Details of the Target
General Information of Target
| Target ID | LDTP05274 | |||||
|---|---|---|---|---|---|---|
| Target Name | Nucleolysin TIAR (TIAL1) | |||||
| Gene Name | TIAL1 | |||||
| Gene ID | 7073 | |||||
| Synonyms |
Nucleolysin TIAR; TIA-1-related protein |
|||||
| 3D Structure | ||||||
| Sequence |
MMEDDGQPRTLYVGNLSRDVTEVLILQLFSQIGPCKSCKMITEHTSNDPYCFVEFYEHRD
AAAALAAMNGRKILGKEVKVNWATTPSSQKKDTSNHFHVFVGDLSPEITTEDIKSAFAPF GKISDARVVKDMATGKSKGYGFVSFYNKLDAENAIVHMGGQWLGGRQIRTNWATRKPPAP KSTQENNTKQLRFEDVVNQSSPKNCTVYCGGIASGLTDQLMRQTFSPFGQIMEIRVFPEK GYSFVRFSTHESAAHAIVSVNGTTIEGHVVKCYWGKESPDMTKNFQQVDYSQWGQWSQVY GNPQQYGQYMANGWQVPPYGVYGQPWNQQGFGVDQSPSAAWMGGFGAQPPQGQAPPPVIP PPNQAGYGMASYQTQ |
|||||
| Target Bioclass |
Other
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
RNA-binding protein involved in alternative pre-RNA splicing and in cytoplasmic stress granules formation. Shows a preference for uridine-rich RNAs. Activates splicing of alternative exons with weak 5' splice sites followed by a U-rich stretch on its own pre-mRNA and on TIA1 mRNA. Promotes the inclusion of TIA1 exon 5 to give rise to the long isoform (isoform a) of TIA1. Acts downstream of the stress-induced phosphorylation of EIF2S1/EIF2A to promote the recruitment of untranslated mRNAs to cytoplasmic stress granules (SG). Possesses nucleolytic activity against cytotoxic lymphocyte target cells. May be involved in apoptosis.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
