Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C-Sul Probe Info |
 |
4.48 | LDD0066 | [1] | |
|
TH211 Probe Info |
 |
Y119(20.00); Y73(9.58); Y101(6.12) | LDD0257 | [2] | |
|
TH214 Probe Info |
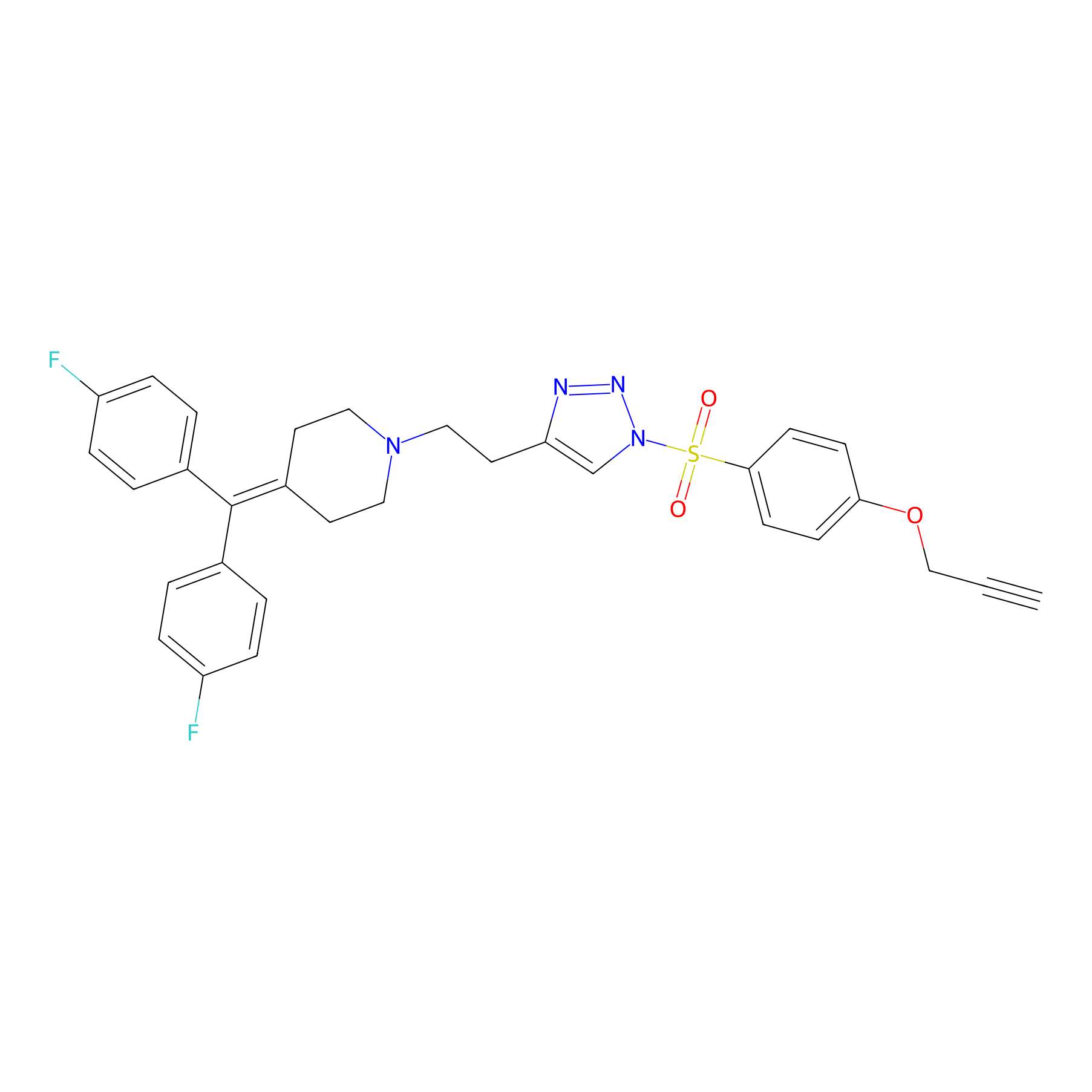 |
Y73(10.64) | LDD0258 | [2] | |
|
TH216 Probe Info |
 |
Y119(20.00); Y73(9.54); Y101(6.59) | LDD0259 | [2] | |
|
ONAyne Probe Info |
 |
K81(10.00) | LDD0274 | [3] | |
|
STPyne Probe Info |
 |
K103(5.43); K122(10.00); K51(7.82); K81(2.63) | LDD0277 | [3] | |
|
AZ-9 Probe Info |
 |
