Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
P1 Probe Info |
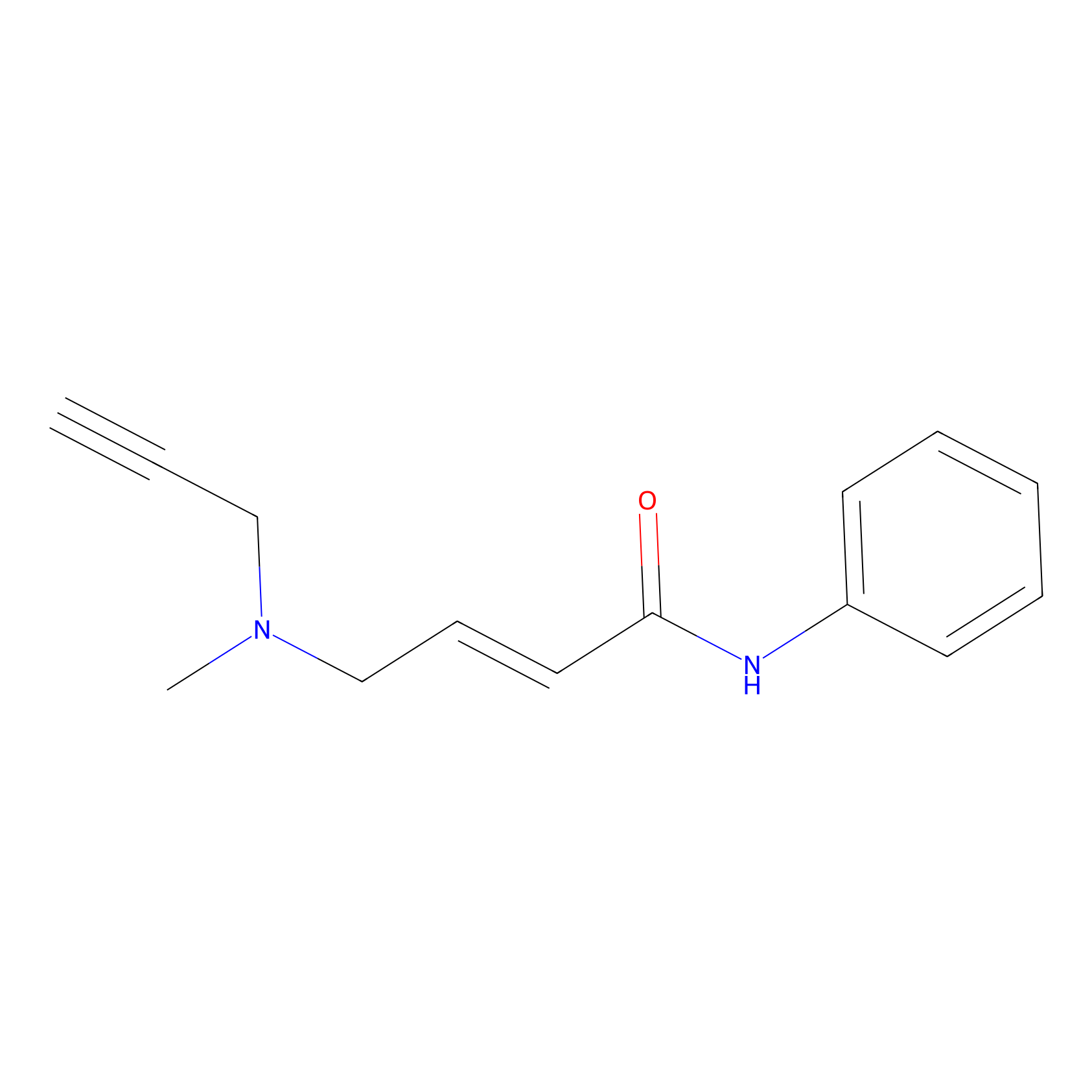 |
2.62 | LDD0448 | [1] | |
|
P2 Probe Info |
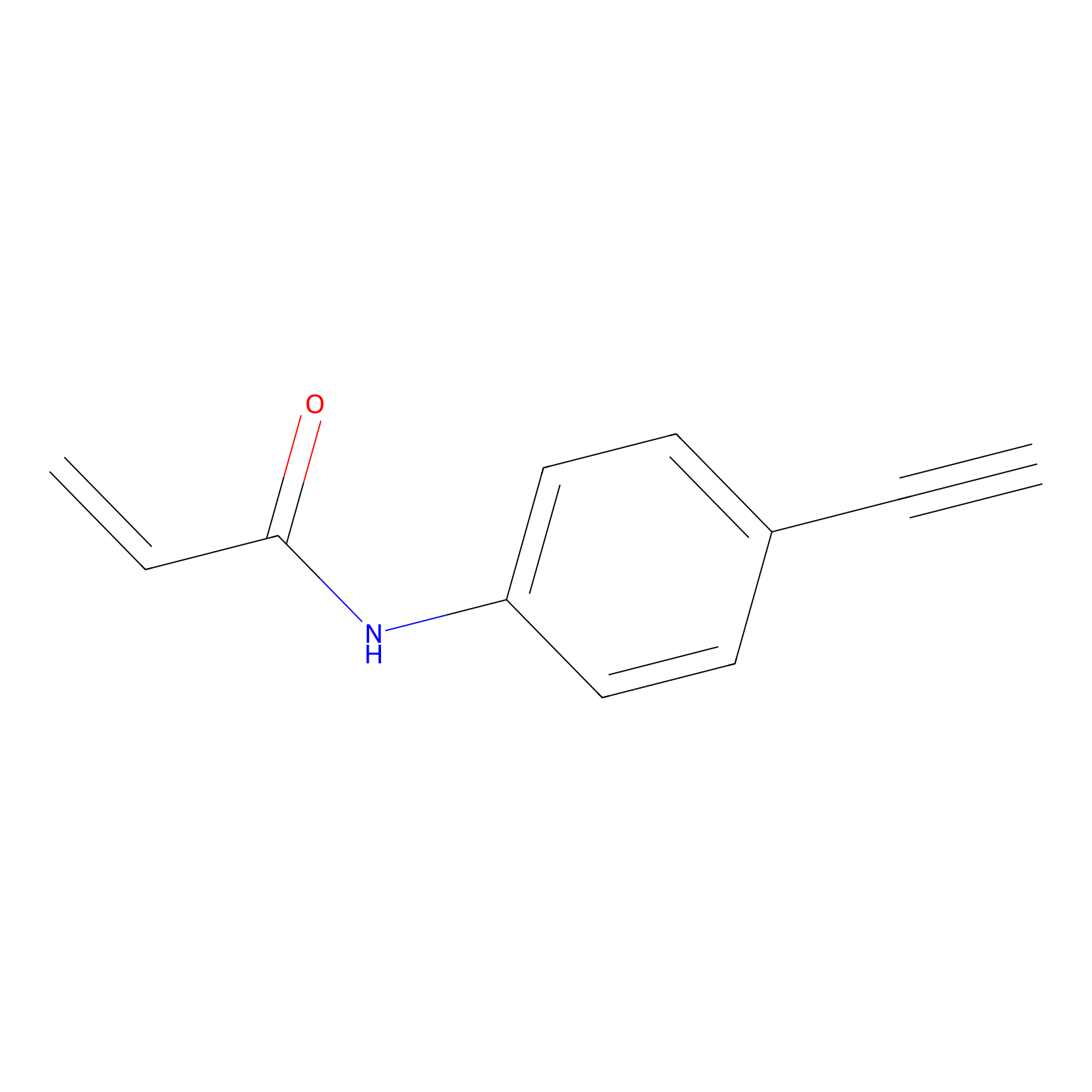 |