ILS-1 Probe Info |
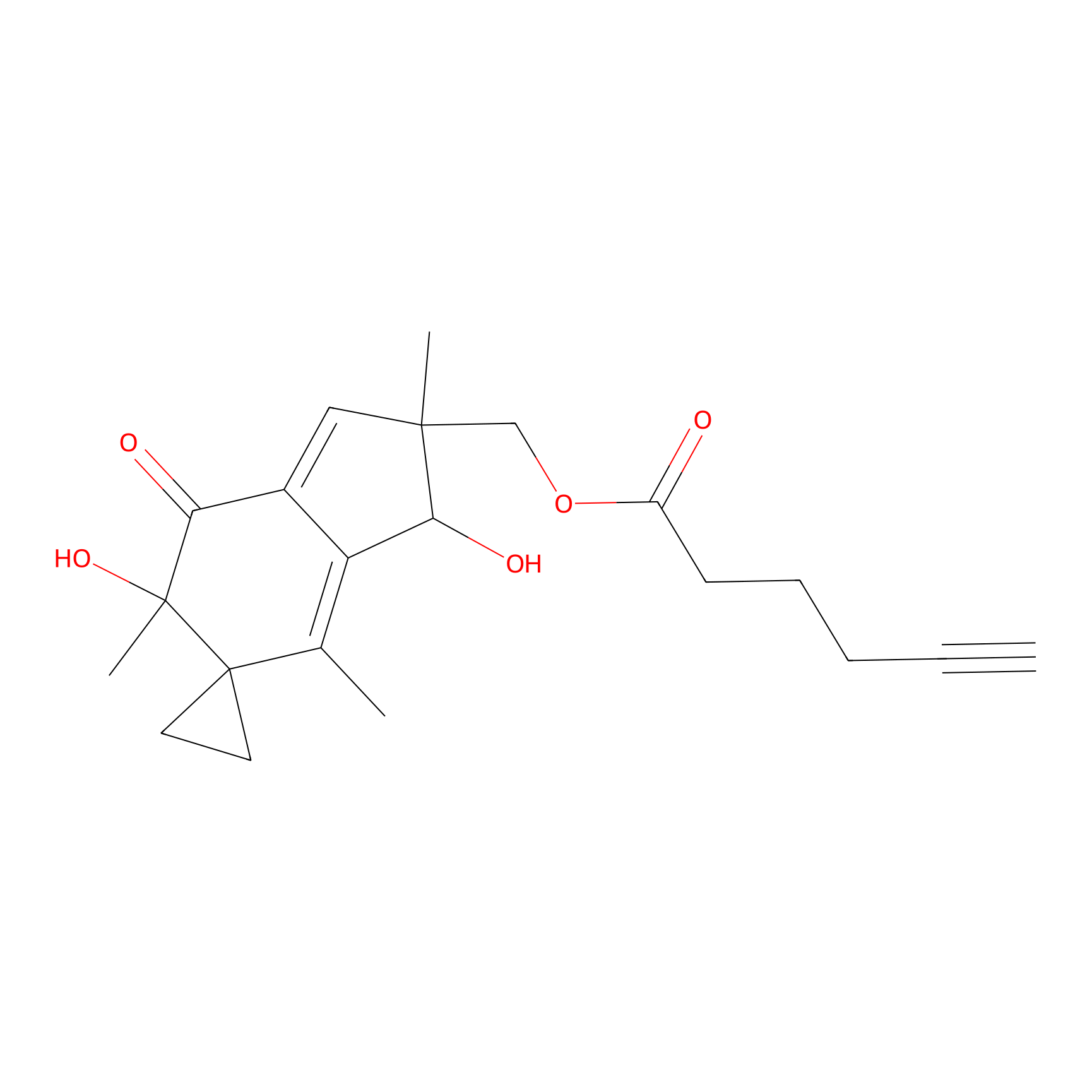 |
2.18 | LDD0415 | [1] | |
|
TH211 Probe Info |
 |
Y242(20.00) | LDD0257 | [2] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [3] | |
|
DBIA Probe Info |
 |
C35(1.25) | LDD3377 | [4] | |
|
HHS-482 Probe Info |
 |
Y140(1.22) | LDD0285 | [5] | |
|
HHS-475 Probe Info |
 |
Y140(0.91) | LDD0264 | [6] | |
|
HHS-465 Probe Info |
 |
Y140(10.00); Y242(2.98) | LDD2237 | [7] | |
|
Acrolein Probe Info |
 |
H157(0.00); C209(0.00) | LDD0221 | [8] | |
|
5E-2FA Probe Info |
 |
H255(0.00); H157(0.00) | LDD2235 | [9] | |
|
ATP probe Probe Info |
 |
K79(0.00); K138(0.00) | LDD0199 | [10] | |
|
m-APA Probe Info |
 |
H255(0.00); H157(0.00) | LDD2231 | [9] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C51(0.00); C35(0.00) | LDD0038 | [11] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0032 | [12] | |
|
Lodoacetamide azide Probe Info |
 |
C35(0.00); C51(0.00) | LDD0037 | [11] | |
|
NHS Probe Info |
 |
K79(0.00); K122(0.00) | LDD0010 | [13] | |
|
SF Probe Info |
 |
Y12(0.00); Y146(0.00); K176(0.00); Y140(0.00) | LDD0028 | [14] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [13] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [15] | |
|
1d-yne Probe Info |
 |
K114(0.00); K148(0.00) | LDD0357 | [15] | |
|
NAIA_5 Probe Info |
 |
C35(0.00); C51(0.00) | LDD2223 | [16] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C165 Probe Info |
