Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
FBPP2 Probe Info |
 |
2.25 | LDD0318 | [2] | |
|
SAA-alkyne Probe Info |
 |
1.10 | LDD0252 | [3] | |
|
AZ-9 Probe Info |
 |
E80(10.00); E102(10.00) | LDD2209 | [4] | |
|
BTD Probe Info |
 |
C70(0.08) | LDD2096 | [5] | |
|
DA-P3 Probe Info |
 |
4.71 | LDD0183 | [6] | |
|
EA-probe Probe Info |
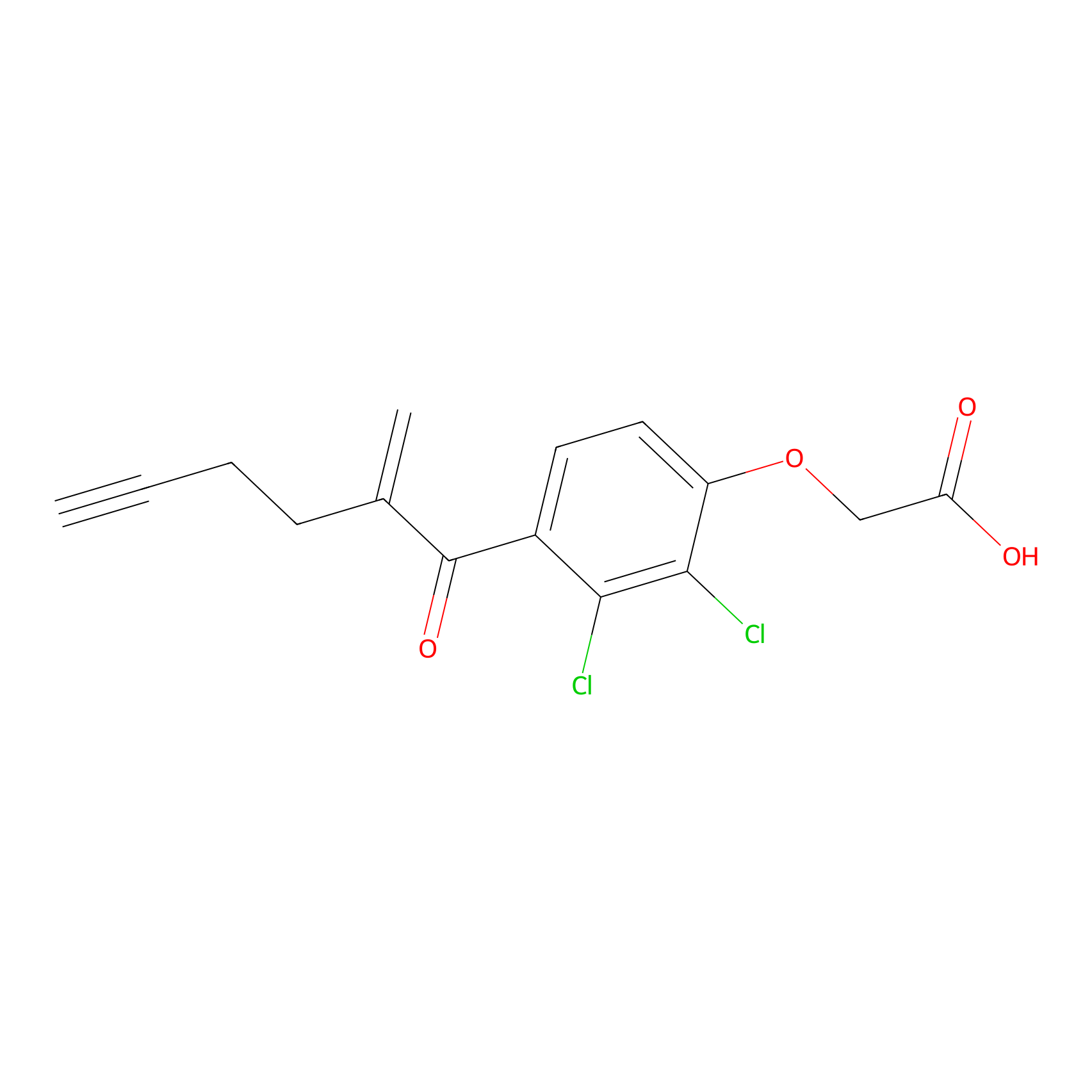 |