5.85 | LDD0449 | [1] | |
|
P3 Probe Info |
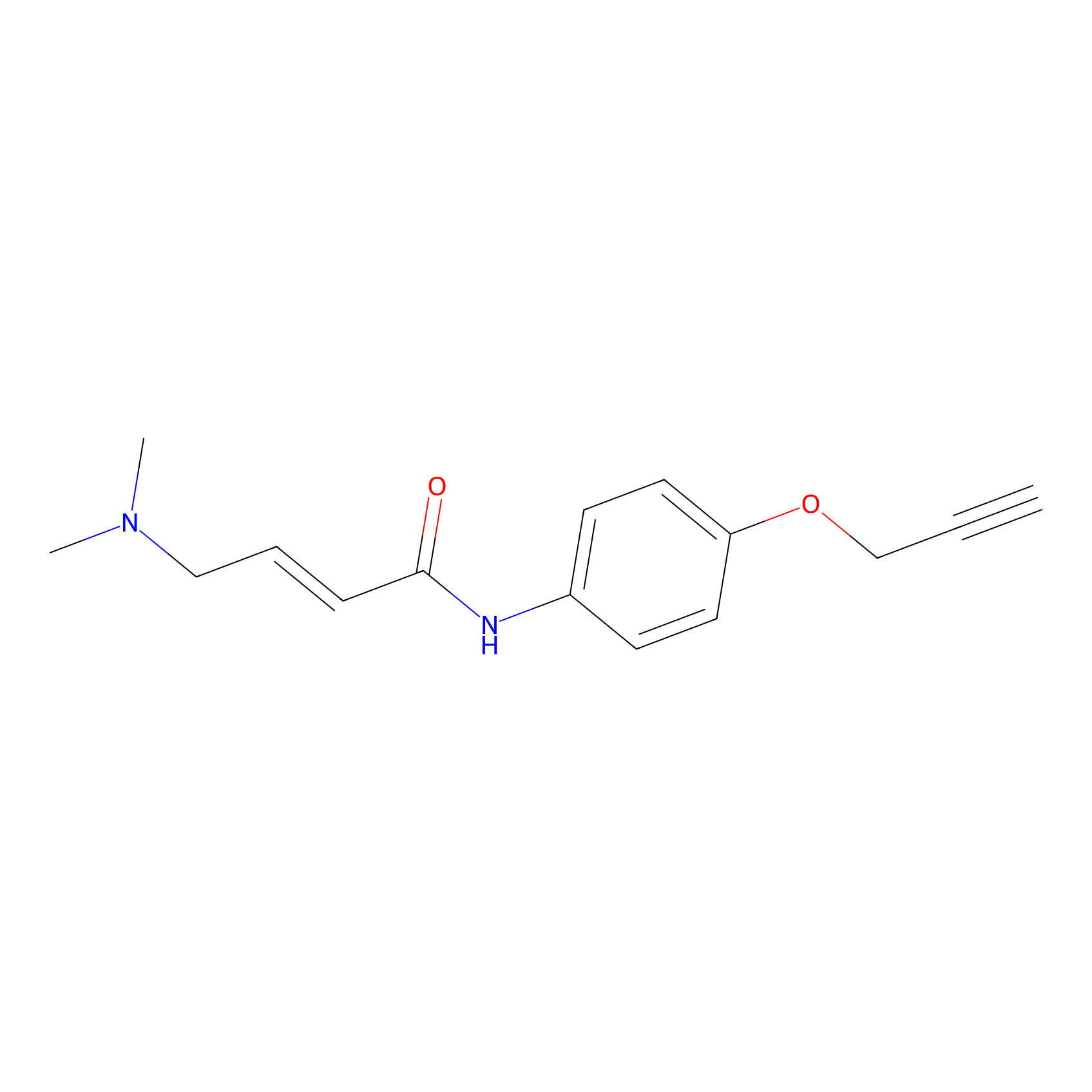 |
2.08 | LDD0450 | [1] | |
|
P8 Probe Info |
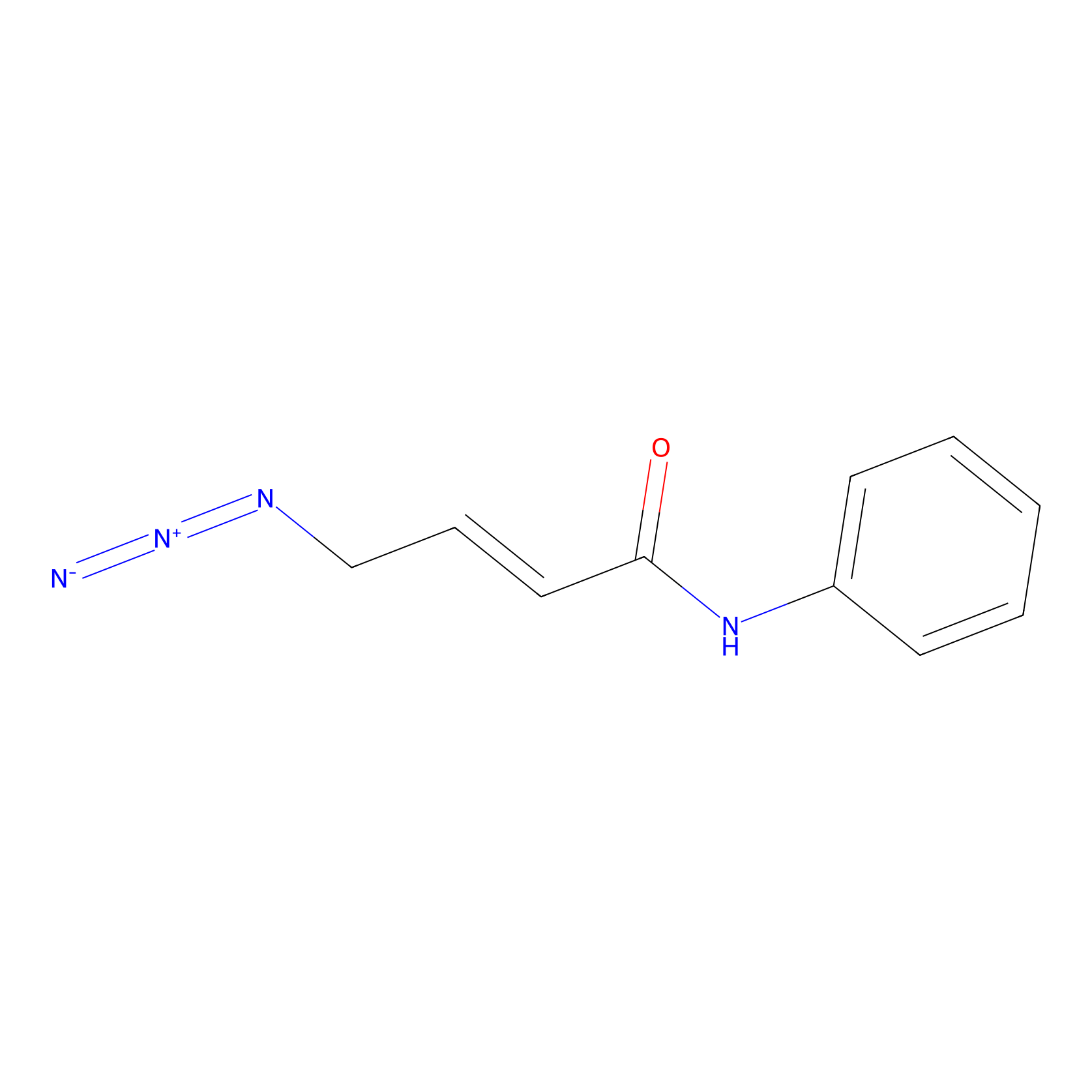 |
2.37 | LDD0451 | [1] | |
|
C-Sul Probe Info |