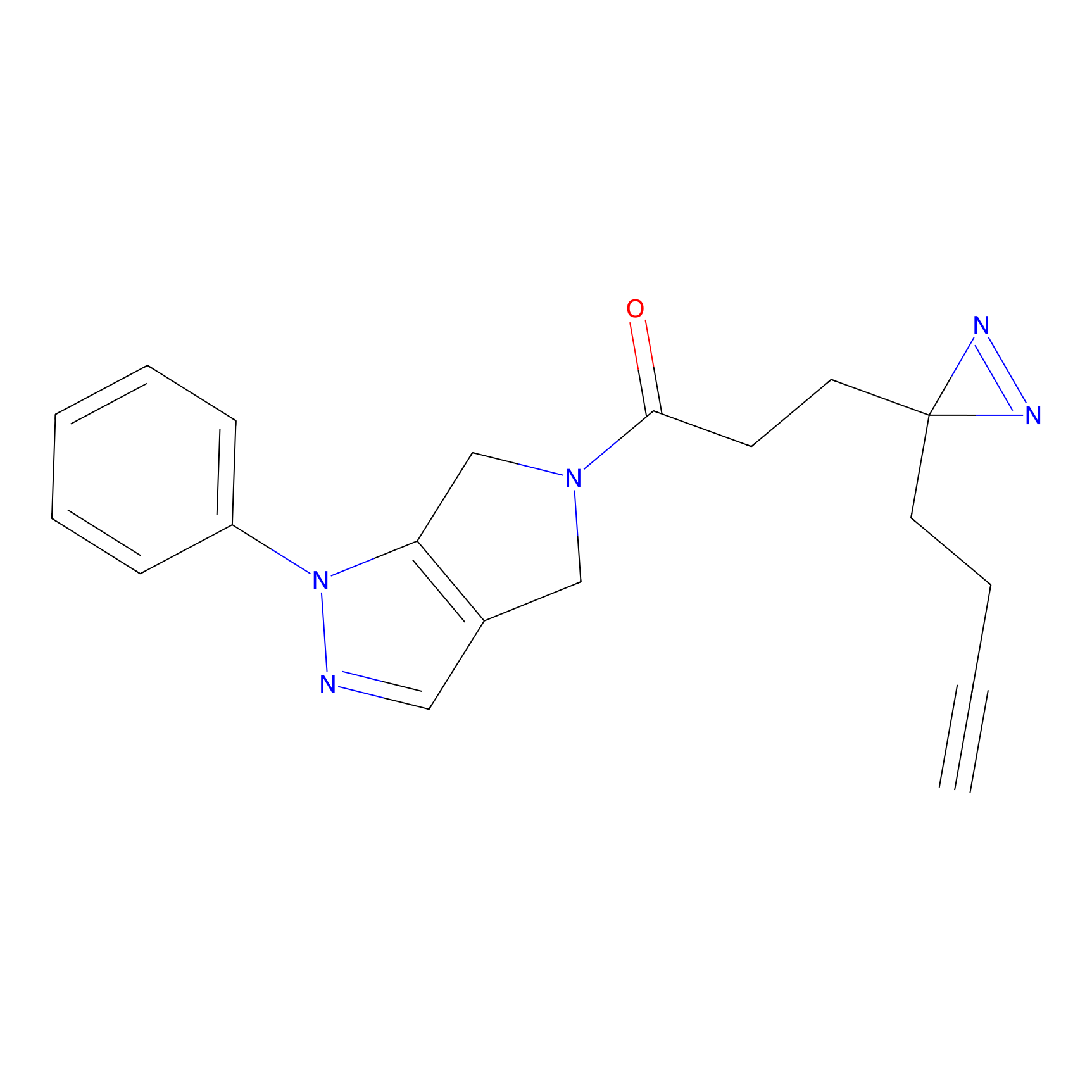 |
13.64 | LDD1845 | [17] | |
|
C191 Probe Info |
 |
10.85 | LDD1868 | [17] | |
|
C232 Probe Info |
 |
35.02 | LDD1905 | [17] | |
|
FFF probe11 Probe Info |
 |
9.15 | LDD0471 | [18] | |
|
FFF probe13 Probe Info |
 |
20.00 | LDD0475 | [18] | |
|
FFF probe2 Probe Info |
 |
14.60 | LDD0463 | [18] | |
|
FFF probe4 Probe Info |
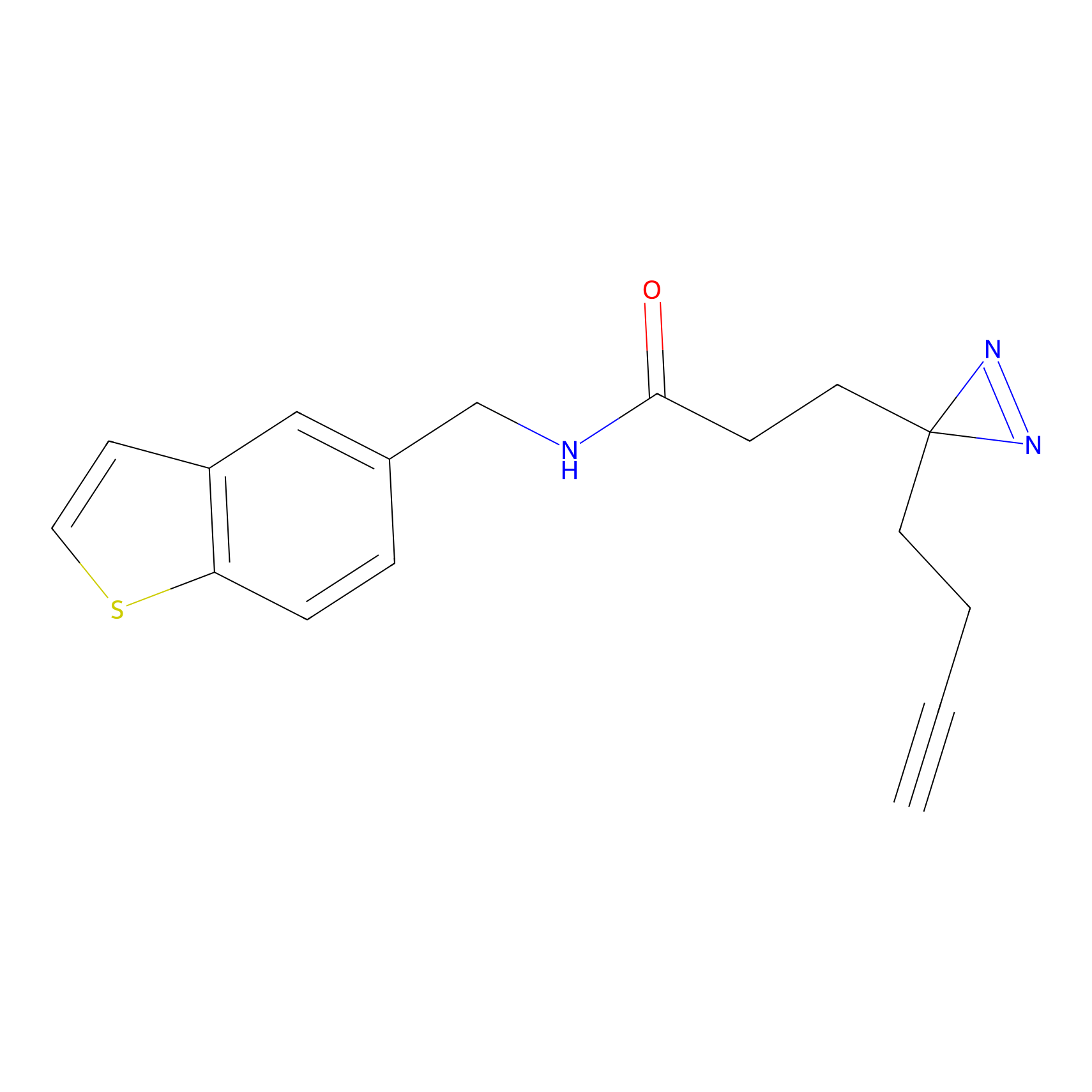 |
20.00 | LDD0466 | [18] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0259 | AC14 | HEK-293T | C51(1.11) | LDD1512 | [19] |
| LDCM0270 | AC15 | HEK-293T | C51(1.00) | LDD1513 | [19] |
| LDCM0282 | AC22 | HEK-293T | C51(0.95) | LDD1521 | [19] |
| LDCM0283 | AC23 | HEK-293T | C51(1.16) | LDD1522 | [19] |
| LDCM0284 | AC24 | HEK-293T | C209(0.92) | LDD1523 | [19] |
| LDCM0291 | AC30 | HEK-293T | C51(0.88) | LDD1530 | [19] |
| LDCM0292 | AC31 | HEK-293T | C51(1.32) | LDD1531 | [19] |
| LDCM0293 | AC32 | HEK-293T | C209(1.17) | LDD1532 | [19] |
| LDCM0299 | AC38 | HEK-293T | C51(1.00) | LDD1538 | [19] |
| LDCM0300 | AC39 | HEK-293T | C51(1.07) | LDD1539 | [19] |
| LDCM0302 | AC40 | HEK-293T | C209(0.99) | LDD1541 | [19] |
| LDCM0308 | AC46 | HEK-293T | C51(0.69) | LDD1547 | [19] |
| LDCM0309 | AC47 | HEK-293T | C51(1.07) | LDD1548 | [19] |
| LDCM0310 | AC48 | HEK-293T | C209(0.94) | LDD1549 | [19] |
| LDCM0317 | AC54 | HEK-293T | C51(1.00) | LDD1556 | [19] |
| LDCM0318 | AC55 | HEK-293T | C51(0.95) | LDD1557 | [19] |
