Details of the Target
General Information of Target
| Target ID | LDTP14069 | |||||
|---|---|---|---|---|---|---|
| Target Name | WD repeat domain phosphoinositide-interacting protein 2 (WIPI2) | |||||
| Gene Name | WIPI2 | |||||
| Gene ID | 26100 | |||||
| Synonyms |
WD repeat domain phosphoinositide-interacting protein 2; WIPI-2; WIPI49-like protein 2 |
|||||
| 3D Structure | ||||||
| Sequence |
MDAATLTYDTLRFAEFEDFPETSEPVWILGRKYSIFTEKDEILSDVASRLWFTYRKNFPA
IGGTGPTSDTGWGCMLRCGQMIFAQALVCRHLGRDWRWTQRKRQPDSYFSVLNAFIDRKD SYYSIHQIAQMGVGEGKSIGQWYGPNTVAQVLKKLAVFDTWSSLAVHIAMDNTVVMEEIR RLCRTSVPCAGATAFPADSDRHCNGFPAGAEVTNRPSPWRPLVLLIPLRLGLTDINEAYV ETLKHCFMMPQSLGVIGGKPNSAHYFIGYVGEELIYLDPHTTQPAVEPTDGCFIPDESFH CQHPPCRMSIAELDPSIAVGFFCKTEDDFNDWCQQVKKLSLLGGALPMFELVELQPSHLA CPDVLNLSLDSSDVERLERFFDSEDEDFEILSL |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
WD repeat PROPPIN family
|
|||||
| Subcellular location |
Preautophagosomal structure membrane
|
|||||
| Function |
Component of the autophagy machinery that controls the major intracellular degradation process by which cytoplasmic materials are packaged into autophagosomes and delivered to lysosomes for degradation. Involved in an early step of the formation of preautophagosomal structures. Binds and is activated by phosphatidylinositol 3-phosphate (PtdIns3P) forming on membranes of the endoplasmic reticulum upon activation of the upstream ULK1 and PI3 kinases. Mediates ER-isolation membranes contacts by interacting with the ULK1:RB1CC1 complex and PtdIns3P. Once activated, WIPI2 recruits at phagophore assembly sites the ATG12-ATG5-ATG16L1 complex that directly controls the elongation of the nascent autophagosomal membrane.; [Isoform 4]: Recruits the ATG12-ATG5-ATG16L1 complex to omegasomes and preautophagosomal structures, resulting in ATG8 family proteins lipidation and starvation-induced autophagy. Isoform 4 is also required for autophagic clearance of pathogenic bacteria. Isoform 4 binds the membrane surrounding Salmonella and recruits the ATG12-5-16L1 complex, initiating LC3 conjugation, autophagosomal membrane formation, and engulfment of Salmonella.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| C32 | Substitution: p.P285S | . | |||
| HEC1 | SNV: p.A350T | DBIA Probe Info | |||
| HT115 | SNV: p.F56V | . | |||
| SW948 | SNV: p.A255G | DBIA Probe Info | |||
| TOV21G | SNV: p.P284L | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
11.21 | LDD0402 | [1] | |
|
STPyne Probe Info |
 |
K105(7.86); K223(6.67) | LDD0277 | [2] | |
|
AHL-Pu-1 Probe Info |
 |
C430(2.23) | LDD0168 | [3] | |
|
EA-probe Probe Info |
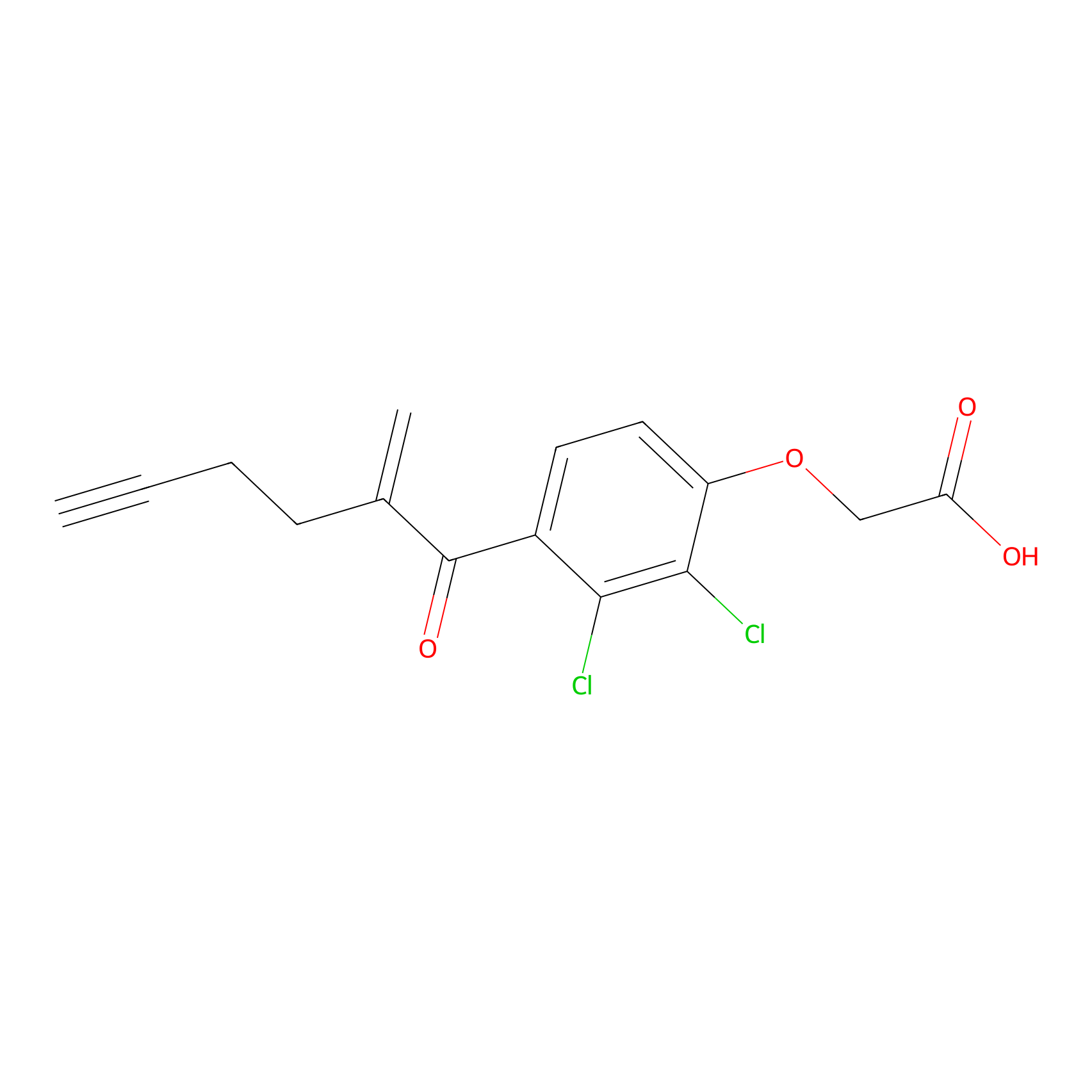 |
C393(1.25) | LDD2210 | [4] | |
|
DBIA Probe Info |
 |
C328(1.03) | LDD0078 | [5] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [6] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C328(0.00); C334(0.00) | LDD0038 | [7] | |
|
IA-alkyne Probe Info |
 |
C328(0.00); C334(0.00) | LDD0036 | [7] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [7] | |
|
NAIA_4 Probe Info |
 |
C393(0.00); C430(0.00) | LDD2226 | [8] | |
|
Compound 10 Probe Info |
 |
N.A. | LDD2216 | [9] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [9] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [10] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [11] | |
|
AOyne Probe Info |
 |
13.10 | LDD0443 | [12] | |
|
NAIA_5 Probe Info |
 |
C328(0.00); C334(0.00) | LDD2223 | [8] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | HEK-293T | C430(2.23) | LDD0168 | [3] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C328(2.18); C430(2.33) | LDD0169 | [3] |
| LDCM0214 | AC1 | HCT 116 | C70(1.20); C77(1.20); C111(1.11); C328(0.70) | LDD0531 | [5] |
| LDCM0215 | AC10 | HCT 116 | C70(1.10); C77(1.10); C111(1.15); C328(1.14) | LDD0532 | [5] |
| LDCM0216 | AC100 | HCT 116 | C111(1.04); C328(1.03); C334(0.73) | LDD0533 | [5] |
| LDCM0217 | AC101 | HCT 116 | C111(0.91); C328(1.06); C334(0.64) | LDD0534 | [5] |
| LDCM0218 | AC102 | HCT 116 | C111(0.97); C328(1.16); C334(0.71) | LDD0535 | [5] |
| LDCM0219 | AC103 | HCT 116 | C111(0.88); C328(1.19); C334(0.64) | LDD0536 | [5] |