 |
5.39 | LDD0066 | [2] | |
|
AZ-9 Probe Info |
 |
D72(0.95); E42(10.00); E74(10.00) | LDD2208 | [3] | |
|
ONAyne Probe Info |
 |
N.A. | LDD0273 | [4] | |
|
AF-1 Probe Info |
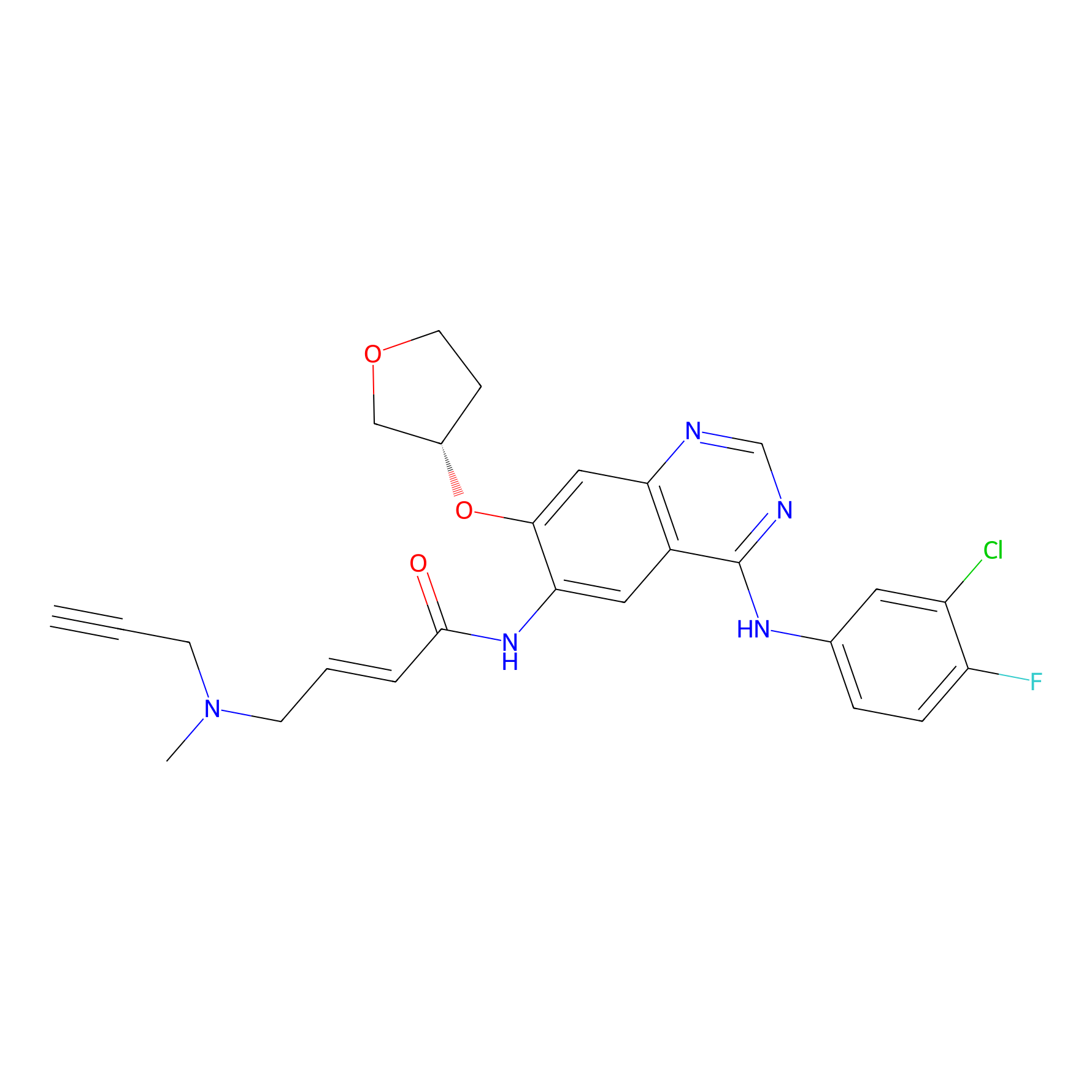 |
3.58 | LDD0421 | [5] | |
|
W1 Probe Info |
 |
R54(0.58) | LDD0237 | [6] | |
|
ATP probe Probe Info |
 |
K140(0.00); K148(0.00); K149(0.00); K168(0.00) | LDD0199 | [7] | |
|
aONE Probe Info |