| LDCM0319 | AC56 | HEK-293T | C209(1.00) | LDD1558 | [19] |
| LDCM0323 | AC6 | HEK-293T | C51(1.00) | LDD1562 | [19] |
| LDCM0326 | AC62 | HEK-293T | C51(0.78) | LDD1565 | [19] |
| LDCM0327 | AC63 | HEK-293T | C51(1.15) | LDD1566 | [19] |
| LDCM0328 | AC64 | HEK-293T | C209(1.13) | LDD1567 | [19] |
| LDCM0334 | AC7 | HEK-293T | C51(0.91) | LDD1568 | [19] |
| LDCM0345 | AC8 | HEK-293T | C209(0.89) | LDD1569 | [19] |
| LDCM0275 | AKOS034007705 | HEK-293T | C209(0.98) | LDD1514 | [19] |
| LDCM0108 | Chloroacetamide | HeLa | C51(0.00); H157(0.00); C209(0.00) | LDD0222 | [8] |
| LDCM0367 | CL1 | HEK-293T | C51(1.03) | LDD1571 | [19] |
| LDCM0368 | CL10 | HEK-293T | C51(1.27) | LDD1572 | [19] |
| LDCM0370 | CL101 | HEK-293T | C51(1.03) | LDD1574 | [19] |
| LDCM0374 | CL105 | HEK-293T | C51(0.90) | LDD1578 | [19] |
| LDCM0378 | CL109 | HEK-293T | C51(1.01) | LDD1582 | [19] |
| LDCM0379 | CL11 | HEK-293T | C51(0.96) | LDD1583 | [19] |
| LDCM0383 | CL113 | HEK-293T | C51(0.98) | LDD1587 | [19] |
| LDCM0387 | CL117 | HEK-293T | C51(1.20) | LDD1591 | [19] |
| LDCM0390 | CL12 | HEK-293T | C209(0.84) | LDD1594 | [19] |
| LDCM0392 | CL121 | HEK-293T | C51(1.20) | LDD1596 | [19] |
| LDCM0396 | CL125 | HEK-293T | C51(0.98) | LDD1600 | [19] |
| LDCM0400 | CL13 | HEK-293T | C51(0.95) | LDD1604 | [19] |
| LDCM0410 | CL22 | HEK-293T | C51(0.92) | LDD1614 | [19] |
| LDCM0411 | CL23 | HEK-293T | C51(1.09) | LDD1615 | [19] |
