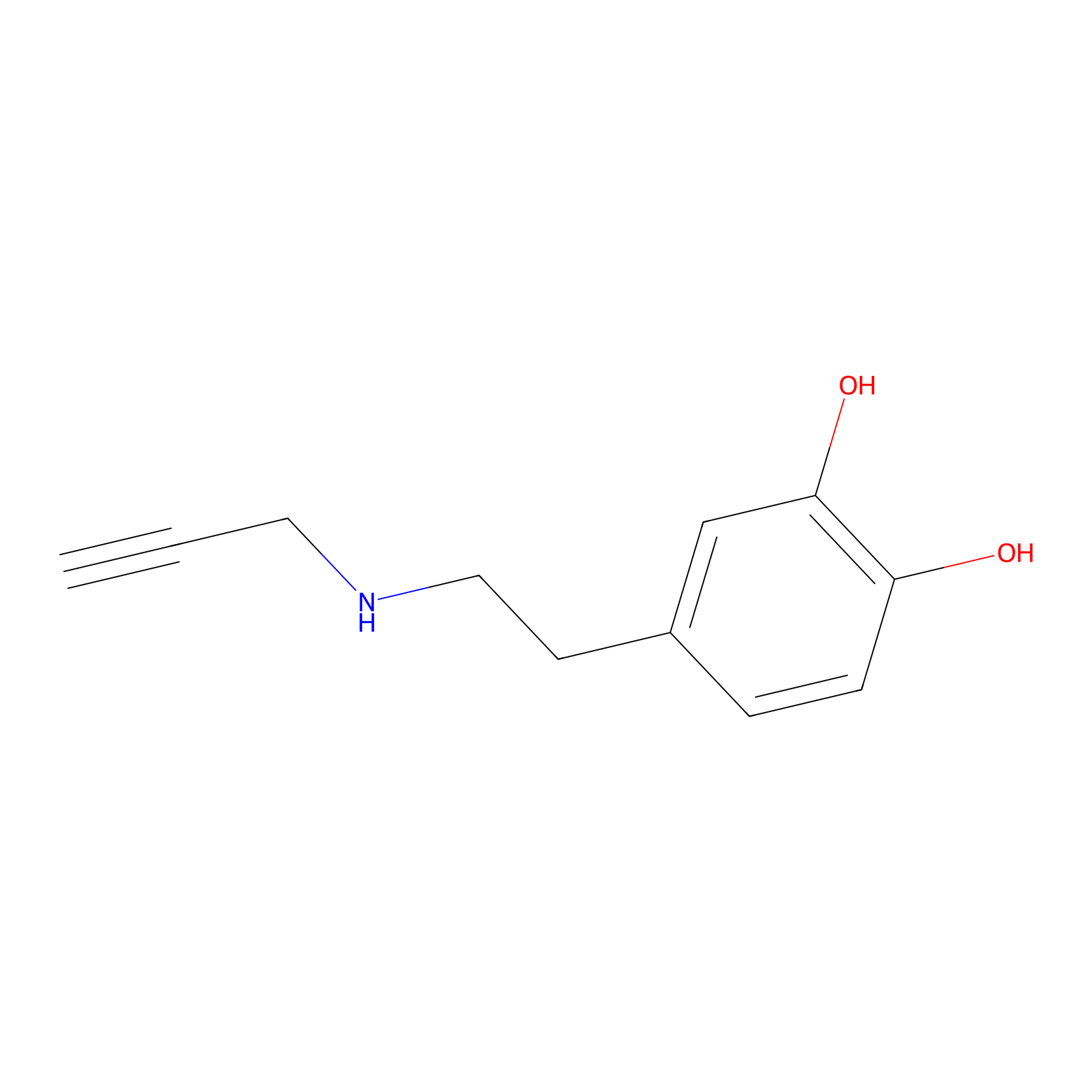 |
3.15 | LDD0261 | [2] | |
|
FBPP2 Probe Info |
 |
2.17 | LDD0318 | [3] | |
|
TH211 Probe Info |
 |
Y23(20.00); Y92(17.99) | LDD0260 | [4] | |
|
C-Sul Probe Info |
 |
9.08 | LDD0066 | [5] | |
|
STPyne Probe Info |
 |
K162(10.00); K36(8.71); K52(2.00) | LDD0277 | [6] | |
|
ONAyne Probe Info |
 |
N.A. | LDD0273 | [6] | |
|
Probe 1 Probe Info |
 |
Y23(13.80); Y44(35.41); Y110(19.96) | LDD3495 | [7] | |
|
HHS-482 Probe Info |
 |
Y44(0.96) | LDD0285 | [8] | |
|
HHS-475 Probe Info |
 |
Y23(0.38); Y110(0.88); Y44(1.05); Y92(1.16) | LDD0264 | [9] | |
|
HHS-465 Probe Info |
