Details of the Target
General Information of Target
| Target ID | LDTP03811 | |||||
|---|---|---|---|---|---|---|
| Target Name | V-type proton ATPase subunit E 1 (ATP6V1E1) | |||||
| Gene Name | ATP6V1E1 | |||||
| Gene ID | 529 | |||||
| Synonyms |
ATP6E; ATP6E2; V-type proton ATPase subunit E 1; V-ATPase subunit E 1; V-ATPase 31 kDa subunit; p31; Vacuolar proton pump subunit E 1 |
|||||
| 3D Structure | ||||||
| Sequence |
MALSDADVQKQIKHMMAFIEQEANEKAEEIDAKAEEEFNIEKGRLVQTQRLKIMEYYEKK
EKQIEQQKKIQMSNLMNQARLKVLRARDDLITDLLNEAKQRLSKVVKDTTRYQVLLDGLV LQGLYQLLEPRMIVRCRKQDFPLVKAAVQKAIPMYKIATKNDVDVQIDQESYLPEDIAGG VEIYNGDRKIKVSNTLESRLDLIAQQMMPEVRGALFGANANRKFLD |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
V-ATPase E subunit family
|
|||||
| Subcellular location |
Apical cell membrane
|
|||||
| Function |
Subunit of the V1 complex of vacuolar(H+)-ATPase (V-ATPase), a multisubunit enzyme composed of a peripheral complex (V1) that hydrolyzes ATP and a membrane integral complex (V0) that translocates protons. V-ATPase is responsible for acidifying and maintaining the pH of intracellular compartments and in some cell types, is targeted to the plasma membrane, where it is responsible for acidifying the extracellular environment.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K107(4.00); K138(10.00); K145(0.17); K156(10.00) | LDD0277 | [1] | |
|
Probe 1 Probe Info |
 |
Y56(43.43); Y57(34.57) | LDD3495 | [2] | |
|