| LDCM0220 | AC104 | HCT 116 | C111(0.95); C328(0.92); C334(0.63) | LDD0537 | [5] |
| LDCM0221 | AC105 | HCT 116 | C111(0.89); C328(1.02); C334(0.59) | LDD0538 | [5] |
| LDCM0222 | AC106 | HCT 116 | C111(0.95); C328(0.93); C334(0.66) | LDD0539 | [5] |
| LDCM0223 | AC107 | HCT 116 | C111(0.91); C328(0.93); C334(1.18) | LDD0540 | [5] |
| LDCM0224 | AC108 | HCT 116 | C111(1.12); C328(0.82); C334(0.91) | LDD0541 | [5] |
| LDCM0225 | AC109 | HCT 116 | C111(0.97); C328(0.85); C334(0.84) | LDD0542 | [5] |
| LDCM0226 | AC11 | HCT 116 | C70(1.59); C77(1.59); C111(1.18); C328(1.09) | LDD0543 | [5] |
| LDCM0227 | AC110 | HCT 116 | C111(1.06); C328(0.96); C334(0.81) | LDD0544 | [5] |
| LDCM0228 | AC111 | HCT 116 | C111(1.18); C328(0.97); C334(0.85) | LDD0545 | [5] |
| LDCM0229 | AC112 | HCT 116 | C111(0.99); C328(0.97); C334(0.90) | LDD0546 | [5] |
| LDCM0230 | AC113 | HCT 116 | C70(0.63); C77(0.63); C111(0.93); C328(0.99) | LDD0547 | [5] |
| LDCM0231 | AC114 | HCT 116 | C70(1.02); C77(0.93); C111(0.79); C328(1.13) | LDD0548 | [5] |
| LDCM0232 | AC115 | HCT 116 | C70(1.05); C77(1.03); C111(0.93); C328(1.28) | LDD0549 | [5] |
| LDCM0233 | AC116 | HCT 116 | C70(1.14); C77(1.67); C111(0.77); C328(1.15) | LDD0550 | [5] |
| LDCM0234 | AC117 | HCT 116 | C70(1.03); C77(1.14); C111(0.75); C328(1.09) | LDD0551 | [5] |
| LDCM0235 | AC118 | HCT 116 | C70(0.87); C77(1.10); C111(0.86); C328(1.05) | LDD0552 | [5] |
| LDCM0236 | AC119 | HCT 116 | C70(0.94); C77(1.24); C111(0.93); C328(1.11) | LDD0553 | [5] |
| LDCM0237 | AC12 | HCT 116 | C70(1.12); C77(1.12); C111(1.19); C328(0.95) | LDD0554 | [5] |
| LDCM0238 | AC120 | HCT 116 | C70(0.94); C77(0.87); C111(0.91); C328(1.24) | LDD0555 | [5] |
| LDCM0239 | AC121 | HCT 116 | C70(0.92); C77(0.96); C111(0.91); C328(0.99) | LDD0556 | [5] |
| LDCM0240 | AC122 | HCT 116 | C70(0.95); C77(1.25); C111(1.01); C328(1.07) | LDD0557 | [5] |
| LDCM0241 | AC123 | HCT 116 | C70(0.76); C77(0.81); C111(1.00); C328(1.07) | LDD0558 | [5] |
| LDCM0242 | AC124 | HCT 116 | C70(0.80); C77(0.89); C111(0.99); C328(1.00) | LDD0559 | [5] |
| LDCM0243 | AC125 | HCT 116 | C70(0.99); C77(1.19); C111(0.98); C328(1.02) | LDD0560 | [5] |
| LDCM0244 | AC126 | HCT 116 | C70(0.97); C77(1.39); C111(0.91); C328(1.13) | LDD0561 | [5] |
| LDCM0245 | AC127 | HCT 116 | C70(0.88); C77(0.88); C111(0.89); C328(1.07) | LDD0562 | [5] |
| LDCM0246 | AC128 | HCT 116 | C70(1.23); C77(1.23); C111(1.02); C328(1.04) | LDD0563 | [5] |
| LDCM0247 | AC129 | HCT 116 | C70(1.46); C77(1.46); C111(1.28); C328(0.93) | LDD0564 | [5] |
| LDCM0249 | AC130 | HCT 116 | C70(1.65); C77(1.65); C111(1.02); C328(1.01) | LDD0566 | [5] |
| LDCM0250 | AC131 | HCT 116 | C70(1.14); C77(1.14); C111(0.96); C328(0.94) | LDD0567 | [5] |
| LDCM0251 | AC132 | HCT 116 | C70(1.61); C77(1.61); C111(1.63); C328(0.92) | LDD0568 | [5] |
| LDCM0252 | AC133 | HCT 116 | C70(1.27); C77(1.27); C111(1.08); C328(1.01) | LDD0569 | [5] |
| LDCM0253 | AC134 | HCT 116 | C70(1.39); C77(1.39); C111(1.25); C328(1.07) | LDD0570 | [5] |
| LDCM0254 | AC135 | HCT 116 | C70(1.27); C77(1.27); C111(1.29); C328(1.06) | LDD0571 | [5] |
| LDCM0255 | AC136 | HCT 116 | C70(1.32); C77(1.32); C111(1.14); C328(1.02) | LDD0572 | [5] |
| LDCM0256 | AC137 | HCT 116 | C70(1.34); C77(1.34); C111(1.22); C328(1.05) | LDD0573 | [5] |
| LDCM0257 | AC138 | HCT 116 | C70(0.96); C77(0.96); C111(1.40); C328(1.17) | LDD0574 | [5] |
| LDCM0258 | AC139 | HCT 116 | C70(1.22); C77(1.22); C111(1.31); C328(0.97) | LDD0575 | [5] |
| LDCM0259 | AC14 | HCT 116 | C70(1.00); C77(1.00); C111(1.08); C328(1.06) | LDD0576 | [5] |
| LDCM0260 | AC140 | HCT 116 | C70(1.68); C77(1.68); C111(1.57); C328(1.13) | LDD0577 | [5] |
| LDCM0261 | AC141 | HCT 116 | C70(1.41); C77(1.41); C111(1.53); C328(1.13) | LDD0578 | [5] |
| LDCM0262 | AC142 | HCT 116 | C70(1.56); C77(1.56); C111(1.21); C328(0.97) | LDD0579 | [5] |
| LDCM0263 | AC143 | HCT 116 | C70(1.45); C77(1.45); C334(0.68); C328(1.07) | LDD0580 | [5] |
| LDCM0264 | AC144 | HCT 116 | C334(0.68); C70(0.76); C77(0.76); C111(1.06) | LDD0581 | [5] |