 |
Y110(9.84); Y115(8.37); Y23(7.71); Y44(9.32) | LDD2237 | [10] | |
|
5E-2FA Probe Info |
 |
H99(0.00); H100(0.00) | LDD2235 | [11] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [12] | |
|
m-APA Probe Info |
 |
H99(0.00); H100(0.00) | LDD2231 | [11] | |
|
SF Probe Info |
 |
Y3(0.00); Y110(0.00); Y115(0.00); Y92(0.00) | LDD0028 | [13] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [14] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [15] | |
|
Crotonaldehyde Probe Info |
 |
H99(0.00); H100(0.00) | LDD0219 | [15] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C087 Probe Info |
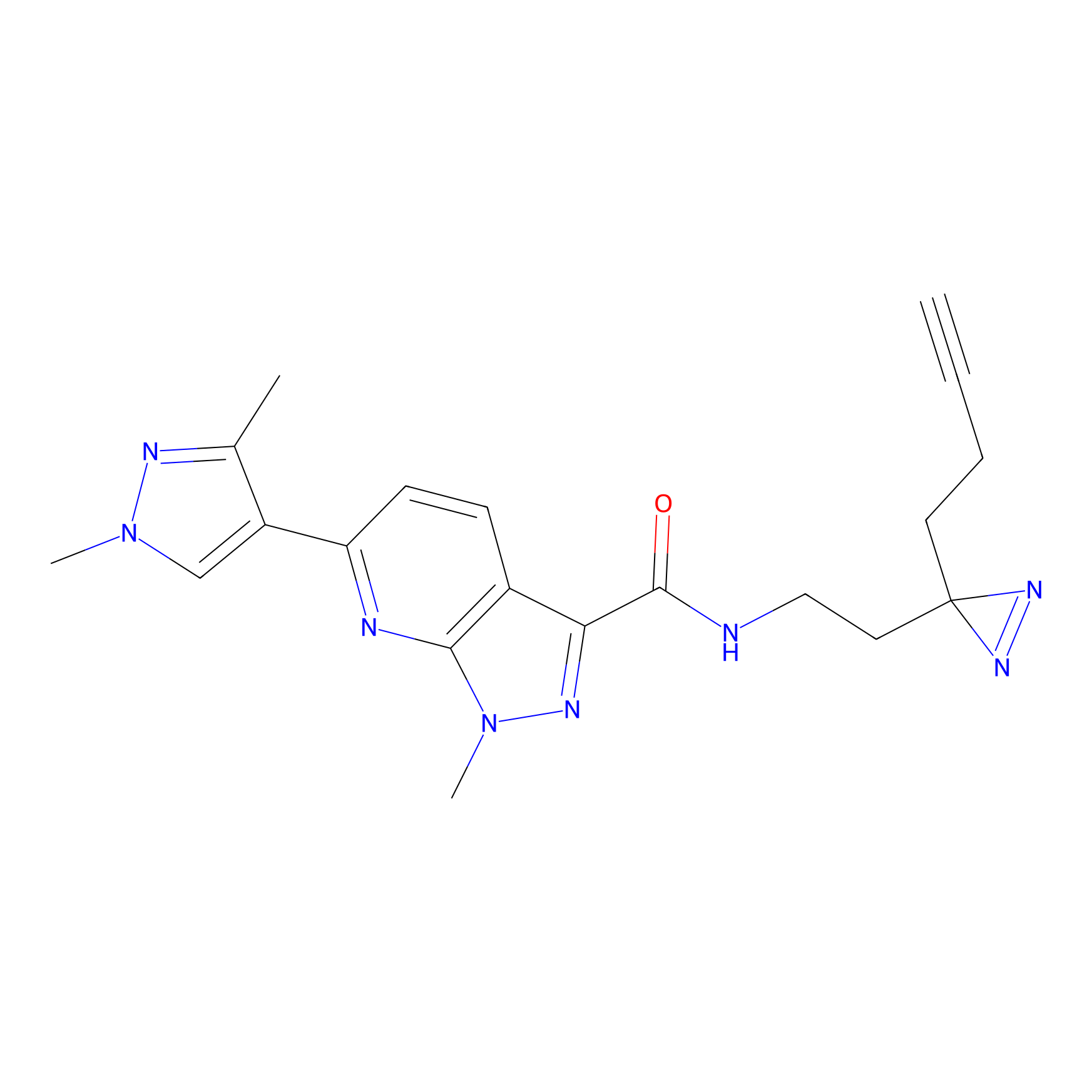 |
7.84 | LDD1779 | [16] | |
|
C145 Probe Info |
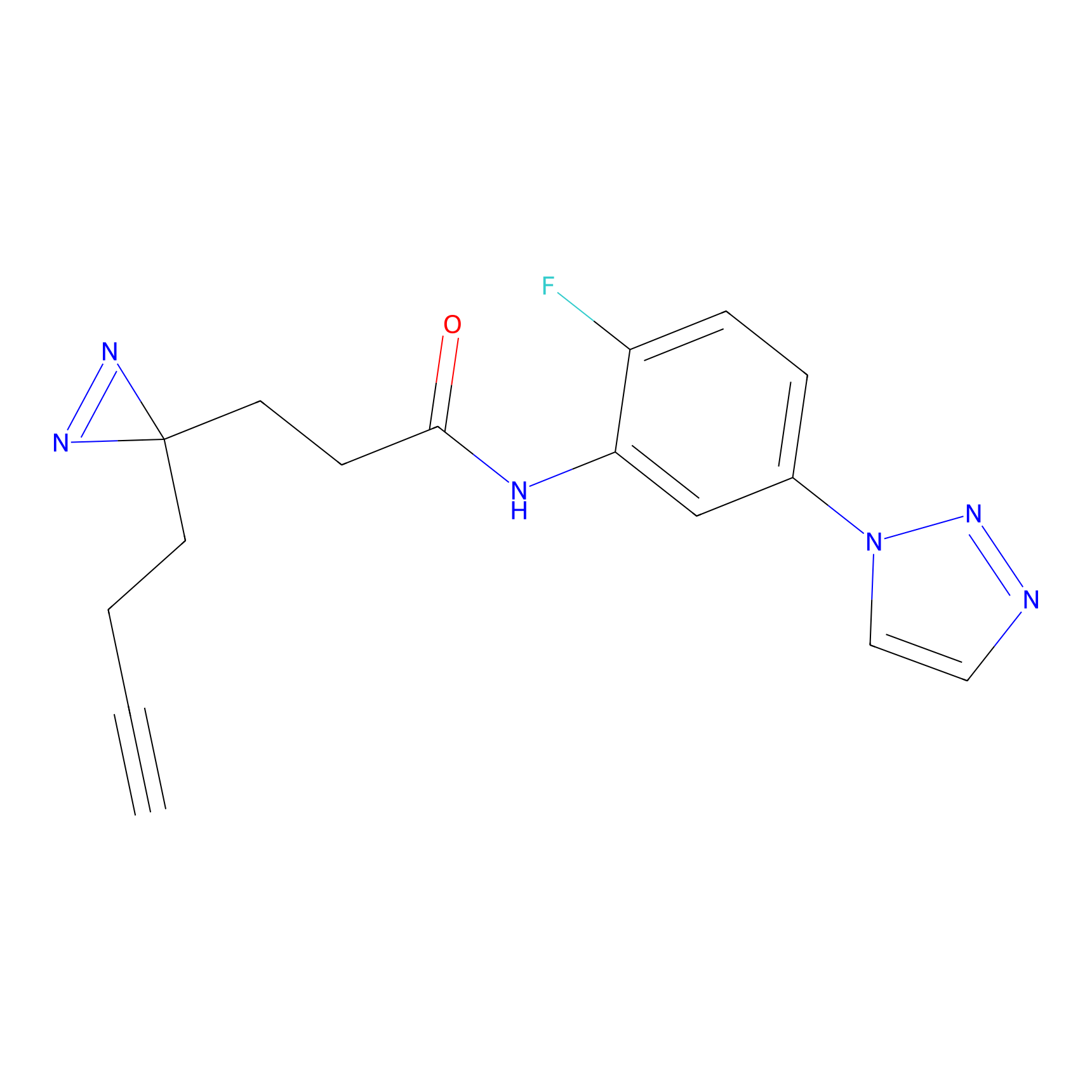 |
9.85 | LDD1827 | [16] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0138 | [17] | |
|
Staurosporine capture compound Probe Info |
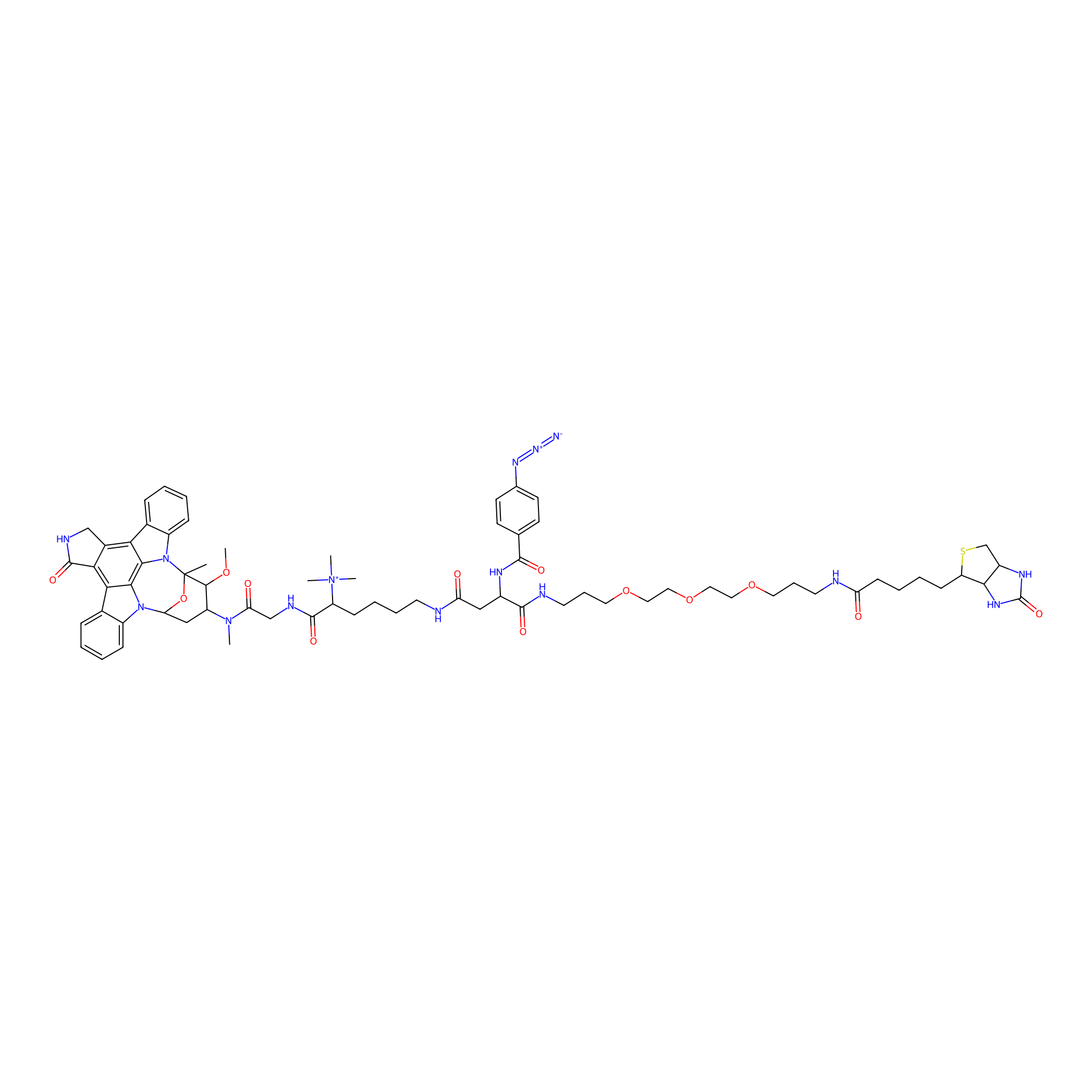 |
3.00 | LDD0083 | [18] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [15] |
| LDCM0116 | HHS-0101 | DM93 | Y23(0.38); Y110(0.88); Y44(1.05); Y92(1.16) | LDD0264 | [9] |
| LDCM0117 | HHS-0201 | DM93 | Y23(0.51); Y92(0.83); Y44(0.91); Y110(0.94) | LDD0265 | [9] |