| LDCM0412 | CL24 | HEK-293T | C209(0.96) | LDD1616 | [19] |
| LDCM0413 | CL25 | HEK-293T | C51(1.10) | LDD1617 | [19] |
| LDCM0423 | CL34 | HEK-293T | C51(0.93) | LDD1627 | [19] |
| LDCM0424 | CL35 | HEK-293T | C51(1.01) | LDD1628 | [19] |
| LDCM0425 | CL36 | HEK-293T | C209(0.75) | LDD1629 | [19] |
| LDCM0426 | CL37 | HEK-293T | C51(0.85) | LDD1630 | [19] |
| LDCM0436 | CL46 | HEK-293T | C51(0.93) | LDD1640 | [19] |
| LDCM0437 | CL47 | HEK-293T | C51(0.95) | LDD1641 | [19] |
| LDCM0438 | CL48 | HEK-293T | C209(0.91) | LDD1642 | [19] |
| LDCM0439 | CL49 | HEK-293T | C51(1.17) | LDD1643 | [19] |
| LDCM0449 | CL58 | HEK-293T | C51(1.18) | LDD1652 | [19] |
| LDCM0450 | CL59 | HEK-293T | C51(0.97) | LDD1653 | [19] |
| LDCM0452 | CL60 | HEK-293T | C209(0.86) | LDD1655 | [19] |
| LDCM0453 | CL61 | HEK-293T | C51(1.21) | LDD1656 | [19] |
| LDCM0463 | CL70 | HEK-293T | C51(0.88) | LDD1666 | [19] |
| LDCM0464 | CL71 | HEK-293T | C51(1.20) | LDD1667 | [19] |
| LDCM0465 | CL72 | HEK-293T | C209(0.90) | LDD1668 | [19] |
| LDCM0466 | CL73 | HEK-293T | C51(0.95) | LDD1669 | [19] |
| LDCM0476 | CL82 | HEK-293T | C51(0.83) | LDD1679 | [19] |
| LDCM0477 | CL83 | HEK-293T | C51(1.08) | LDD1680 | [19] |
| LDCM0478 | CL84 | HEK-293T | C209(0.78) | LDD1681 | [19] |
| LDCM0479 | CL85 | HEK-293T | C51(0.93) | LDD1682 | [19] |
| LDCM0489 | CL94 | HEK-293T | C51(0.89) | LDD1692 | [19] |
| LDCM0490 | CL95 | HEK-293T | C51(1.15) | LDD1693 | [19] |