| LDCM0265 | AC145 | HCT 116 | C334(0.76); C70(0.79); C77(0.79); C111(0.94) | LDD0582 | [5] |
| LDCM0266 | AC146 | HCT 116 | C70(0.86); C77(0.86); C334(1.07); C111(1.15) | LDD0583 | [5] |
| LDCM0267 | AC147 | HCT 116 | C70(0.82); C77(0.82); C334(0.84); C111(1.01) | LDD0584 | [5] |
| LDCM0268 | AC148 | HCT 116 | C334(0.88); C70(0.93); C77(0.93); C111(1.33) | LDD0585 | [5] |
| LDCM0269 | AC149 | HCT 116 | C334(0.68); C70(1.05); C77(1.05); C111(1.23) | LDD0586 | [5] |
| LDCM0270 | AC15 | HCT 116 | C70(0.90); C77(0.90); C334(1.00); C328(1.02) | LDD0587 | [5] |
| LDCM0271 | AC150 | HCT 116 | C111(0.93); C328(1.00); C70(1.30); C77(1.30) | LDD0588 | [5] |
| LDCM0272 | AC151 | HCT 116 | C111(0.84); C334(0.87); C70(0.88); C77(0.88) | LDD0589 | [5] |
| LDCM0273 | AC152 | HCT 116 | C70(0.88); C77(0.88); C111(1.05); C334(1.06) | LDD0590 | [5] |
| LDCM0274 | AC153 | HCT 116 | C334(0.75); C70(0.89); C77(0.89); C111(1.34) | LDD0591 | [5] |
| LDCM0621 | AC154 | HCT 116 | C70(0.84); C77(0.84); C111(1.09); C328(1.33) | LDD2158 | [5] |
| LDCM0622 | AC155 | HCT 116 | C70(1.10); C77(1.10); C111(1.21); C328(1.10) | LDD2159 | [5] |
| LDCM0623 | AC156 | HCT 116 | C70(1.41); C77(1.41); C111(1.07); C328(0.91) | LDD2160 | [5] |
| LDCM0624 | AC157 | HCT 116 | C70(1.43); C77(1.43); C111(0.85); C328(1.13) | LDD2161 | [5] |
| LDCM0276 | AC17 | HCT 116 | C328(0.79); C70(0.94); C77(0.94); C334(0.96) | LDD0593 | [5] |
| LDCM0277 | AC18 | HCT 116 | C328(0.69); C70(0.85); C77(0.85); C334(0.86) | LDD0594 | [5] |
| LDCM0278 | AC19 | HCT 116 | C328(0.68); C334(1.00); C70(1.07); C77(1.07) | LDD0595 | [5] |
| LDCM0279 | AC2 | HCT 116 | C328(0.62); C334(0.92); C111(1.14); C70(1.38) | LDD0596 | [5] |
| LDCM0280 | AC20 | HCT 116 | C328(0.81); C70(0.96); C77(0.96); C111(1.01) | LDD0597 | [5] |
| LDCM0281 | AC21 | HCT 116 | C328(0.75); C334(0.95); C70(0.99); C77(0.99) | LDD0598 | [5] |
| LDCM0282 | AC22 | HCT 116 | C328(0.80); C70(0.88); C77(0.88); C111(0.96) | LDD0599 | [5] |
| LDCM0283 | AC23 | HCT 116 | C328(0.75); C70(0.94); C77(0.94); C111(0.97) | LDD0600 | [5] |
| LDCM0284 | AC24 | HCT 116 | C328(0.70); C334(1.03); C111(1.06); C70(1.16) | LDD0601 | [5] |
| LDCM0285 | AC25 | HCT 116 | C328(0.61); C334(1.04); C70(1.06); C77(1.06) | LDD0602 | [5] |
| LDCM0286 | AC26 | HCT 116 | C328(0.60); C334(0.92); C111(1.19); C70(1.36) | LDD0603 | [5] |
| LDCM0287 | AC27 | HCT 116 | C328(0.62); C334(1.02); C70(1.10); C77(1.10) | LDD0604 | [5] |
| LDCM0288 | AC28 | HCT 116 | C328(0.59); C334(1.09); C111(1.36); C70(1.40) | LDD0605 | [5] |
| LDCM0289 | AC29 | HCT 116 | C328(0.65); C334(0.94); C70(1.15); C77(1.15) | LDD0606 | [5] |
| LDCM0290 | AC3 | HCT 116 | C328(0.58); C334(0.94); C111(1.13); C70(1.48) | LDD0607 | [5] |
| LDCM0291 | AC30 | HCT 116 | C328(0.80); C334(0.91); C70(0.98); C77(0.98) | LDD0608 | [5] |
| LDCM0292 | AC31 | HCT 116 | C328(0.55); C334(0.96); C111(1.19); C70(1.26) | LDD0609 | [5] |
| LDCM0293 | AC32 | HCT 116 | C328(0.53); C334(0.93); C70(1.19); C77(1.19) | LDD0610 | [5] |
| LDCM0294 | AC33 | HCT 116 | C328(0.54); C334(0.90); C70(1.27); C77(1.27) | LDD0611 | [5] |
| LDCM0295 | AC34 | HCT 116 | C328(0.58); C334(0.94); C70(1.06); C77(1.06) | LDD0612 | [5] |
| LDCM0296 | AC35 | HCT 116 | C70(0.89); C77(0.89); C328(0.94); C111(1.10) | LDD0613 | [5] |
| LDCM0297 | AC36 | HCT 116 | C70(0.69); C77(0.69); C111(0.81); C334(1.00) | LDD0614 | [5] |
| LDCM0298 | AC37 | HCT 116 | C111(0.85); C328(0.91); C70(0.93); C77(0.93) | LDD0615 | [5] |
| LDCM0299 | AC38 | HCT 116 | C328(0.84); C70(0.93); C77(0.93); C334(0.96) | LDD0616 | [5] |
| LDCM0300 | AC39 | HCT 116 | C70(0.80); C77(0.80); C328(0.87); C111(0.95) | LDD0617 | [5] |
| LDCM0301 | AC4 | HCT 116 | C328(0.58); C334(0.94); C111(1.16); C70(1.27) | LDD0618 | [5] |
| LDCM0302 | AC40 | HCT 116 | C70(0.72); C77(0.72); C111(0.85); C334(0.93) | LDD0619 | [5] |
| LDCM0303 | AC41 | HCT 116 | C328(0.89); C334(0.94); C111(0.98); C70(1.01) | LDD0620 | [5] |
| LDCM0304 | AC42 | HCT 116 | C328(0.90); C70(0.92); C77(0.92); C334(0.96) | LDD0621 | [5] |
| LDCM0305 | AC43 | HCT 116 | C328(0.89); C70(0.94); C77(0.94); C334(0.96) | LDD0622 | [5] |