D77(10.00); E71(10.00) | LDD2209 | [4] | |
|
OPA-S-S-alkyne Probe Info |
 |
K81(4.93) | LDD3494 | [5] | |
|
Probe 1 Probe Info |
 |
Y73(30.52); Y119(120.75) | LDD3495 | [6] | |
|
HHS-482 Probe Info |
 |
Y101(0.75); Y119(1.19); Y73(0.96); Y99(0.90) | LDD0285 | [7] | |
|
HHS-475 Probe Info |
 |
Y99(0.97); Y101(1.03); Y73(1.05); Y119(2.76) | LDD0264 | [8] | |
|
HHS-465 Probe Info |
 |
Y119(10.00); Y73(10.00) | LDD2237 | [9] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0221 | [10] | |
|
AMP probe Probe Info |
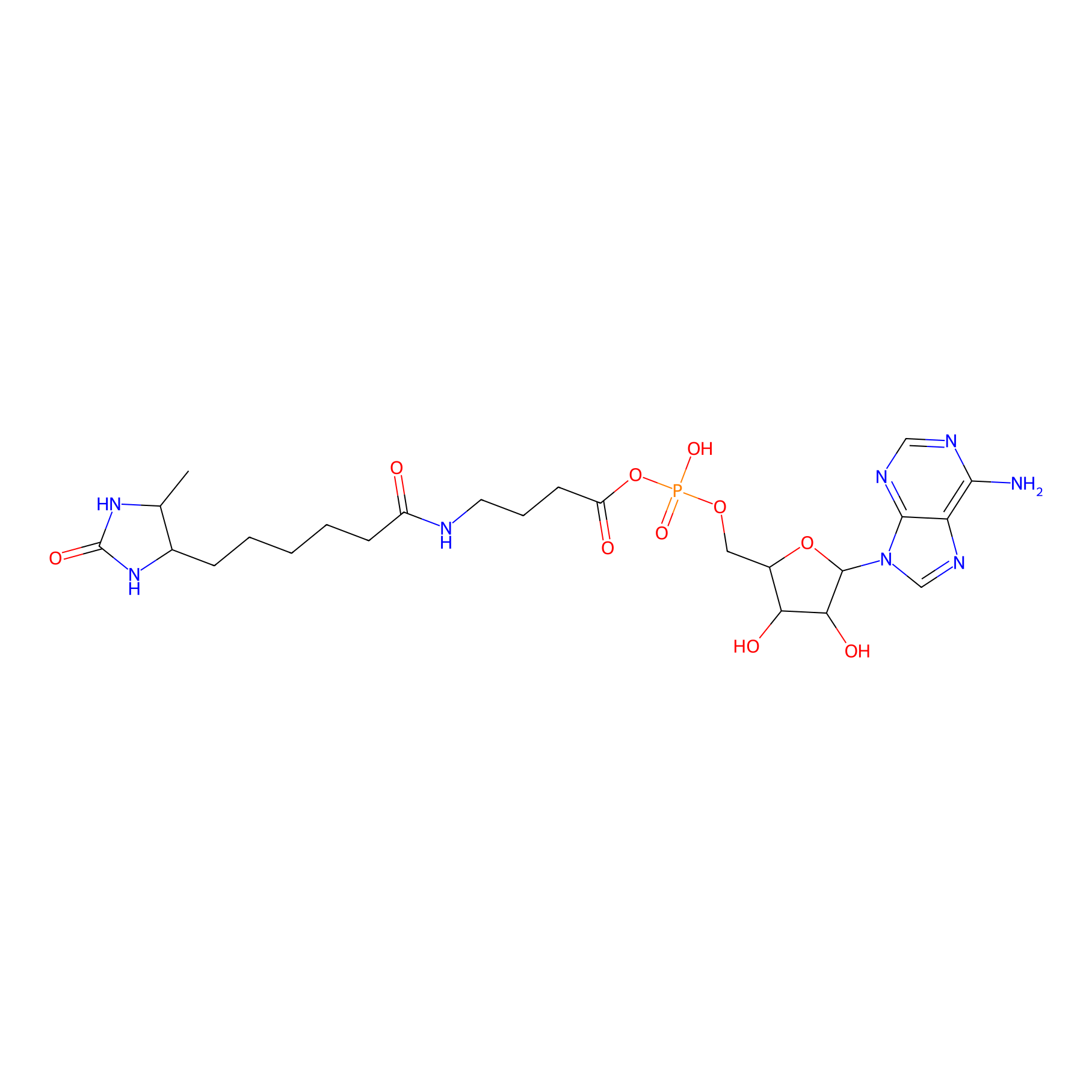 |
N.A. | LDD0200 | [11] | |
|
ATP probe Probe Info |
 |
K122(0.00); K81(0.00); K103(0.00); K104(0.00) | LDD0199 | [11] | |
|
Alkyne tyramide Probe Info |
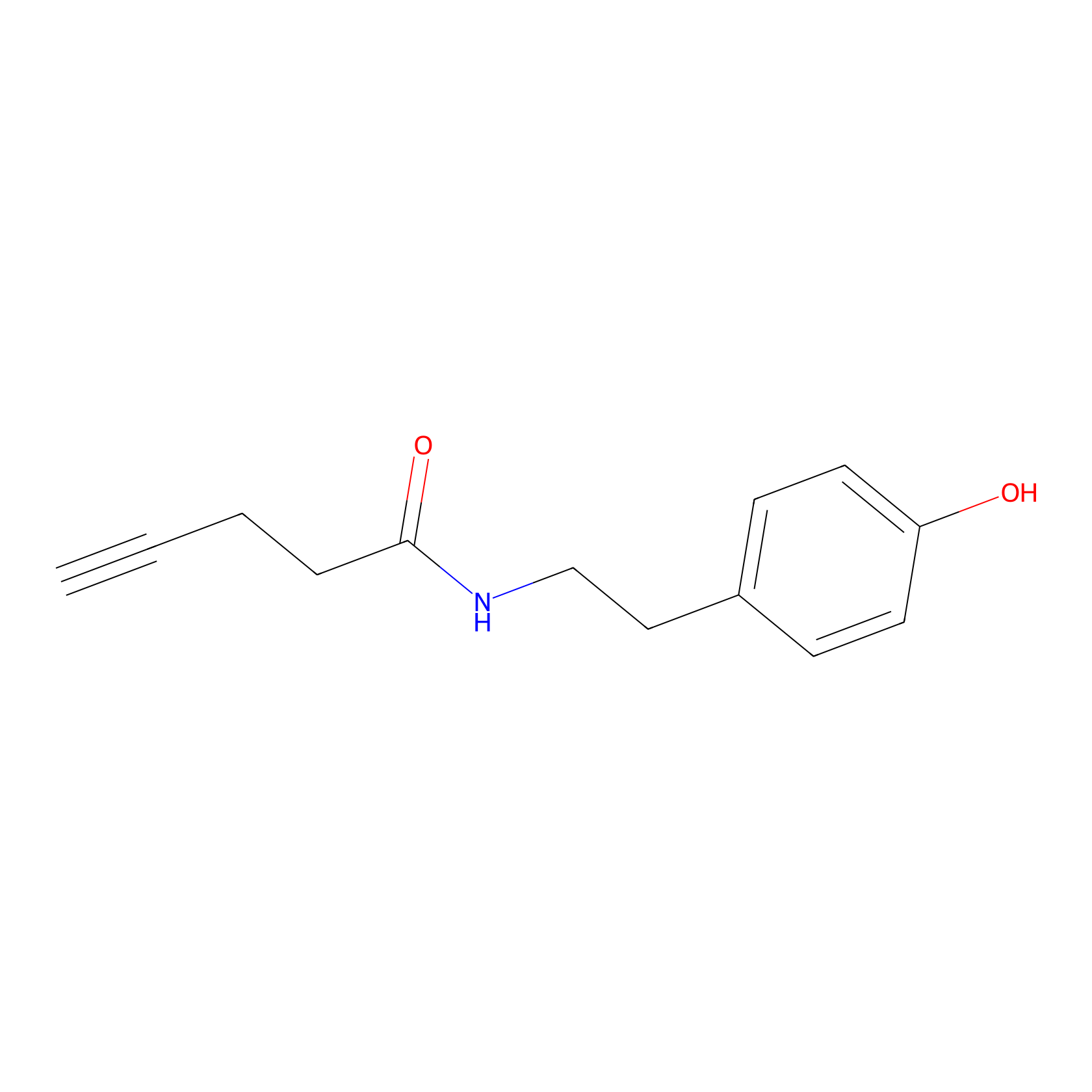 |
Y119(0.00); Y73(0.00) | LDD0003 | [12] | |
|
1d-yne Probe Info |
 |
N.A. | LDD0356 | [13] | |
|
NHS Probe Info |
 |
K122(0.00); K81(0.00) | LDD0010 | [12] | |
|
OSF Probe Info |
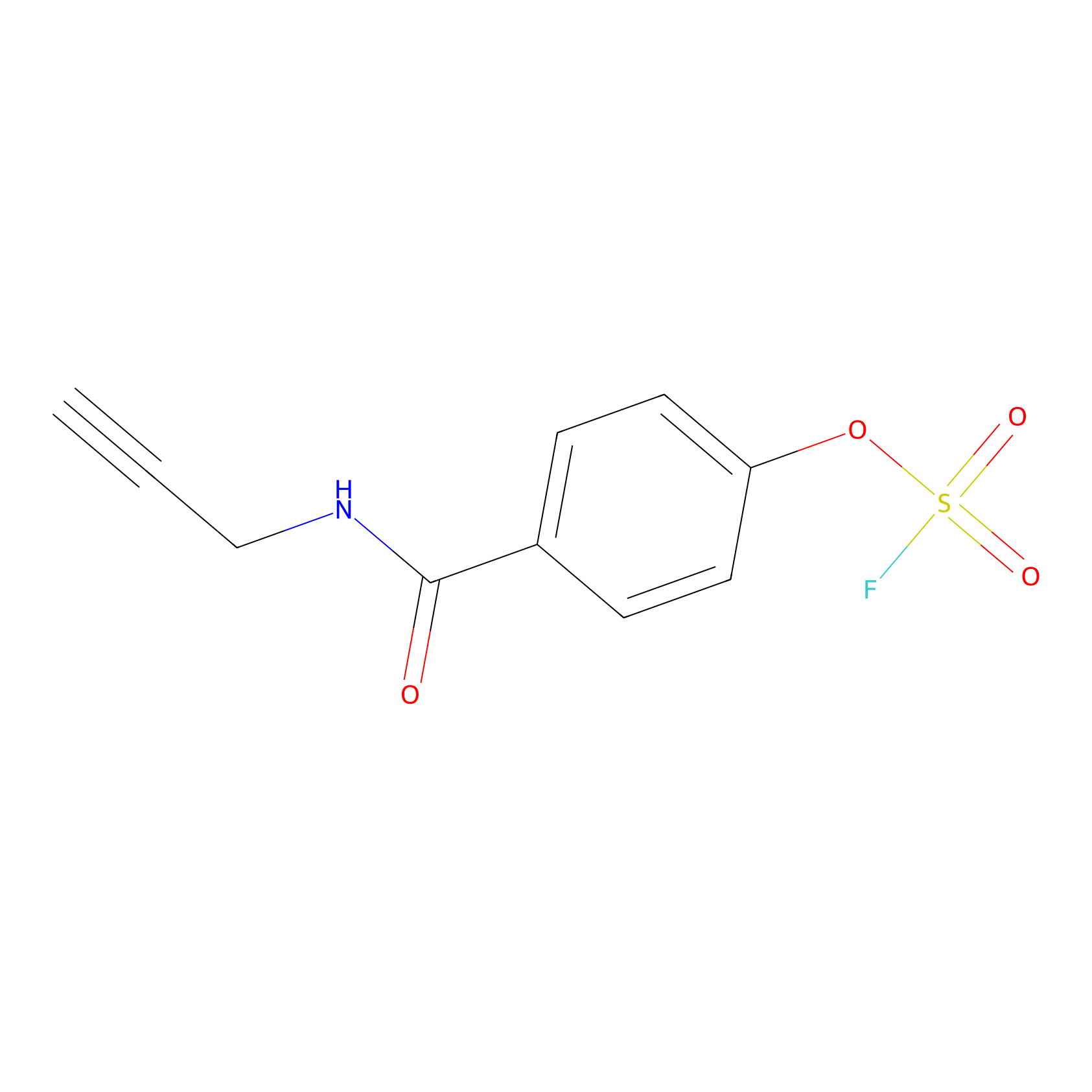 |
N.A. | LDD0029 | [14] | |
|
SF Probe Info |
 |