N.A. | LDD0440 | [7] | |
|
ATP probe Probe Info |
 |
K18(0.00); K94(0.00); K136(0.00); K10(0.00) | LDD0199 | [8] | |
|
1d-yne Probe Info |
 |
N.A. | LDD0358 | [9] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [10] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [11] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [12] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [11] | |
|
1c-yne Probe Info |
 |
K84(0.00); K43(0.00); K94(0.00); K18(0.00) | LDD0228 | [9] | |
|
HHS-465 Probe Info |
 |
N.A. | LDD2240 | [13] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C087 Probe Info |
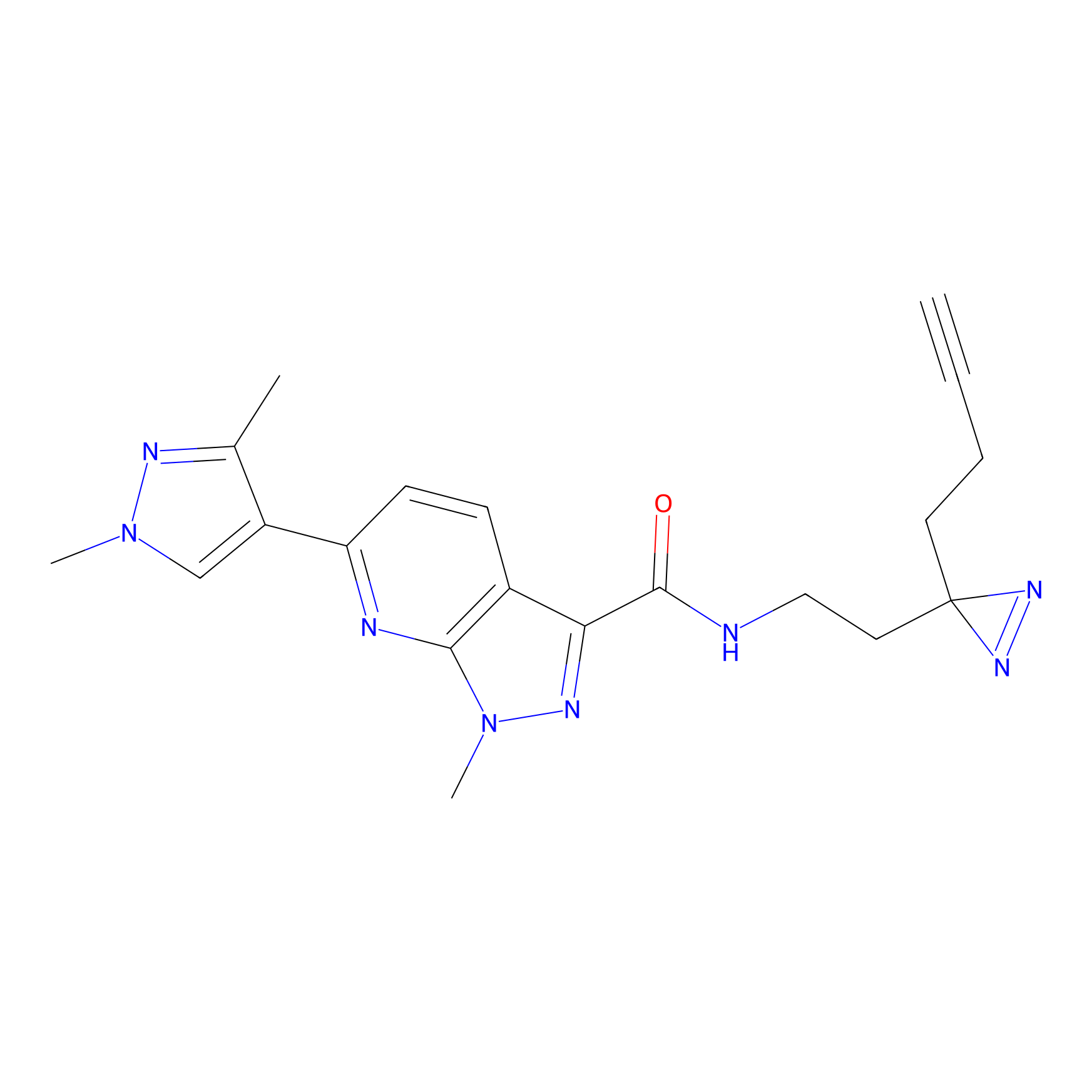 |
12.64 | LDD1779 | [14] | |
|
C249 Probe Info |
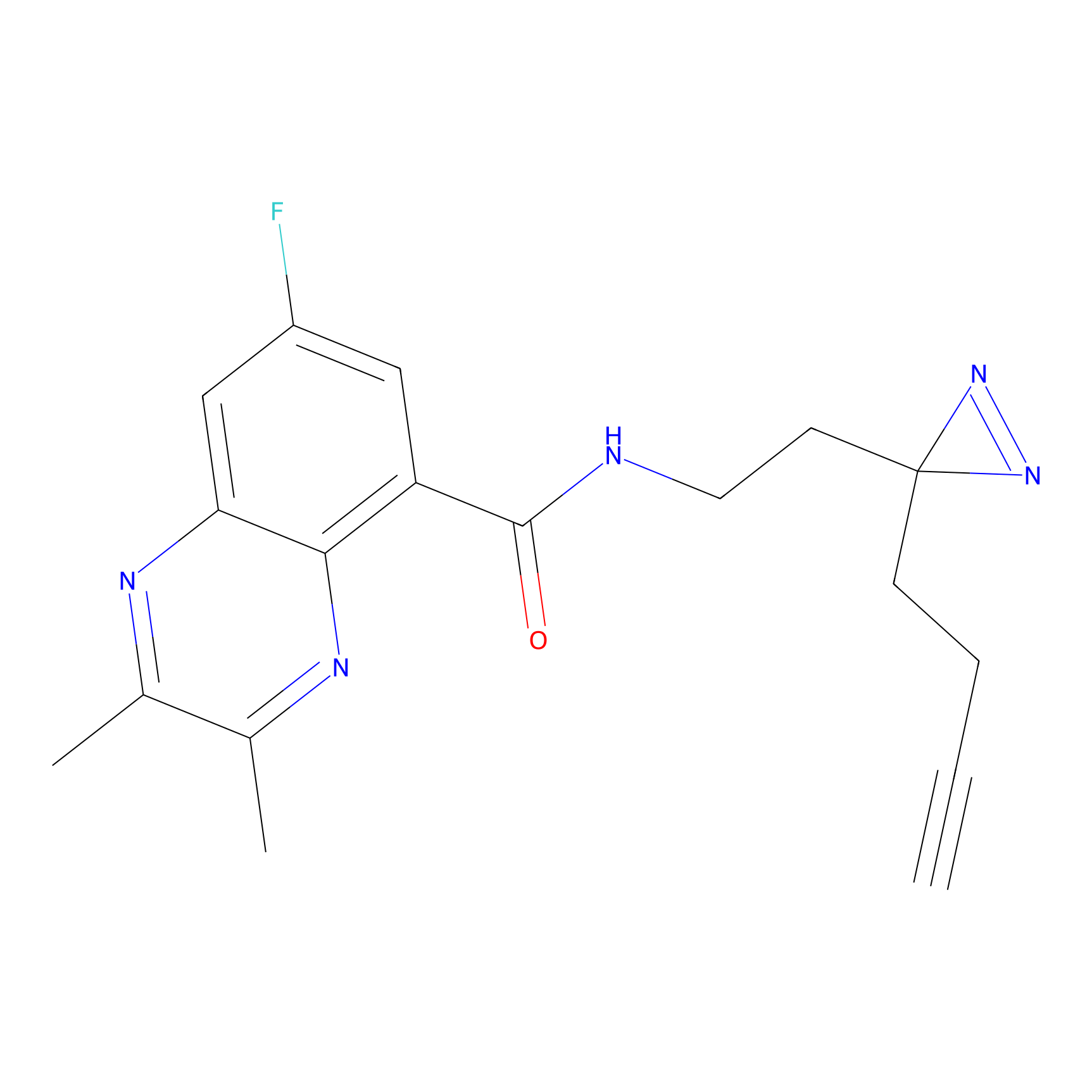 |
14.42 | LDD1922 | [14] | |
|
C269 Probe Info |
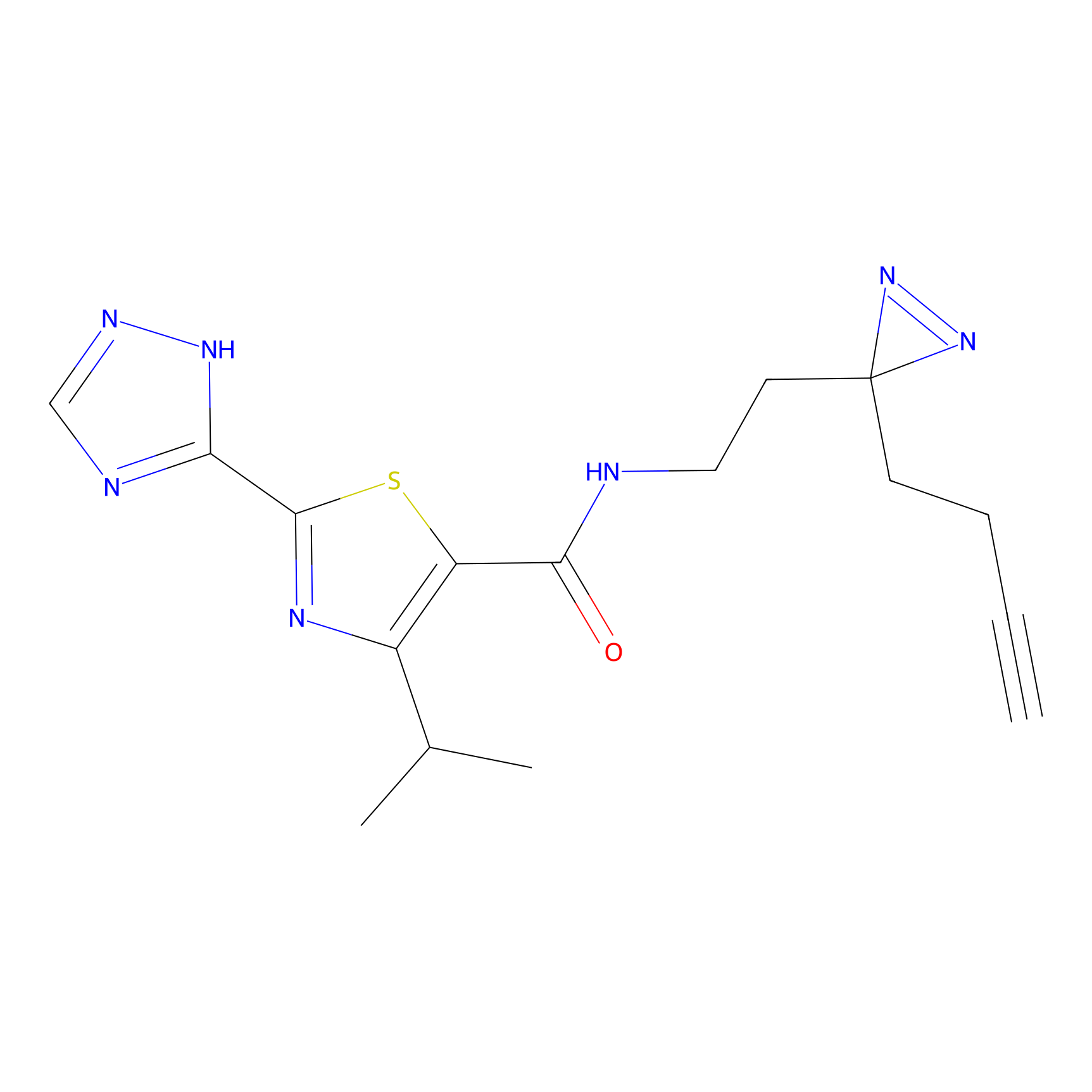 |