| LDCM0306 | AC44 | HCT 116 | C328(0.90); C111(0.94); C334(0.99); C70(1.11) | LDD0623 | [5] |
| LDCM0307 | AC45 | HCT 116 | C70(0.79); C77(0.79); C111(0.94); C334(0.94) | LDD0624 | [5] |
| LDCM0308 | AC46 | HCT 116 | C70(0.82); C77(0.82); C111(0.94); C334(0.98) | LDD0625 | [5] |
| LDCM0309 | AC47 | HCT 116 | C70(0.92); C77(0.92); C111(0.99); C328(1.00) | LDD0626 | [5] |
| LDCM0310 | AC48 | HCT 116 | C328(0.90); C111(1.00); C334(1.06); C70(1.09) | LDD0627 | [5] |
| LDCM0311 | AC49 | HCT 116 | C70(0.88); C77(0.88); C334(0.95); C328(1.08) | LDD0628 | [5] |
| LDCM0312 | AC5 | HCT 116 | C328(0.57); C334(0.95); C111(1.12); C70(1.34) | LDD0629 | [5] |
| LDCM0313 | AC50 | HCT 116 | C70(0.98); C77(0.98); C334(1.04); C328(1.07) | LDD0630 | [5] |
| LDCM0314 | AC51 | HCT 116 | C328(0.82); C70(0.86); C77(0.86); C111(0.90) | LDD0631 | [5] |
| LDCM0315 | AC52 | HCT 116 | C70(0.80); C77(0.80); C328(0.97); C111(1.01) | LDD0632 | [5] |
| LDCM0316 | AC53 | HCT 116 | C70(0.85); C77(0.85); C328(0.98); C111(1.01) | LDD0633 | [5] |
| LDCM0317 | AC54 | HCT 116 | C70(0.91); C77(0.91); C328(1.02); C111(1.06) | LDD0634 | [5] |
| LDCM0318 | AC55 | HCT 116 | C70(0.76); C77(0.76); C334(1.03); C111(1.07) | LDD0635 | [5] |
| LDCM0319 | AC56 | HCT 116 | C70(0.86); C77(0.86); C334(1.00); C111(1.17) | LDD0636 | [5] |
| LDCM0320 | AC57 | HCT 116 | C328(1.08); C111(1.21); C70(1.25); C77(1.25) | LDD0637 | [5] |
| LDCM0321 | AC58 | HCT 116 | C328(1.12); C111(1.22); C70(1.39); C77(1.39) | LDD0638 | [5] |
| LDCM0322 | AC59 | HCT 116 | C328(1.08); C70(1.09); C77(1.09); C111(1.15) | LDD0639 | [5] |
| LDCM0323 | AC6 | HCT 116 | C334(0.96); C70(1.01); C77(1.01); C328(1.14) | LDD0640 | [5] |
| LDCM0324 | AC60 | HCT 116 | C70(1.07); C77(1.07); C328(1.10); C111(1.14) | LDD0641 | [5] |
| LDCM0325 | AC61 | HCT 116 | C70(0.94); C77(0.94); C111(1.01); C328(1.02) | LDD0642 | [5] |
| LDCM0326 | AC62 | HCT 116 | C70(1.12); C77(1.12); C328(1.21); C111(1.22) | LDD0643 | [5] |
| LDCM0327 | AC63 | HCT 116 | C70(1.04); C77(1.04); C328(1.08); C111(1.09) | LDD0644 | [5] |
| LDCM0328 | AC64 | HCT 116 | C70(0.98); C77(0.98); C328(1.04); C111(1.33) | LDD0645 | [5] |
| LDCM0329 | AC65 | HCT 116 | C328(1.02); C70(1.16); C77(1.16); C111(1.45) | LDD0646 | [5] |
| LDCM0330 | AC66 | HCT 116 | C328(0.96); C70(1.38); C77(1.38); C111(1.46) | LDD0647 | [5] |
| LDCM0331 | AC67 | HCT 116 | C70(1.00); C77(1.00); C328(1.05); C111(1.27) | LDD0648 | [5] |
| LDCM0332 | AC68 | HCT 116 | C334(0.76); C111(0.83); C70(0.92); C77(0.92) | LDD0649 | [5] |
| LDCM0333 | AC69 | HCT 116 | C334(0.76); C70(0.87); C77(0.87); C111(0.95) | LDD0650 | [5] |
| LDCM0334 | AC7 | HCT 116 | C334(1.01); C111(1.06); C70(1.13); C77(1.13) | LDD0651 | [5] |
| LDCM0335 | AC70 | HCT 116 | C334(0.74); C70(0.89); C77(0.89); C111(1.01) | LDD0652 | [5] |
| LDCM0336 | AC71 | HCT 116 | C111(0.79); C70(0.87); C77(0.87); C334(0.97) | LDD0653 | [5] |
| LDCM0337 | AC72 | HCT 116 | C334(0.86); C111(0.90); C70(0.97); C77(0.97) | LDD0654 | [5] |
| LDCM0338 | AC73 | HCT 116 | C70(0.97); C77(0.97); C334(0.76); C111(1.06) | LDD0655 | [5] |
| LDCM0339 | AC74 | HCT 116 | C70(0.86); C77(0.86); C111(0.93); C334(1.03) | LDD0656 | [5] |
| LDCM0340 | AC75 | HCT 116 | C334(0.78); C70(0.94); C77(0.94); C111(1.00) | LDD0657 | [5] |
| LDCM0341 | AC76 | HCT 116 | C70(0.85); C77(0.85); C111(0.87); C334(0.93) | LDD0658 | [5] |
| LDCM0342 | AC77 | HCT 116 | C334(0.78); C70(0.86); C77(0.86); C111(0.90) | LDD0659 | [5] |
| LDCM0343 | AC78 | HCT 116 | C334(0.87); C111(0.87); C70(0.95); C77(0.95) | LDD0660 | [5] |
| LDCM0344 | AC79 | HCT 116 | C334(0.73); C328(1.02); C111(1.03); C70(1.10) | LDD0661 | [5] |
| LDCM0345 | AC8 | HCT 116 | C334(0.89); C70(1.07); C77(1.07); C328(1.13) | LDD0662 | [5] |
| LDCM0346 | AC80 | HCT 116 | C111(0.93); C334(1.02); C328(1.03); C70(1.10) | LDD0663 | [5] |
| LDCM0347 | AC81 | HCT 116 | C111(0.78); C334(0.85); C328(1.06); C70(1.48) | LDD0664 | [5] |
| LDCM0348 | AC82 | HCT 116 | C334(0.87); C70(0.88); C77(0.88); C111(1.21) | LDD0665 | [5] |
| LDCM0349 | AC83 | HCT 116 | C70(0.66); C77(0.66); C111(0.90); C328(1.31) | LDD0666 | [5] |