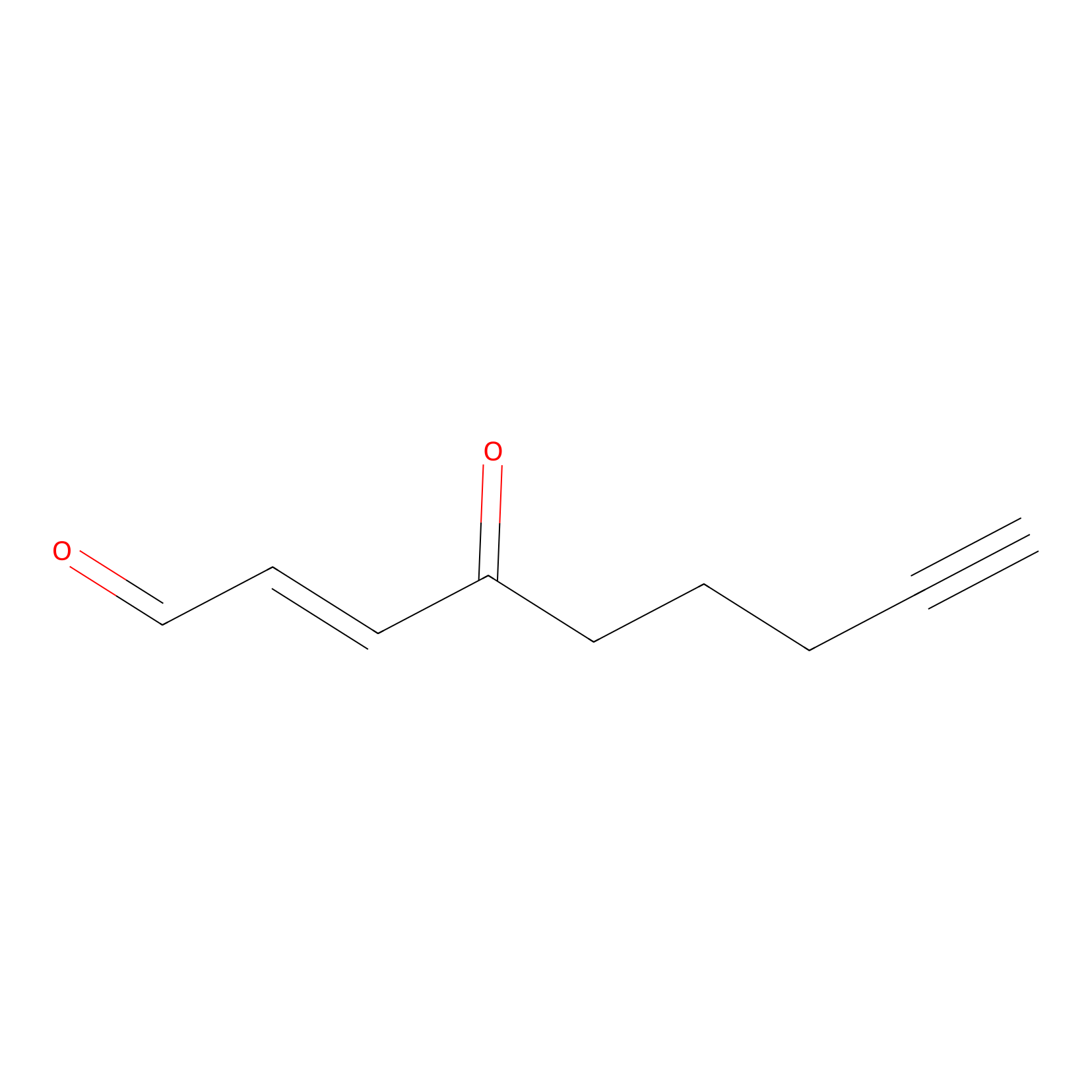 |
N.A. | LDD0002 | [8] | |
|
NHS Probe Info |
 |
K34(0.00); K97(0.00); K75(0.00); K46(0.00) | LDD0010 | [8] | |
|
SF Probe Info |
 |