m-APA Probe Info |
 |
9.48 | LDD0403 | [3] | |
|
HHS-482 Probe Info |
 |
Y56(0.77) | LDD0285 | [4] | |
|
HHS-475 Probe Info |
 |
Y184(0.36); Y56(0.85) | LDD0264 | [5] | |
|
HHS-465 Probe Info |
 |
Y56(10.00) | LDD2237 | [6] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [7] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [8] | |
|
SF Probe Info |
 |
Y57(0.00); Y56(0.00) | LDD0028 | [9] | |
|
1c-yne Probe Info |
 |
K191(0.00); K10(0.00) | LDD0228 | [10] | |
|
MPP-AC Probe Info |
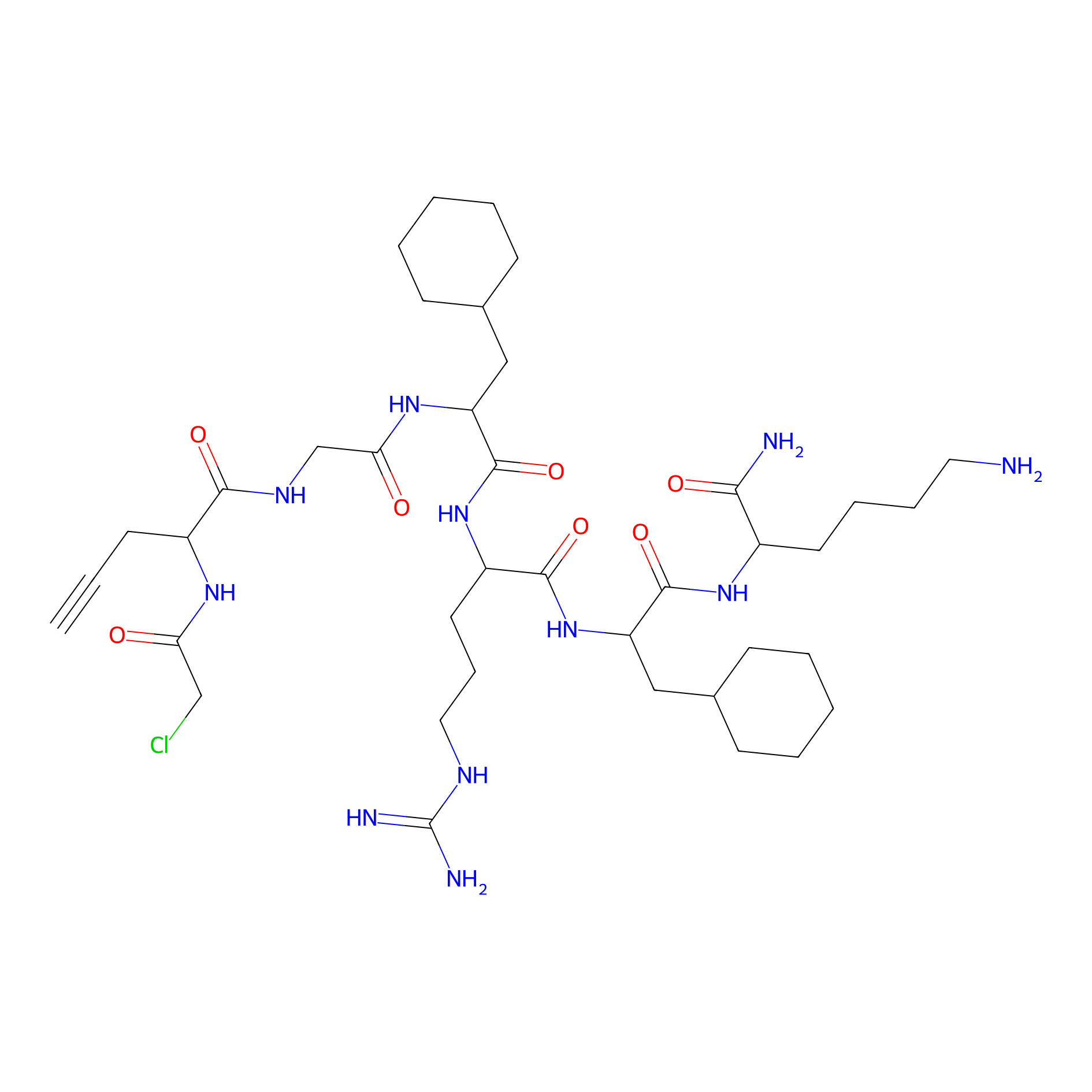 |
N.A. | LDD0428 | [11] | |
|
TER-AC Probe Info |
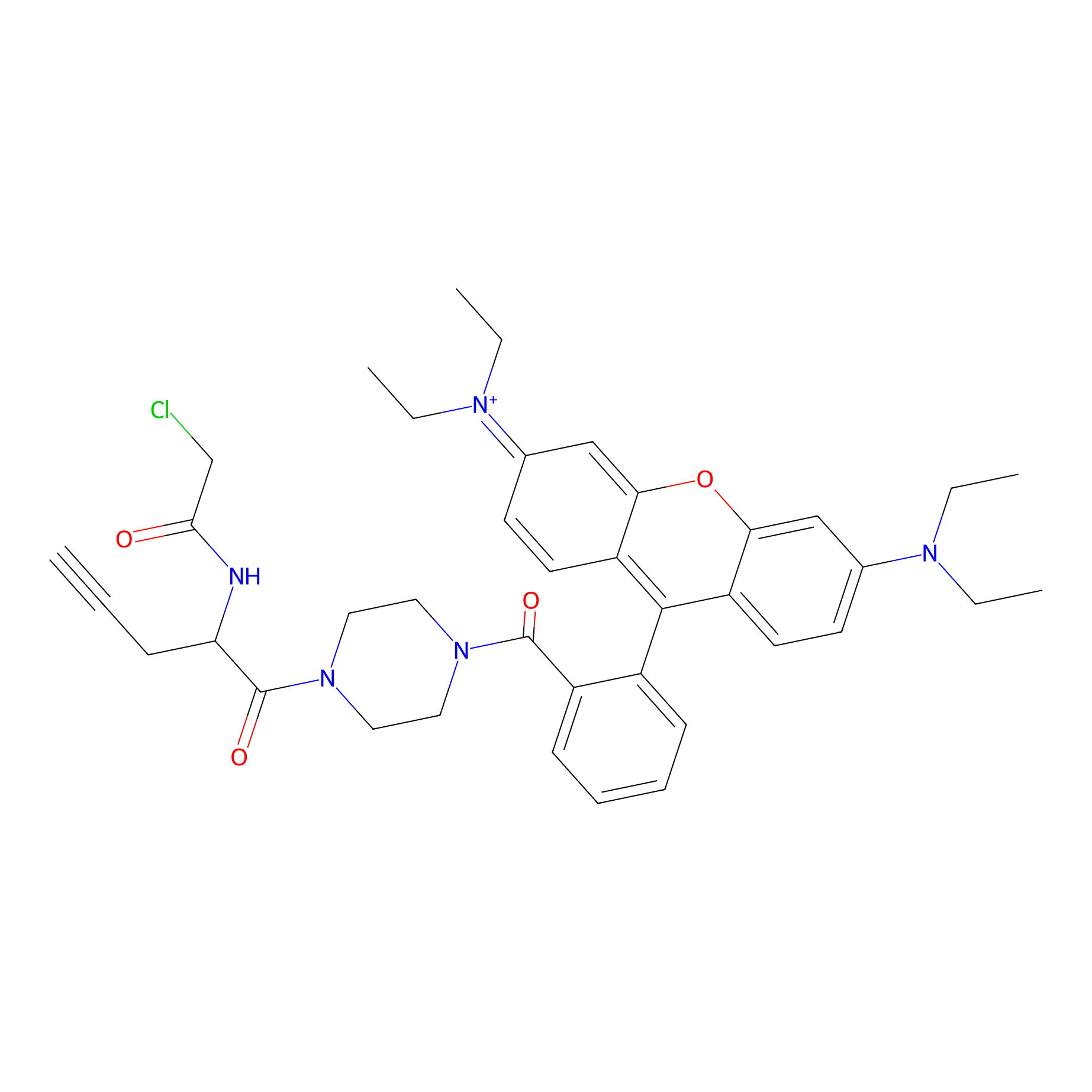 |
N.A. | LDD0426 | [11] | |
|
TPP-AC Probe Info |
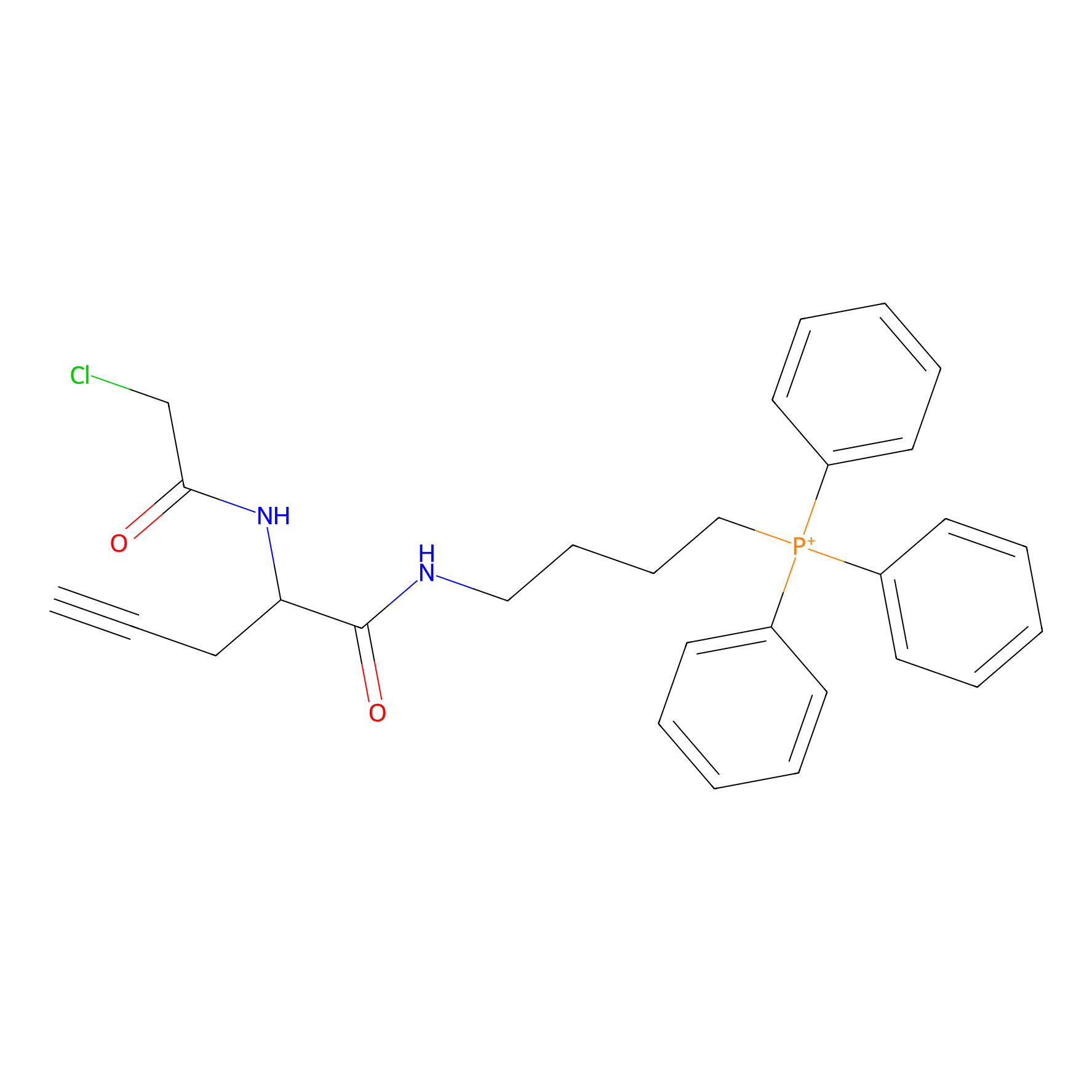 |