| LDCM0350 | AC84 | HCT 116 | C111(0.76); C70(1.13); C77(1.13); C328(1.31) | LDD0667 | [5] |
| LDCM0351 | AC85 | HCT 116 | C111(0.87); C70(0.90); C77(0.90); C328(1.14) | LDD0668 | [5] |
| LDCM0352 | AC86 | HCT 116 | C70(0.59); C77(0.59); C111(1.00); C328(1.08) | LDD0669 | [5] |
| LDCM0353 | AC87 | HCT 116 | C70(0.70); C77(0.70); C111(0.91); C328(1.01) | LDD0670 | [5] |
| LDCM0354 | AC88 | HCT 116 | C111(0.85); C70(1.03); C77(1.03); C328(1.03) | LDD0671 | [5] |
| LDCM0355 | AC89 | HCT 116 | C70(0.75); C77(0.75); C111(0.91); C328(1.07) | LDD0672 | [5] |
| LDCM0357 | AC90 | HCT 116 | C328(0.85); C111(1.04); C70(1.58); C77(1.58) | LDD0674 | [5] |
| LDCM0358 | AC91 | HCT 116 | C70(0.74); C77(0.74); C111(0.85); C328(1.11) | LDD0675 | [5] |
| LDCM0359 | AC92 | HCT 116 | C70(0.55); C77(0.55); C111(1.02); C328(1.06) | LDD0676 | [5] |
| LDCM0360 | AC93 | HCT 116 | C70(0.65); C77(0.65); C111(0.89); C328(1.04) | LDD0677 | [5] |
| LDCM0361 | AC94 | HCT 116 | C70(0.56); C77(0.56); C328(0.91); C111(1.10) | LDD0678 | [5] |
| LDCM0362 | AC95 | HCT 116 | C70(0.38); C77(0.38); C328(0.85); C111(0.85) | LDD0679 | [5] |
| LDCM0363 | AC96 | HCT 116 | C70(0.67); C77(0.67); C111(0.93); C328(0.99) | LDD0680 | [5] |
| LDCM0364 | AC97 | HCT 116 | C70(0.72); C77(0.72); C111(0.97); C328(1.05) | LDD0681 | [5] |
| LDCM0365 | AC98 | HCT 116 | C334(0.41); C111(1.07); C328(1.15) | LDD0682 | [5] |
| LDCM0366 | AC99 | HCT 116 | C334(0.69); C111(1.00); C328(1.14) | LDD0683 | [5] |
| LDCM0248 | AKOS034007472 | HCT 116 | C70(1.38); C77(1.38); C111(1.09); C328(0.95) | LDD0565 | [5] |
| LDCM0356 | AKOS034007680 | HCT 116 | C334(0.94); C111(1.08); C328(1.09); C70(1.15) | LDD0673 | [5] |
| LDCM0275 | AKOS034007705 | HCT 116 | C334(0.89); C70(0.92); C77(0.92); C328(1.28) | LDD0592 | [5] |
| LDCM0156 | Aniline | NCI-H1299 | 9.54 | LDD0403 | [1] |
| LDCM0020 | ARS-1620 | HCC44 | C328(1.03) | LDD0078 | [5] |
| LDCM0630 | CCW28-3 | 231MFP | C393(1.49) | LDD2214 | [13] |
| LDCM0367 | CL1 | HCT 116 | C328(0.88); C111(0.88); C334(1.02); C70(1.39) | LDD0684 | [5] |
| LDCM0368 | CL10 | HCT 116 | C334(0.87); C111(1.06); C328(1.14); C70(11.85) | LDD0685 | [5] |
| LDCM0369 | CL100 | HCT 116 | C334(0.69); C328(0.71); C111(1.12); C70(1.32) | LDD0686 | [5] |
| LDCM0370 | CL101 | HCT 116 | C334(0.96); C70(1.01); C77(1.01); C111(1.15) | LDD0687 | [5] |
| LDCM0371 | CL102 | HCT 116 | C334(1.01); C328(1.01); C111(1.03); C70(1.64) | LDD0688 | [5] |
| LDCM0372 | CL103 | HCT 116 | C328(0.98); C334(1.02); C111(1.06); C70(1.36) | LDD0689 | [5] |
| LDCM0373 | CL104 | HCT 116 | C111(0.93); C334(0.98); C328(1.03); C70(1.08) | LDD0690 | [5] |
| LDCM0374 | CL105 | HCT 116 | C328(0.85); C334(1.00); C111(1.13); C70(1.14) | LDD0691 | [5] |
| LDCM0375 | CL106 | HCT 116 | C70(0.82); C77(0.82); C328(0.83); C334(0.91) | LDD0692 | [5] |
| LDCM0376 | CL107 | HCT 116 | C328(0.88); C334(0.91); C70(1.00); C77(1.00) | LDD0693 | [5] |
| LDCM0377 | CL108 | HCT 116 | C328(0.77); C334(0.98); C70(1.18); C77(1.18) | LDD0694 | [5] |
| LDCM0378 | CL109 | HCT 116 | C328(0.73); C334(0.87); C70(1.04); C77(1.04) | LDD0695 | [5] |
| LDCM0379 | CL11 | HCT 116 | C334(0.89); C70(0.97); C77(0.97); C328(1.06) | LDD0696 | [5] |
| LDCM0380 | CL110 | HCT 116 | C328(0.81); C334(0.97); C111(1.15); C70(1.22) | LDD0697 | [5] |
| LDCM0381 | CL111 | HCT 116 | C328(0.77); C334(0.83); C70(0.95); C77(0.95) | LDD0698 | [5] |
| LDCM0382 | CL112 | HCT 116 | C328(0.65); C334(1.02); C70(1.13); C77(1.13) | LDD0699 | [5] |
| LDCM0383 | CL113 | HCT 116 | C328(0.51); C334(0.97); C70(1.02); C77(1.02) | LDD0700 | [5] |
| LDCM0384 | CL114 | HCT 116 | C328(0.41); C334(1.06); C70(1.07); C77(1.07) | LDD0701 | [5] |
| LDCM0385 | CL115 | HCT 116 | C328(0.48); C334(1.04); C111(1.22); C70(1.45) | LDD0702 | [5] |
| LDCM0386 | CL116 | HCT 116 | C328(0.48); C334(0.95); C70(1.09); C77(1.09) | LDD0703 | [5] |
| LDCM0387 | CL117 | HCT 116 | C111(0.70); C334(0.84); C70(0.87); C77(0.87) | LDD0704 | [5] |
| LDCM0388 | CL118 | HCT 116 | C70(0.85); C77(0.85); C328(0.91); C334(0.95) | LDD0705 | [5] |
| LDCM0389 | CL119 | HCT 116 | C328(0.92); C334(1.02); C70(1.04); C77(1.04) | LDD0706 | [5] |