Y73(0.00); Y101(0.00); Y99(0.00) | LDD0028 | [14] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [15] | |
|
1c-yne Probe Info |
 |
K113(0.00); K51(0.00) | LDD0228 | [13] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C087 Probe Info |
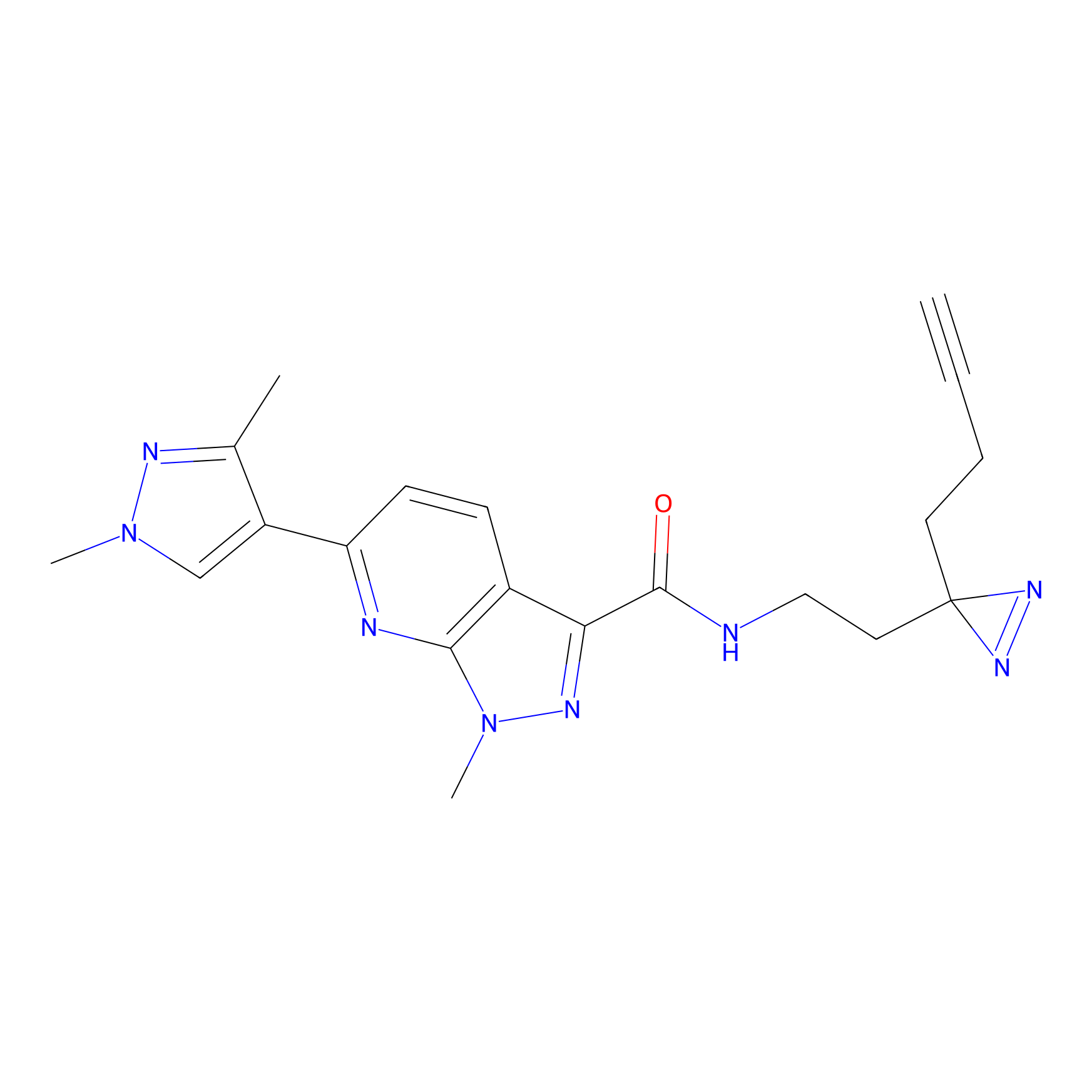 |
8.94 | LDD1779 | [16] | |
|
C210 Probe Info |
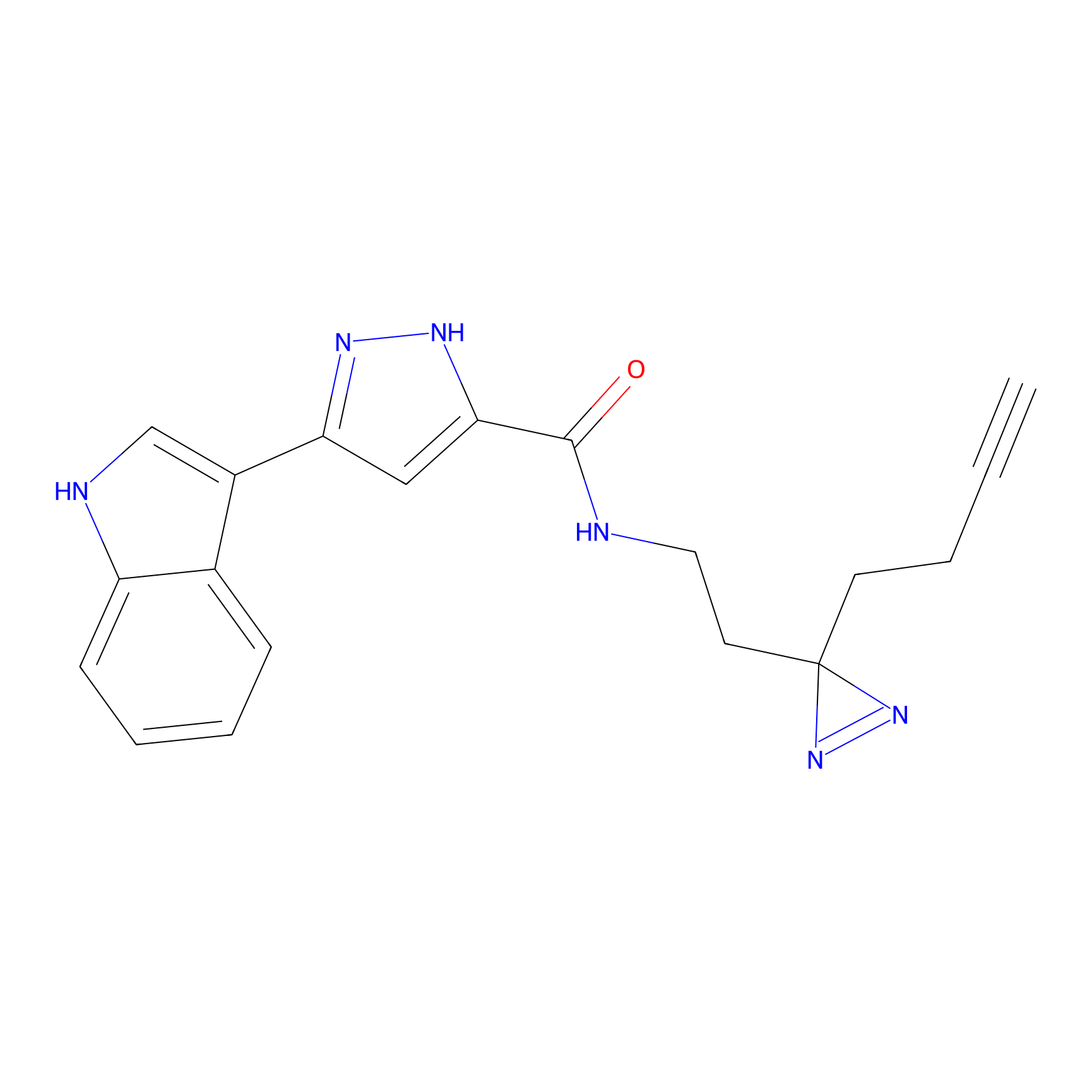 |
61.39 | LDD1884 | [16] | |
|
C338 Probe Info |
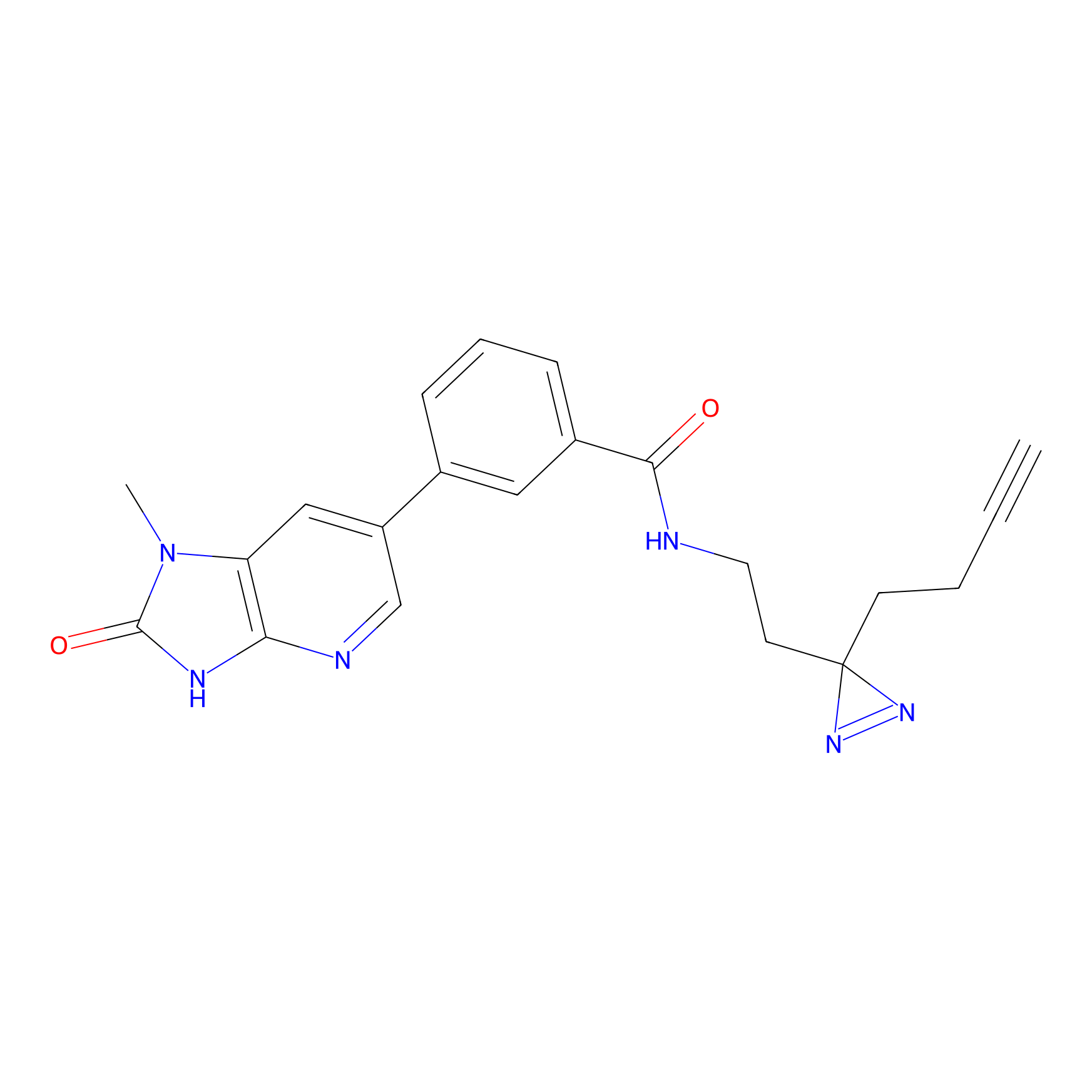 |
11.39 | LDD2001 | [16] | |
|
C355 Probe Info |
 |
22.63 | LDD2016 | [16] | |
|
FFF probe13 Probe Info |
 |