5.66 | LDD1939 | [14] | |
|
C284 Probe Info |
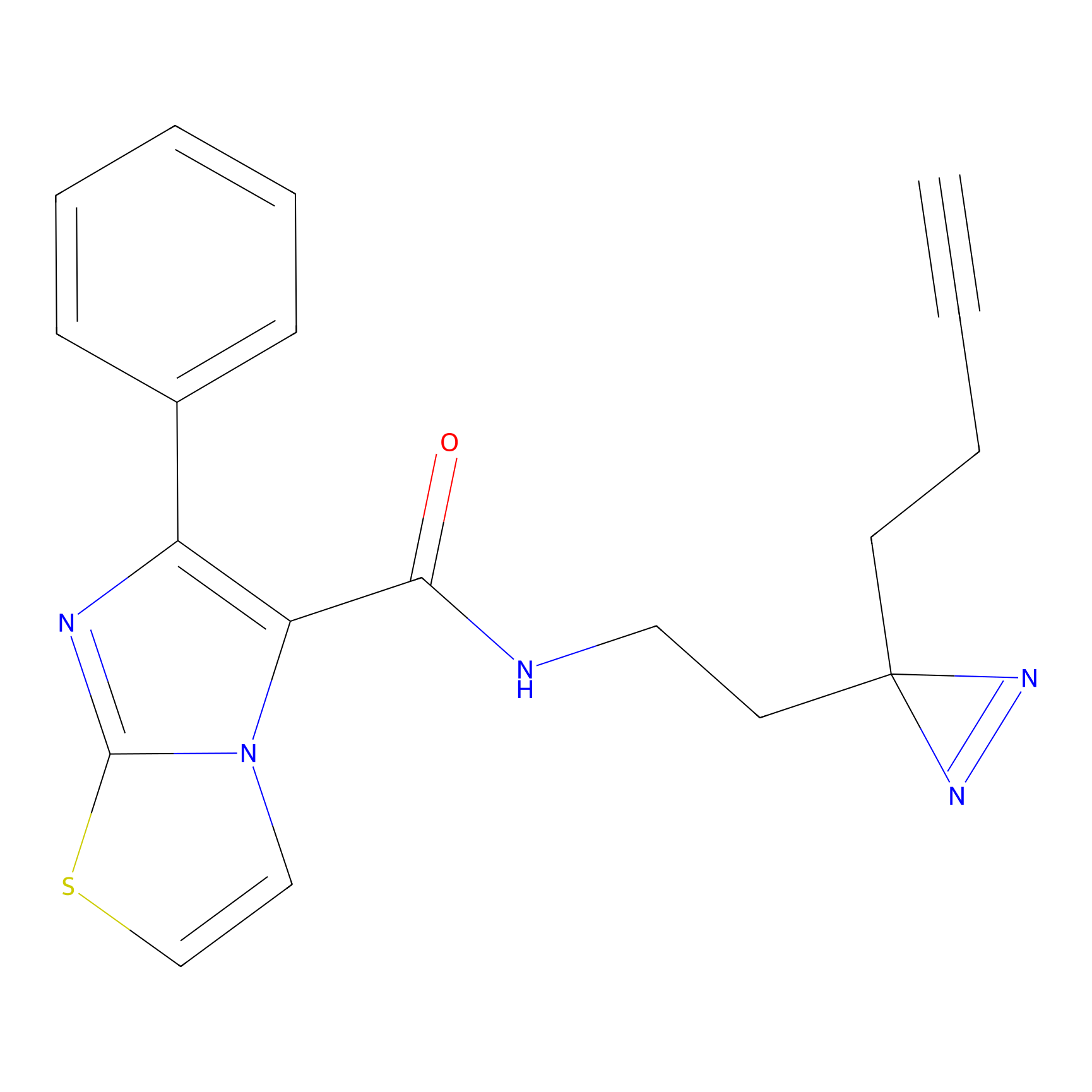 |
26.17 | LDD1954 | [14] | |
|
C305 Probe Info |
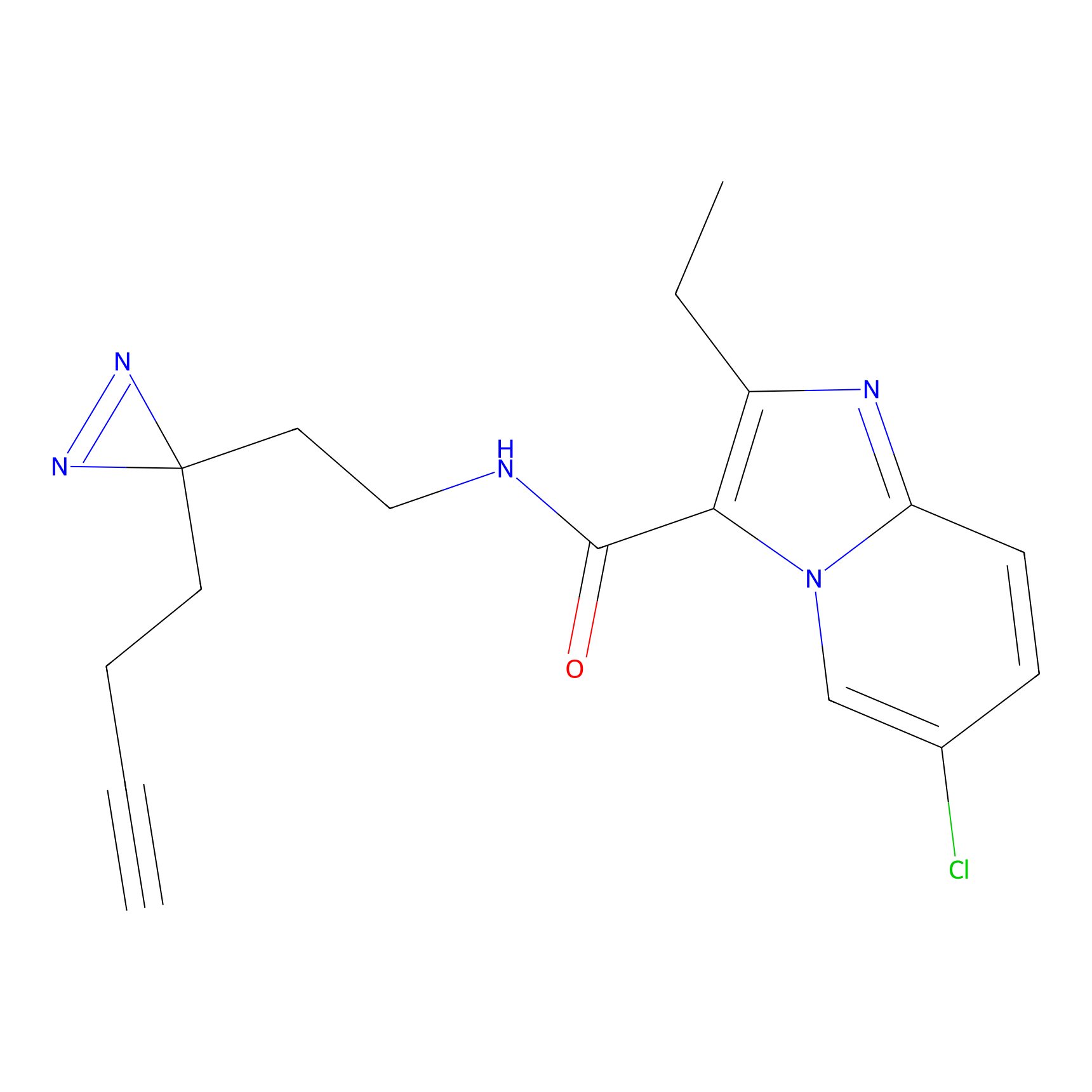 |
10.48 | LDD1974 | [14] | |
|
C313 Probe Info |
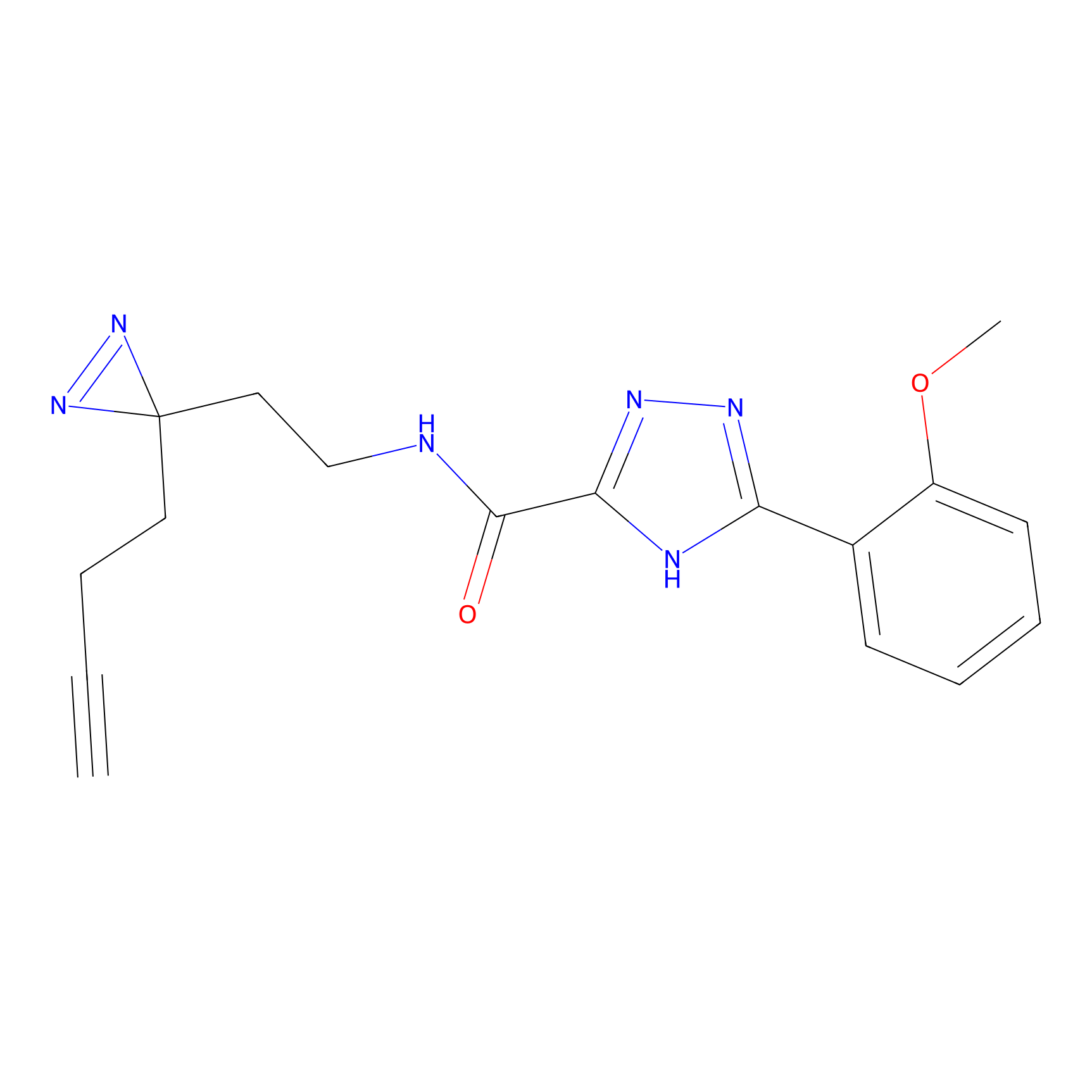 |
14.03 | LDD1980 | [14] | |
|
C354 Probe Info |
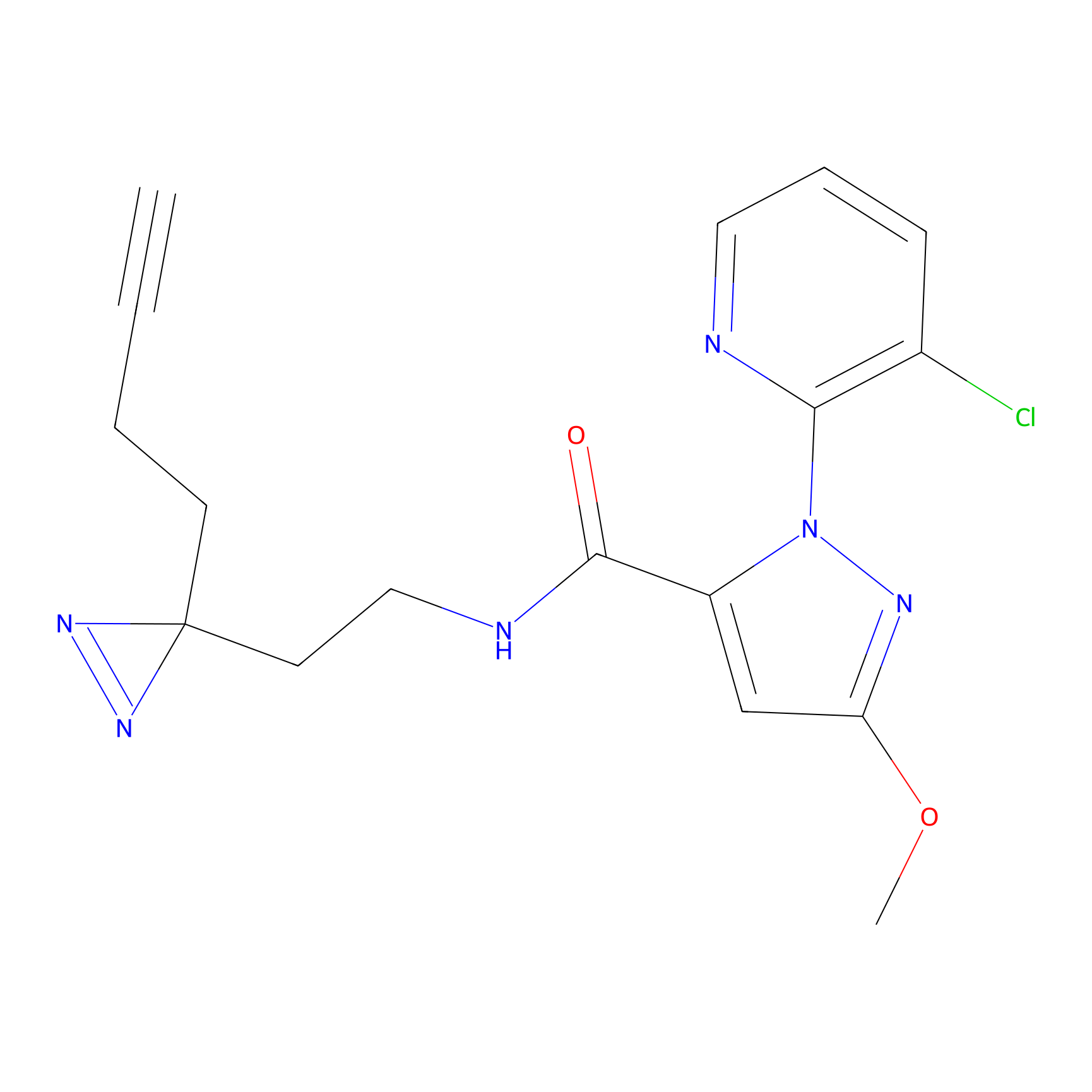 |
7.01 | LDD2015 | [14] | |
|