K196(0.00); K63(0.00); K106(0.00); K129(0.00) | LDD0028 | [9] | |
|
STPyne Probe Info |
 |
K106(0.00); K46(0.00); K129(0.00); K90(0.00) | LDD0009 | [8] | |
|
1c-yne Probe Info |
 |
K17(0.00); K97(0.00); K34(0.00); K46(0.00) | LDD0228 | [10] | |
|
MPP-AC Probe Info |
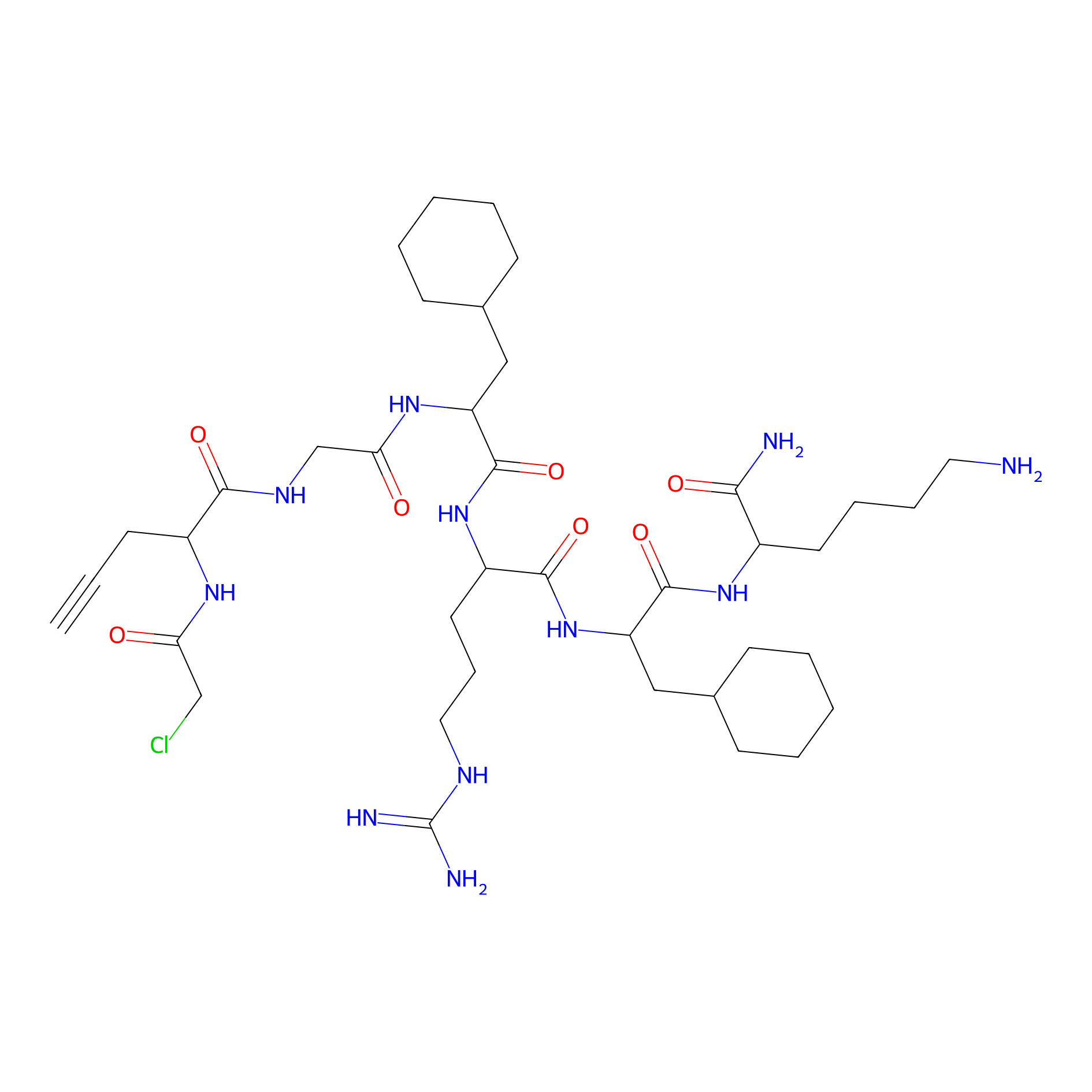 |
N.A. | LDD0428 | [11] | |
|
TER-AC Probe Info |
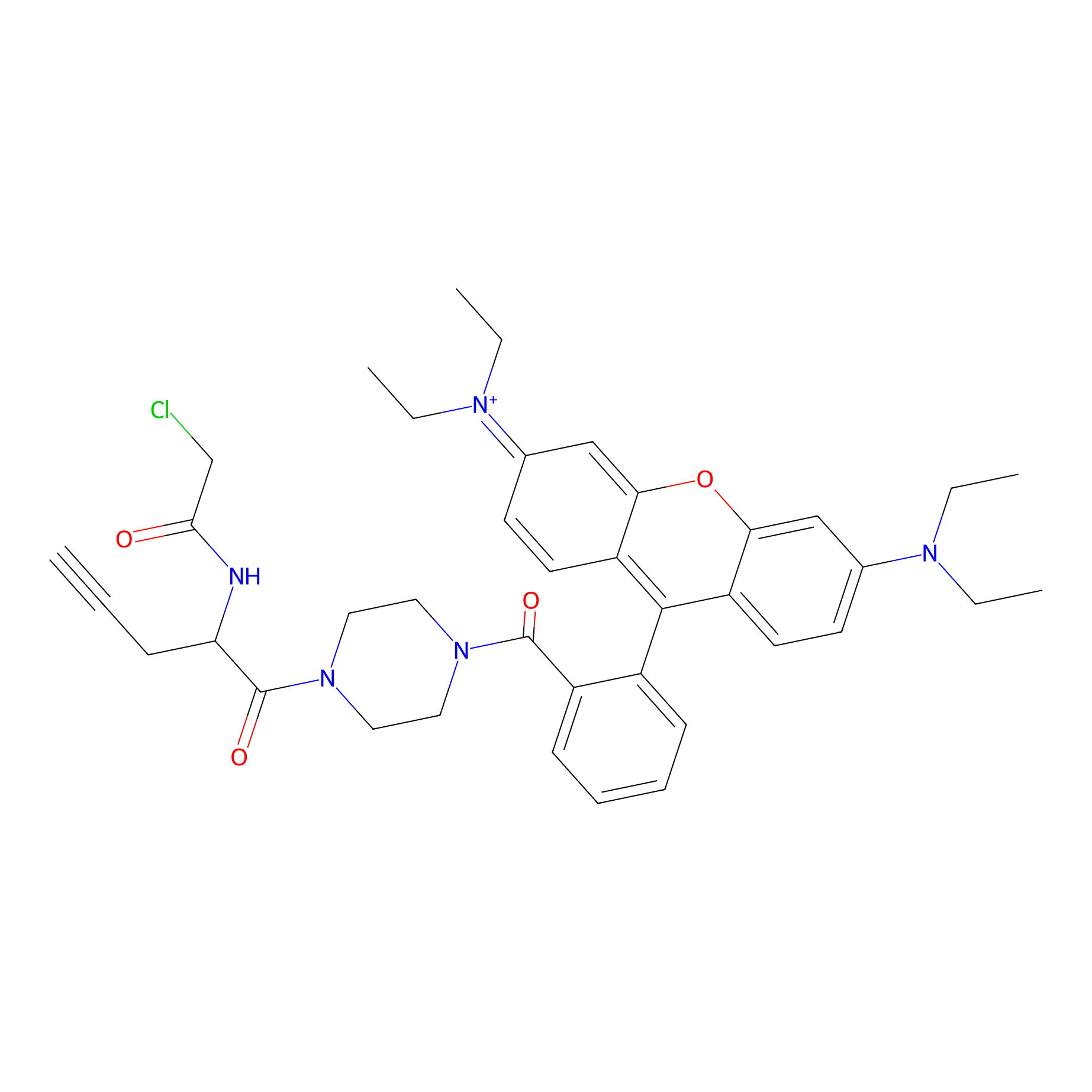 |
N.A. | LDD0426 | [11] | |
|