| LDCM0390 | CL12 | HCT 116 | C70(0.88); C77(0.88); C334(0.96); C111(1.09) | LDD0707 | [5] |
| LDCM0391 | CL120 | HCT 116 | C328(0.83); C70(0.85); C77(0.85); C111(0.94) | LDD0708 | [5] |
| LDCM0392 | CL121 | HCT 116 | C111(1.01); C328(1.08); C70(1.10); C77(1.10) | LDD0709 | [5] |
| LDCM0393 | CL122 | HCT 116 | C334(0.90); C111(0.96); C70(1.08); C77(1.08) | LDD0710 | [5] |
| LDCM0394 | CL123 | HCT 116 | C334(1.03); C328(1.05); C111(1.05); C70(1.12) | LDD0711 | [5] |
| LDCM0395 | CL124 | HCT 116 | C70(0.90); C77(0.90); C334(1.04); C111(1.07) | LDD0712 | [5] |
| LDCM0396 | CL125 | HCT 116 | C328(0.92); C111(1.04); C70(1.08); C77(1.08) | LDD0713 | [5] |
| LDCM0397 | CL126 | HCT 116 | C328(1.07); C111(1.12); C70(1.47); C77(1.47) | LDD0714 | [5] |
| LDCM0398 | CL127 | HCT 116 | C328(1.00); C111(1.08); C70(1.23); C77(1.23) | LDD0715 | [5] |
| LDCM0399 | CL128 | HCT 116 | C70(0.90); C77(0.90); C328(1.07); C111(1.26) | LDD0716 | [5] |
| LDCM0400 | CL13 | HCT 116 | C70(0.75); C77(0.75); C334(0.76); C111(0.90) | LDD0717 | [5] |
| LDCM0401 | CL14 | HCT 116 | C328(0.89); C111(0.91); C70(0.99); C77(0.99) | LDD0718 | [5] |
| LDCM0402 | CL15 | HCT 116 | C111(0.89); C70(0.92); C77(0.92); C334(0.95) | LDD0719 | [5] |
| LDCM0403 | CL16 | HCT 116 | C334(0.73); C70(0.96); C77(0.96); C328(1.11) | LDD0720 | [5] |
| LDCM0404 | CL17 | HCT 116 | C70(0.95); C77(0.95); C328(1.05); C334(1.19) | LDD0721 | [5] |
| LDCM0405 | CL18 | HCT 116 | C70(0.73); C77(0.73); C334(0.92); C111(0.99) | LDD0722 | [5] |
| LDCM0406 | CL19 | HCT 116 | C70(0.90); C77(0.90); C334(1.07); C328(1.10) | LDD0723 | [5] |
| LDCM0407 | CL2 | HCT 116 | C111(0.84); C334(0.89); C328(0.96); C70(0.98) | LDD0724 | [5] |
| LDCM0408 | CL20 | HCT 116 | C70(0.98); C77(0.98); C334(1.01); C111(1.20) | LDD0725 | [5] |
| LDCM0409 | CL21 | HCT 116 | C334(1.09); C70(1.16); C77(1.16); C328(1.18) | LDD0726 | [5] |
| LDCM0410 | CL22 | HCT 116 | C334(0.91); C70(1.10); C77(1.10); C111(1.31) | LDD0727 | [5] |
| LDCM0411 | CL23 | HCT 116 | C70(0.83); C77(0.83); C334(0.90); C328(1.02) | LDD0728 | [5] |
| LDCM0412 | CL24 | HCT 116 | C70(0.84); C77(0.84); C111(1.56); C328(1.07) | LDD0729 | [5] |
| LDCM0413 | CL25 | HCT 116 | C70(1.04); C77(1.04); C111(1.55); C328(1.02) | LDD0730 | [5] |
| LDCM0414 | CL26 | HCT 116 | C70(0.91); C77(0.91); C111(1.23); C328(1.00) | LDD0731 | [5] |
| LDCM0415 | CL27 | HCT 116 | C70(1.08); C77(1.08); C111(1.17); C328(0.82) | LDD0732 | [5] |
| LDCM0416 | CL28 | HCT 116 | C70(0.96); C77(0.96); C111(2.24); C328(0.91) | LDD0733 | [5] |
| LDCM0417 | CL29 | HCT 116 | C70(1.33); C77(1.33); C111(1.26); C328(0.97) | LDD0734 | [5] |
| LDCM0418 | CL3 | HCT 116 | C70(0.98); C77(0.98); C111(0.81); C328(0.94) | LDD0735 | [5] |
| LDCM0419 | CL30 | HCT 116 | C70(1.10); C77(1.10); C111(1.31); C328(0.86) | LDD0736 | [5] |
| LDCM0420 | CL31 | HCT 116 | C70(0.94); C77(0.94); C111(1.01); C328(0.98) | LDD0737 | [5] |
| LDCM0421 | CL32 | HCT 116 | C70(0.60); C77(0.60); C111(1.09); C328(1.00) | LDD0738 | [5] |
| LDCM0422 | CL33 | HCT 116 | C70(1.00); C77(1.00); C111(0.90); C328(1.15) | LDD0739 | [5] |
| LDCM0423 | CL34 | HCT 116 | C70(1.14); C77(1.14); C111(1.28); C328(1.26) | LDD0740 | [5] |
| LDCM0424 | CL35 | HCT 116 | C70(0.71); C77(0.71); C111(1.37); C328(1.11) | LDD0741 | [5] |
| LDCM0425 | CL36 | HCT 116 | C70(0.75); C77(0.75); C111(1.07); C328(1.20) | LDD0742 | [5] |
| LDCM0426 | CL37 | HCT 116 | C70(0.75); C77(0.75); C111(1.05); C328(1.21) | LDD0743 | [5] |
| LDCM0428 | CL39 | HCT 116 | C70(1.29); C77(1.29); C111(1.03); C328(1.12) | LDD0745 | [5] |
| LDCM0429 | CL4 | HCT 116 | C70(1.07); C77(1.07); C111(0.91); C328(0.99) | LDD0746 | [5] |
| LDCM0430 | CL40 | HCT 116 | C70(0.77); C77(0.77); C111(1.04); C328(1.03) | LDD0747 | [5] |
| LDCM0431 | CL41 | HCT 116 | C70(0.82); C77(0.82); C111(1.34); C328(1.05) | LDD0748 | [5] |
| LDCM0432 | CL42 | HCT 116 | C70(0.99); C77(0.99); C111(1.29); C328(1.33) | LDD0749 | [5] |
| LDCM0433 | CL43 | HCT 116 | C70(0.71); C77(0.71); C111(1.31); C328(1.14) | LDD0750 | [5] |
| LDCM0434 | CL44 | HCT 116 | C70(0.79); C77(0.79); C111(1.32); C328(0.95) | LDD0751 | [5] |