C391 Probe Info |
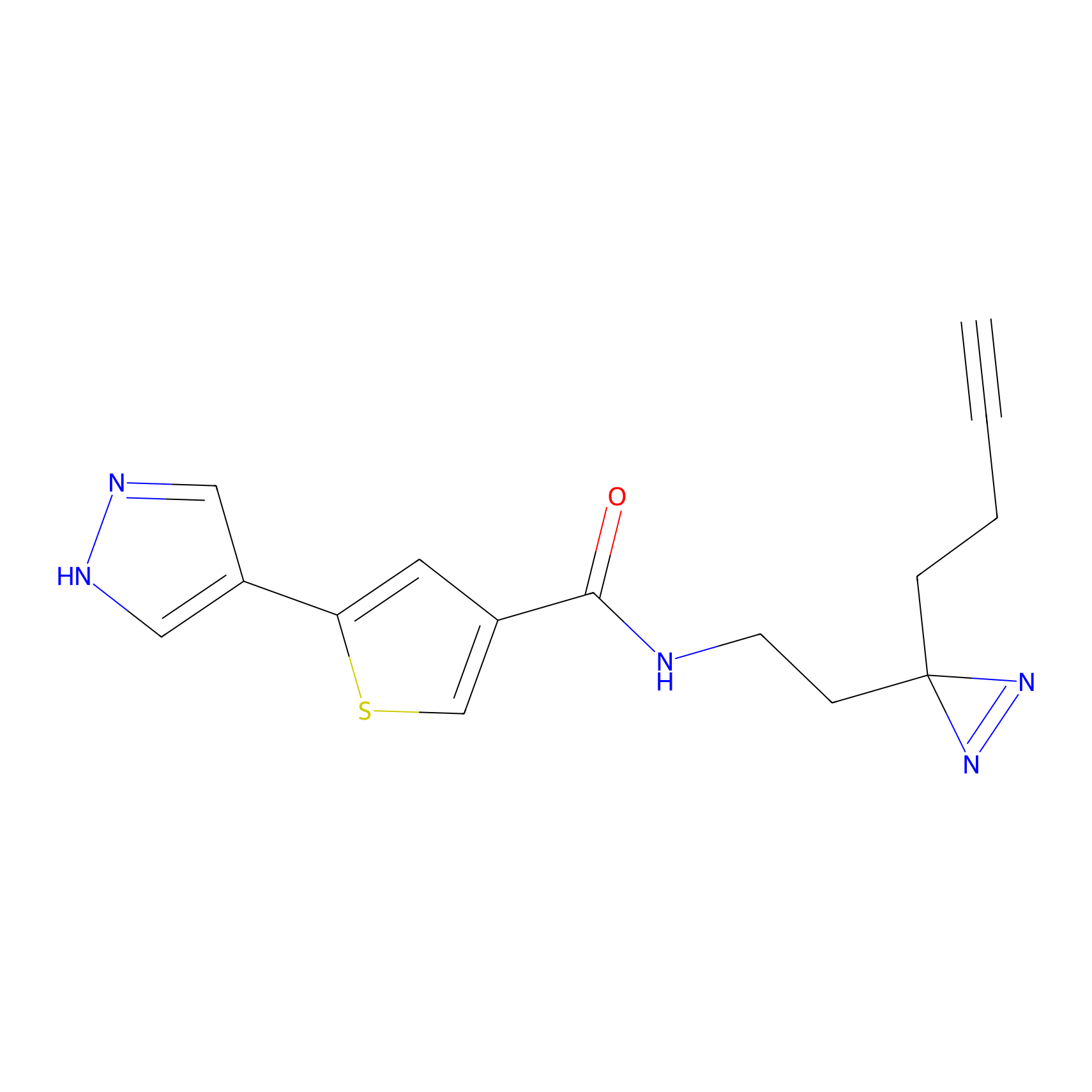 |
12.47 | LDD2050 | [14] | |
|
C403 Probe Info |
 |
14.03 | LDD2061 | [14] | |
|
C407 Probe Info |
 |
11.08 | LDD2064 | [14] | |
|
FFF probe11 Probe Info |
 |
8.42 | LDD0472 | [15] | |
|
FFF probe13 Probe Info |
 |
20.00 | LDD0475 | [15] | |
|
FFF probe14 Probe Info |
 |
20.00 | LDD0477 | [15] | |
|
FFF probe2 Probe Info |
 |