15.12 | LDD0475 | [17] | |
|
FFF probe2 Probe Info |
 |
9.65 | LDD0463 | [17] | |
|
FFF probe3 Probe Info |
 |
8.21 | LDD0464 | [17] | |
|
JN0003 Probe Info |
 |
13.37 | LDD0469 | [17] | |
|
BD-F Probe Info |
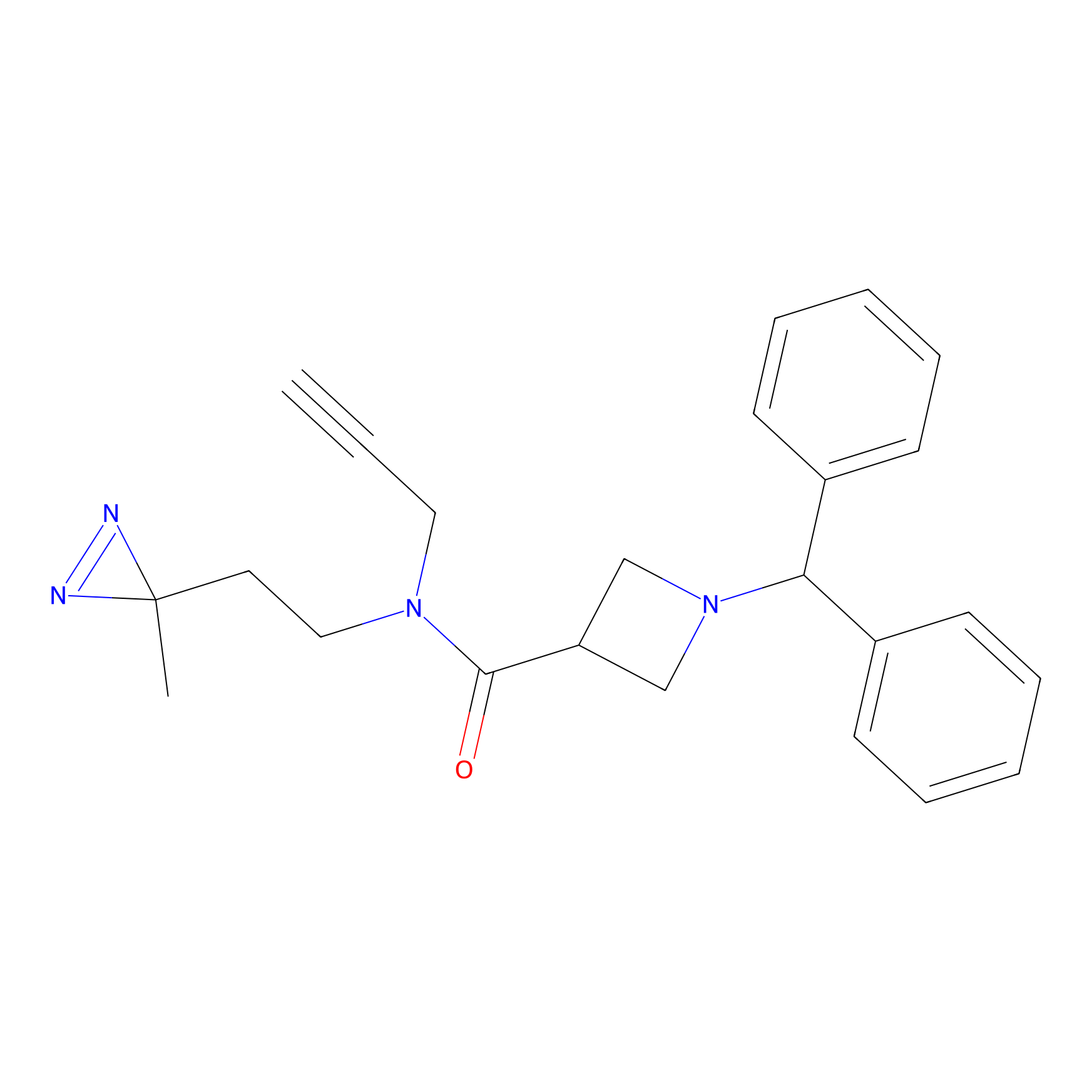 |
N.A. | LDD0024 | [18] | |
|
OEA-DA Probe Info |
 |
5.17 | LDD0046 | [19] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [10] |
| LDCM0116 | HHS-0101 | DM93 | Y99(0.97); Y101(1.03); Y73(1.05); Y119(2.76) | LDD0264 | [8] |
| LDCM0117 | HHS-0201 | DM93 | Y73(0.96); Y99(1.06); Y101(1.98); Y119(2.91) | LDD0265 | [8] |