| LDCM0118 | HHS-0301 | DM93 | Y23(0.50); Y92(0.65); Y110(0.86); Y44(0.95) | LDD0266 | [9] |
| LDCM0119 | HHS-0401 | DM93 | Y23(0.41); Y92(0.80); Y110(0.89); Y44(1.08) | LDD0267 | [9] |
| LDCM0120 | HHS-0701 | DM93 | Y23(0.33); Y110(0.87); Y92(0.88); Y44(0.91) | LDD0268 | [9] |
| LDCM0107 | IAA | HeLa | H99(0.00); H100(0.00) | LDD0221 | [15] |
| LDCM0123 | JWB131 | DM93 | Y44(0.96) | LDD0285 | [8] |
| LDCM0124 | JWB142 | DM93 | Y44(0.65) | LDD0286 | [8] |
| LDCM0125 | JWB146 | DM93 | Y44(1.21) | LDD0287 | [8] |
| LDCM0126 | JWB150 | DM93 | Y44(2.63) | LDD0288 | [8] |
| LDCM0127 | JWB152 | DM93 | Y44(1.60) | LDD0289 | [8] |
| LDCM0128 | JWB198 | DM93 | Y44(0.94) | LDD0290 | [8] |
| LDCM0129 | JWB202 | DM93 | Y44(0.36) | LDD0291 | [8] |
| LDCM0130 | JWB211 | DM93 | Y44(0.91) | LDD0292 | [8] |
| LDCM0109 | NEM | HeLa | H100(0.00); H99(0.00) | LDD0223 | [15] |
| LDCM0019 | Staurosporine | Hep-G2 | 3.00 | LDD0083 | [18] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Histone acetyltransferase KAT5 (KAT5) | MYST (SAS/MOZ) family | Q92993 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Splicing factor U2AF 35 kDa subunit (U2AF1) | Splicing factor SR family | Q01081 | |||
| Corepressor interacting with RBPJ 1 (CIR1) | . | Q86X95 | |||
References