19.19 | LDD0463 | [15] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0156 | Aniline | NCI-H1299 | 13.88 | LDD0403 | [1] |
| LDCM0031 | Epigallocatechin gallate | HEK-293T | 4.71 | LDD0183 | [6] |
| LDCM0175 | Ethacrynic acid | HeLa | N.A. | LDD0440 | [7] |
| LDCM0503 | Nucleophilic fragment 14b | MDA-MB-231 | C70(0.08) | LDD2096 | [5] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C70(0.09) | LDD2106 | [5] |
| LDCM0523 | Nucleophilic fragment 24b | MDA-MB-231 | C70(0.19) | LDD2116 | [5] |
| LDCM0525 | Nucleophilic fragment 25b | MDA-MB-231 | C70(0.15) | LDD2118 | [5] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C70(1.00) | LDD2120 | [5] |
| LDCM0529 | Nucleophilic fragment 27b | MDA-MB-231 | C70(0.14) | LDD2122 | [5] |
| LDCM0531 | Nucleophilic fragment 28b | MDA-MB-231 | C70(0.10) | LDD2124 | [5] |
| LDCM0533 | Nucleophilic fragment 29b | MDA-MB-231 | C70(0.11) | LDD2126 | [5] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C70(0.33) | LDD2128 | [5] |
| LDCM0555 | Nucleophilic fragment 7b | MDA-MB-231 | C70(0.14) | LDD2149 | [5] |
| LDCM0557 | Nucleophilic fragment 8b | MDA-MB-231 | C70(0.12) | LDD2151 | [5] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Other
References