| LDCM0118 | HHS-0301 | DM93 | Y73(0.71); Y99(1.66); Y101(2.01); Y119(3.34) | LDD0266 | [8] |
| LDCM0119 | HHS-0401 | DM93 | Y73(1.02); Y99(1.56); Y101(1.62); Y119(4.22) | LDD0267 | [8] |
| LDCM0120 | HHS-0701 | DM93 | Y73(1.32); Y99(1.47); Y119(5.36); Y101(6.75) | LDD0268 | [8] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [10] |
| LDCM0123 | JWB131 | DM93 | Y101(0.75); Y119(1.19); Y73(0.96); Y99(0.90) | LDD0285 | [7] |
| LDCM0124 | JWB142 | DM93 | Y101(0.99); Y119(1.63); Y73(0.99); Y99(0.88) | LDD0286 | [7] |
| LDCM0125 | JWB146 | DM93 | Y101(0.64); Y119(1.21); Y73(1.11); Y99(0.97) | LDD0287 | [7] |
| LDCM0126 | JWB150 | DM93 | Y101(2.41); Y119(4.01); Y73(3.81); Y99(2.70) | LDD0288 | [7] |
| LDCM0127 | JWB152 | DM93 | Y101(1.19); Y119(3.41); Y73(1.87); Y99(1.98) | LDD0289 | [7] |
| LDCM0128 | JWB198 | DM93 | Y101(1.23); Y119(0.91); Y73(1.26); Y99(1.18) | LDD0290 | [7] |
| LDCM0129 | JWB202 | DM93 | Y101(0.82); Y119(1.10); Y73(0.73); Y99(0.58) | LDD0291 | [7] |
| LDCM0130 | JWB211 | DM93 | Y101(0.73); Y119(1.28); Y73(1.03); Y99(0.90) | LDD0292 | [7] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [10] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| OTU domain-containing protein 6A (OTUD6A) | . | Q7L8S5 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Chorion-specific transcription factor GCMb (GCM2) | . | O75603 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Galectin-7 (LGALS7; LGALS7B) | . | P47929 | |||
| Single-stranded DNA-binding protein, mitochondrial (SSBP1) | . | Q04837 | |||
References