N.A. | LDD0427 | [11] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C040 Probe Info |
 |
9.51 | LDD1740 | [12] | |
|
C210 Probe Info |
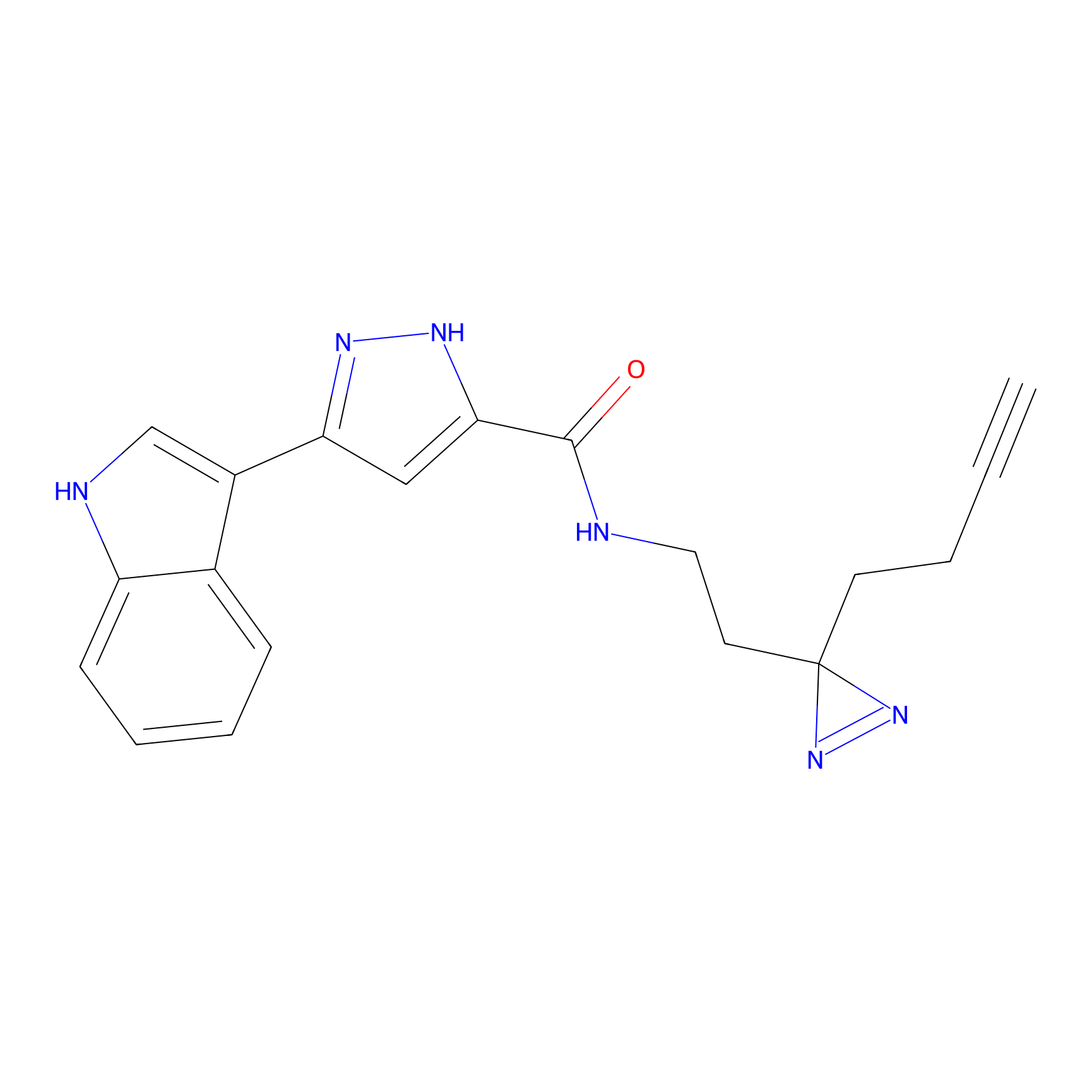 |
62.68 | LDD1884 | [12] | |
|
C244 Probe Info |
 |
17.75 | LDD1917 | [12] | |
|
C310 Probe Info |
 |
7.89 | LDD1977 | [12] | |
|
FFF probe13 Probe Info |
 |
6.54 | LDD0476 | [13] | |
|
FFF probe3 Probe Info |
 |
17.15 | LDD0464 | [13] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [14] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0156 | Aniline | NCI-H1299 | 9.48 | LDD0403 | [3] |
| LDCM0116 | HHS-0101 | DM93 | Y184(0.36); Y56(0.85) | LDD0264 | [5] |
| LDCM0117 | HHS-0201 | DM93 | Y184(0.63); Y56(0.76) | LDD0265 | [5] |
| LDCM0118 | HHS-0301 | DM93 | Y184(0.61); Y56(0.87) | LDD0266 | [5] |
| LDCM0119 | HHS-0401 | DM93 | Y184(0.63); Y56(0.79) | LDD0267 | [5] |
| LDCM0120 | HHS-0701 | DM93 | Y184(0.73); Y56(0.93) | LDD0268 | [5] |
| LDCM0123 | JWB131 | DM93 | Y56(0.77) | LDD0285 | [4] |
| LDCM0124 | JWB142 | DM93 | Y56(0.87) | LDD0286 | [4] |
| LDCM0125 | JWB146 | DM93 | Y56(0.81) | LDD0287 | [4] |
| LDCM0126 | JWB150 | DM93 | Y56(2.22) | LDD0288 | [4] |
| LDCM0127 | JWB152 | DM93 | Y56(1.77) | LDD0289 | [4] |
| LDCM0128 | JWB198 | DM93 | Y56(1.18) | LDD0290 | [4] |
| LDCM0129 | JWB202 | DM93 | Y56(0.76) | LDD0291 | [4] |
| LDCM0130 | JWB211 | DM93 | Y56(1.02) | LDD0292 | [4] |
The Interaction Atlas With This Target
References