| LDCM0435 | CL45 | HCT 116 | C70(0.71); C77(0.71); C111(1.50); C328(0.95) | LDD0752 | [5] |
| LDCM0436 | CL46 | HCT 116 | C70(1.11); C77(1.11); C111(0.92); C328(0.79) | LDD0753 | [5] |
| LDCM0437 | CL47 | HCT 116 | C70(1.07); C77(1.07); C111(0.92); C328(0.86) | LDD0754 | [5] |
| LDCM0438 | CL48 | HCT 116 | C70(1.02); C77(1.02); C111(0.84); C328(0.85) | LDD0755 | [5] |
| LDCM0439 | CL49 | HCT 116 | C70(1.06); C77(1.06); C111(1.05); C328(0.80) | LDD0756 | [5] |
| LDCM0440 | CL5 | HCT 116 | C70(1.04); C77(1.04); C111(0.85); C328(0.94) | LDD0757 | [5] |
| LDCM0441 | CL50 | HCT 116 | C70(1.67); C77(1.67); C111(0.88); C328(0.79) | LDD0758 | [5] |
| LDCM0442 | CL51 | HCT 116 | C70(1.23); C77(1.23); C111(0.93); C328(0.76) | LDD0759 | [5] |
| LDCM0443 | CL52 | HCT 116 | C70(1.14); C77(1.14); C111(0.98); C328(0.82) | LDD0760 | [5] |
| LDCM0444 | CL53 | HCT 116 | C70(1.07); C77(1.07); C111(0.81); C328(0.93) | LDD0761 | [5] |
| LDCM0445 | CL54 | HCT 116 | C70(1.17); C77(1.17); C111(0.92); C328(0.74) | LDD0762 | [5] |
| LDCM0446 | CL55 | HCT 116 | C70(1.10); C77(1.10); C111(0.85); C328(0.73) | LDD0763 | [5] |
| LDCM0447 | CL56 | HCT 116 | C70(1.01); C77(1.01); C111(0.99); C328(0.90) | LDD0764 | [5] |
| LDCM0448 | CL57 | HCT 116 | C70(1.03); C77(1.03); C111(0.92); C328(0.74) | LDD0765 | [5] |
| LDCM0449 | CL58 | HCT 116 | C70(1.05); C77(1.05); C111(0.96); C328(0.70) | LDD0766 | [5] |
| LDCM0450 | CL59 | HCT 116 | C70(1.12); C77(1.12); C111(0.97); C328(0.79) | LDD0767 | [5] |
| LDCM0451 | CL6 | HCT 116 | C70(0.89); C77(0.89); C111(0.97); C328(0.92) | LDD0768 | [5] |
| LDCM0452 | CL60 | HCT 116 | C70(1.08); C77(1.08); C111(1.14); C328(0.73) | LDD0769 | [5] |
| LDCM0453 | CL61 | HCT 116 | C70(0.87); C77(0.87); C111(0.90); C328(0.95) | LDD0770 | [5] |
| LDCM0454 | CL62 | HCT 116 | C70(0.94); C77(0.94); C111(0.89); C328(1.01) | LDD0771 | [5] |
| LDCM0455 | CL63 | HCT 116 | C70(0.96); C77(0.96); C111(0.95); C328(1.16) | LDD0772 | [5] |
| LDCM0456 | CL64 | HCT 116 | C70(1.11); C77(1.11); C111(1.02); C328(1.03) | LDD0773 | [5] |
| LDCM0457 | CL65 | HCT 116 | C70(0.91); C77(0.91); C111(0.95); C328(1.02) | LDD0774 | [5] |
| LDCM0458 | CL66 | HCT 116 | C70(1.04); C77(1.04); C111(1.21); C328(1.16) | LDD0775 | [5] |
| LDCM0459 | CL67 | HCT 116 | C70(0.68); C77(0.68); C111(1.18); C328(1.16) | LDD0776 | [5] |
| LDCM0460 | CL68 | HCT 116 | C70(0.61); C77(0.61); C111(0.97); C328(1.31) | LDD0777 | [5] |
| LDCM0461 | CL69 | HCT 116 | C70(0.96); C77(0.96); C111(0.81); C328(1.02) | LDD0778 | [5] |
| LDCM0462 | CL7 | HCT 116 | C70(0.84); C77(0.84); C111(1.16); C328(1.02) | LDD0779 | [5] |
| LDCM0463 | CL70 | HCT 116 | C70(1.02); C77(1.02); C111(0.89); C328(1.16) | LDD0780 | [5] |
| LDCM0464 | CL71 | HCT 116 | C70(0.89); C77(0.89); C111(0.93); C328(1.27) | LDD0781 | [5] |
| LDCM0465 | CL72 | HCT 116 | C70(0.82); C77(0.82); C111(0.81); C328(1.24) | LDD0782 | [5] |
| LDCM0466 | CL73 | HCT 116 | C70(0.81); C77(0.81); C111(1.14); C328(1.15) | LDD0783 | [5] |
| LDCM0467 | CL74 | HCT 116 | C70(0.81); C77(0.81); C111(0.97); C328(1.15) | LDD0784 | [5] |
| LDCM0469 | CL76 | HCT 116 | C70(0.89); C77(0.89); C111(0.97); C328(1.09) | LDD0786 | [5] |
| LDCM0470 | CL77 | HCT 116 | C70(0.96); C77(0.96); C111(0.98); C328(1.09) | LDD0787 | [5] |
| LDCM0471 | CL78 | HCT 116 | C70(0.82); C77(0.82); C111(0.98); C328(1.06) | LDD0788 | [5] |
| LDCM0472 | CL79 | HCT 116 | C70(0.89); C77(0.89); C111(0.97); C328(1.19) | LDD0789 | [5] |
| LDCM0473 | CL8 | HCT 116 | C70(0.95); C77(0.95); C111(0.98); C328(1.30) | LDD0790 | [5] |
| LDCM0474 | CL80 | HCT 116 | C70(0.82); C77(0.82); C111(0.97); C328(1.05) | LDD0791 | [5] |
| LDCM0475 | CL81 | HCT 116 | C70(0.88); C77(0.88); C111(1.09); C328(1.05) | LDD0792 | [5] |
| LDCM0476 | CL82 | HCT 116 | C70(1.01); C77(1.01); C111(1.17); C328(1.28) | LDD0793 | [5] |
| LDCM0477 | CL83 | HCT 116 | C70(0.94); C77(0.94); C111(1.10); C328(1.25) | LDD0794 | [5] |
| LDCM0478 | CL84 | HCT 116 | C70(1.07); C77(1.07); C111(1.17); C328(1.34) | LDD0795 | [5] |
| LDCM0479 | CL85 | HCT 116 | C70(0.81); C77(0.81); C111(1.03); C328(1.10) | LDD0796 | [5] |