| LDCM0491 | CL96 | HEK-293T | C209(0.91) | LDD1694 | [19] |
| LDCM0492 | CL97 | HEK-293T | C51(0.90) | LDD1695 | [19] |
| LDCM0116 | HHS-0101 | DM93 | Y140(0.91) | LDD0264 | [6] |
| LDCM0117 | HHS-0201 | DM93 | Y140(1.60) | LDD0265 | [6] |
| LDCM0118 | HHS-0301 | DM93 | Y140(1.99) | LDD0266 | [6] |
| LDCM0119 | HHS-0401 | DM93 | Y140(2.08) | LDD0267 | [6] |
| LDCM0120 | HHS-0701 | DM93 | Y140(2.32) | LDD0268 | [6] |
| LDCM0107 | IAA | HeLa | H157(0.00); C209(0.00) | LDD0221 | [8] |
| LDCM0123 | JWB131 | DM93 | Y140(1.22) | LDD0285 | [5] |
| LDCM0124 | JWB142 | DM93 | Y140(1.38) | LDD0286 | [5] |
| LDCM0125 | JWB146 | DM93 | Y140(1.20) | LDD0287 | [5] |
| LDCM0126 | JWB150 | DM93 | Y140(2.55) | LDD0288 | [5] |
| LDCM0127 | JWB152 | DM93 | Y140(1.92) | LDD0289 | [5] |
| LDCM0128 | JWB198 | DM93 | Y140(0.19) | LDD0290 | [5] |
| LDCM0129 | JWB202 | DM93 | Y140(1.14) | LDD0291 | [5] |
| LDCM0130 | JWB211 | DM93 | Y140(1.11) | LDD0292 | [5] |
| LDCM0022 | KB02 | OPM-2 | C35(1.28) | LDD2543 | [4] |
| LDCM0023 | KB03 | OPM-2 | C35(1.25) | LDD2960 | [4] |
| LDCM0024 | KB05 | OPM-2 | C35(1.25) | LDD3377 | [4] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [8] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Protein lifeguard 1 (GRINA) | BI1 family | Q7Z429 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Krueppel-like factor 1 (KLF1) | Krueppel C2H2-type zinc-finger protein family | Q13351 | |||
Other
References