| LDCM0480 | CL86 | HCT 116 | C70(1.07); C77(1.07); C111(0.95); C328(0.95) | LDD0797 | [5] |
| LDCM0481 | CL87 | HCT 116 | C70(1.18); C77(1.18); C111(0.99); C328(0.98) | LDD0798 | [5] |
| LDCM0482 | CL88 | HCT 116 | C70(0.85); C77(0.85); C111(1.05); C328(1.19) | LDD0799 | [5] |
| LDCM0483 | CL89 | HCT 116 | C70(0.91); C77(0.91); C111(1.22); C328(1.55) | LDD0800 | [5] |
| LDCM0484 | CL9 | HCT 116 | C70(0.90); C77(0.90); C111(0.95); C328(1.04) | LDD0801 | [5] |
| LDCM0485 | CL90 | HCT 116 | C70(1.63); C77(1.63); C111(0.74); C328(0.88) | LDD0802 | [5] |
| LDCM0486 | CL91 | HCT 116 | C70(1.40); C77(1.40); C111(1.07); C328(1.03) | LDD0803 | [5] |
| LDCM0487 | CL92 | HCT 116 | C70(1.71); C77(1.71); C111(0.94); C328(0.96) | LDD0804 | [5] |
| LDCM0488 | CL93 | HCT 116 | C70(1.00); C77(1.00); C111(1.00); C328(0.80) | LDD0805 | [5] |
| LDCM0489 | CL94 | HCT 116 | C70(1.60); C77(1.60); C111(1.10); C328(0.90) | LDD0806 | [5] |
| LDCM0490 | CL95 | HCT 116 | C70(1.30); C77(1.30); C111(1.16); C328(0.80) | LDD0807 | [5] |
| LDCM0491 | CL96 | HCT 116 | C70(1.35); C77(1.35); C111(0.98); C328(0.89) | LDD0808 | [5] |
| LDCM0492 | CL97 | HCT 116 | C70(1.45); C77(1.45); C111(1.09); C328(0.79) | LDD0809 | [5] |
| LDCM0493 | CL98 | HCT 116 | C70(1.18); C77(1.18); C111(1.11); C328(0.94) | LDD0810 | [5] |
| LDCM0494 | CL99 | HCT 116 | C70(1.35); C77(1.35); C111(1.02); C328(0.74) | LDD0811 | [5] |
| LDCM0634 | CY-0357 | Hep-G2 | C393(1.11) | LDD2228 | [8] |
| LDCM0495 | E2913 | HEK-293T | C328(0.86); C334(0.97); C111(0.94); C393(1.21) | LDD1698 | [14] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C328(1.78) | LDD1702 | [15] |
| LDCM0175 | Ethacrynic acid | HeLa | C393(1.25) | LDD2210 | [4] |
| LDCM0625 | F8 | Ramos | C393(0.43) | LDD2187 | [16] |
| LDCM0572 | Fragment10 | Ramos | C328(3.60) | LDD2189 | [16] |
| LDCM0573 | Fragment11 | Ramos | C328(20.00); C393(2.66) | LDD2190 | [16] |
| LDCM0574 | Fragment12 | Ramos | C328(5.77); C393(0.60) | LDD2191 | [16] |
| LDCM0575 | Fragment13 | Ramos | C393(1.60) | LDD2192 | [16] |
| LDCM0576 | Fragment14 | Ramos | C328(0.97); C393(0.70) | LDD2193 | [16] |
| LDCM0579 | Fragment20 | Ramos | C393(1.92) | LDD2194 | [16] |
| LDCM0580 | Fragment21 | Ramos | C328(0.76); C393(0.78) | LDD2195 | [16] |
| LDCM0582 | Fragment23 | Ramos | C393(2.11) | LDD2196 | [16] |
| LDCM0578 | Fragment27 | Ramos | C393(0.95) | LDD2197 | [16] |
| LDCM0586 | Fragment28 | Ramos | C328(0.58); C393(0.83) | LDD2198 | [16] |
| LDCM0588 | Fragment30 | Ramos | C393(0.23) | LDD2199 | [16] |
| LDCM0589 | Fragment31 | Ramos | C393(1.17) | LDD2200 | [16] |
| LDCM0468 | Fragment33 | HCT 116 | C70(0.83); C77(0.83); C111(0.99); C328(1.18) | LDD0785 | [5] |
| LDCM0596 | Fragment38 | Ramos | C393(1.53) | LDD2203 | [16] |
| LDCM0566 | Fragment4 | Ramos | C328(1.42); C393(0.97) | LDD2184 | [16] |
| LDCM0427 | Fragment51 | HCT 116 | C70(0.98); C77(0.98); C111(1.36); C328(1.23) | LDD0744 | [5] |
| LDCM0610 | Fragment52 | Ramos | C393(0.49) | LDD2204 | [16] |
| LDCM0614 | Fragment56 | Ramos | C393(0.96) | LDD2205 | [16] |
| LDCM0569 | Fragment7 | Ramos | C328(13.20); C393(0.22) | LDD2186 | [16] |
| LDCM0571 | Fragment9 | Ramos | C393(0.63) | LDD2188 | [16] |
| LDCM0022 | KB02 | HCT 116 | C334(1.55); C328(1.59); C70(2.55); C77(2.55) | LDD0080 | [5] |
| LDCM0023 | KB03 | HCT 116 | C334(1.83); C328(1.43); C70(3.08); C77(3.08) | LDD0081 | [5] |
| LDCM0024 | KB05 | HCT 116 | C334(1.59); C328(1.36); C70(0.88); C77(0.88) | LDD0082 | [5] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C70(1.84) | LDD2206 | [17] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C70(4.22) | LDD2207 | [17] |
| LDCM0131 | RA190 | MM1.R | C393(1.73); C430(1.22) | LDD0304 | [18] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Diacylglycerol O-acyltransferase 2-like protein 6 (DGAT2L6) | Diacylglycerol acyltransferase family | Q6ZPD8 | |||
| Peptidyl-prolyl cis-trans isomerase FKBP7 (FKBP7) | . | Q9Y680 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Prenylated Rab acceptor protein 1 (RABAC1) | PRA1 family | Q9UI14 | |||
| Synaptophysin (SYP) | Synaptophysin/synaptobrevin family | P08247 | |||
Other
References