TPP-AC Probe Info |
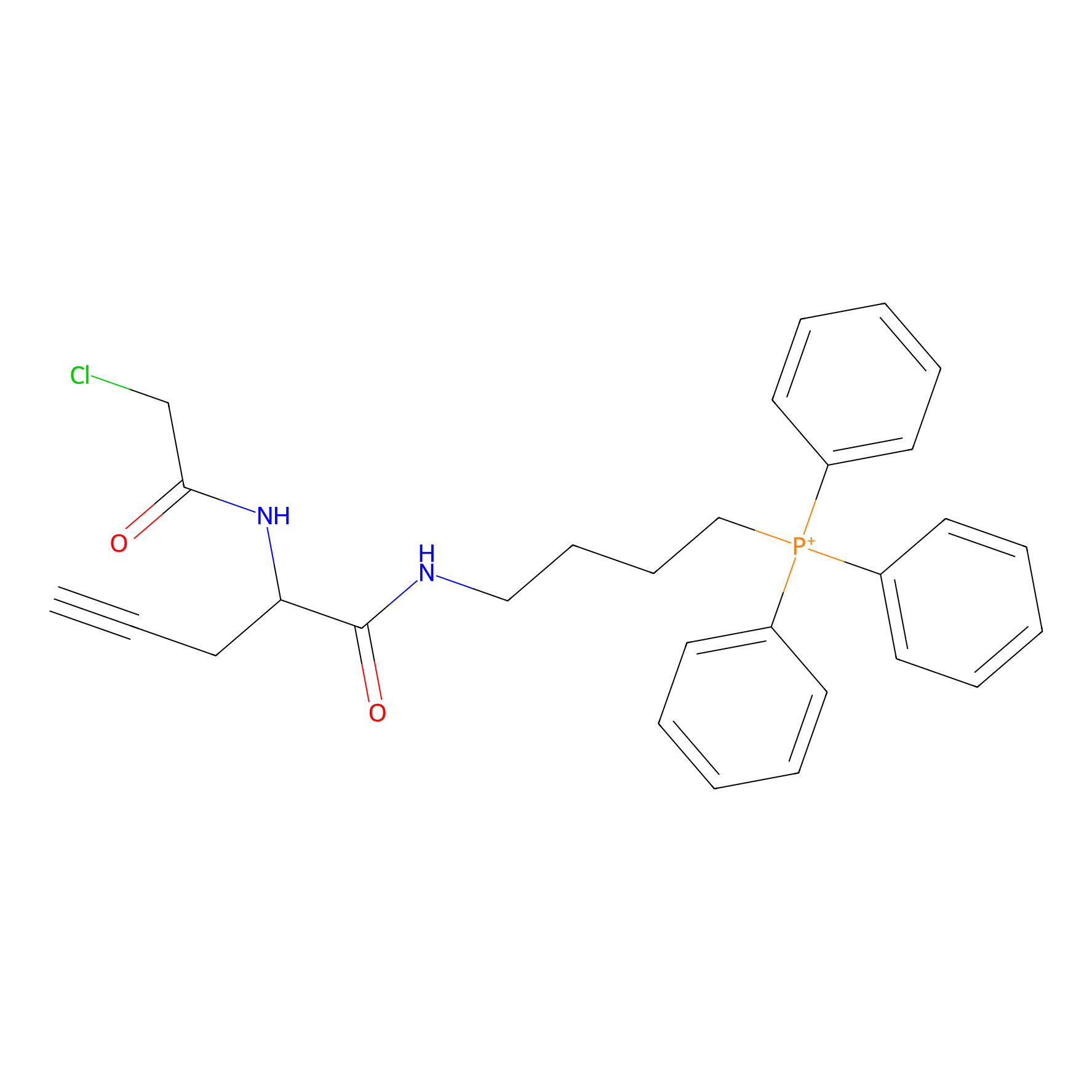 |
N.A. | LDD0427 | [11] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
JN0003 Probe Info |
 |
6.62 | LDD0469 | [12] | |
|
CR-1 Probe Info |
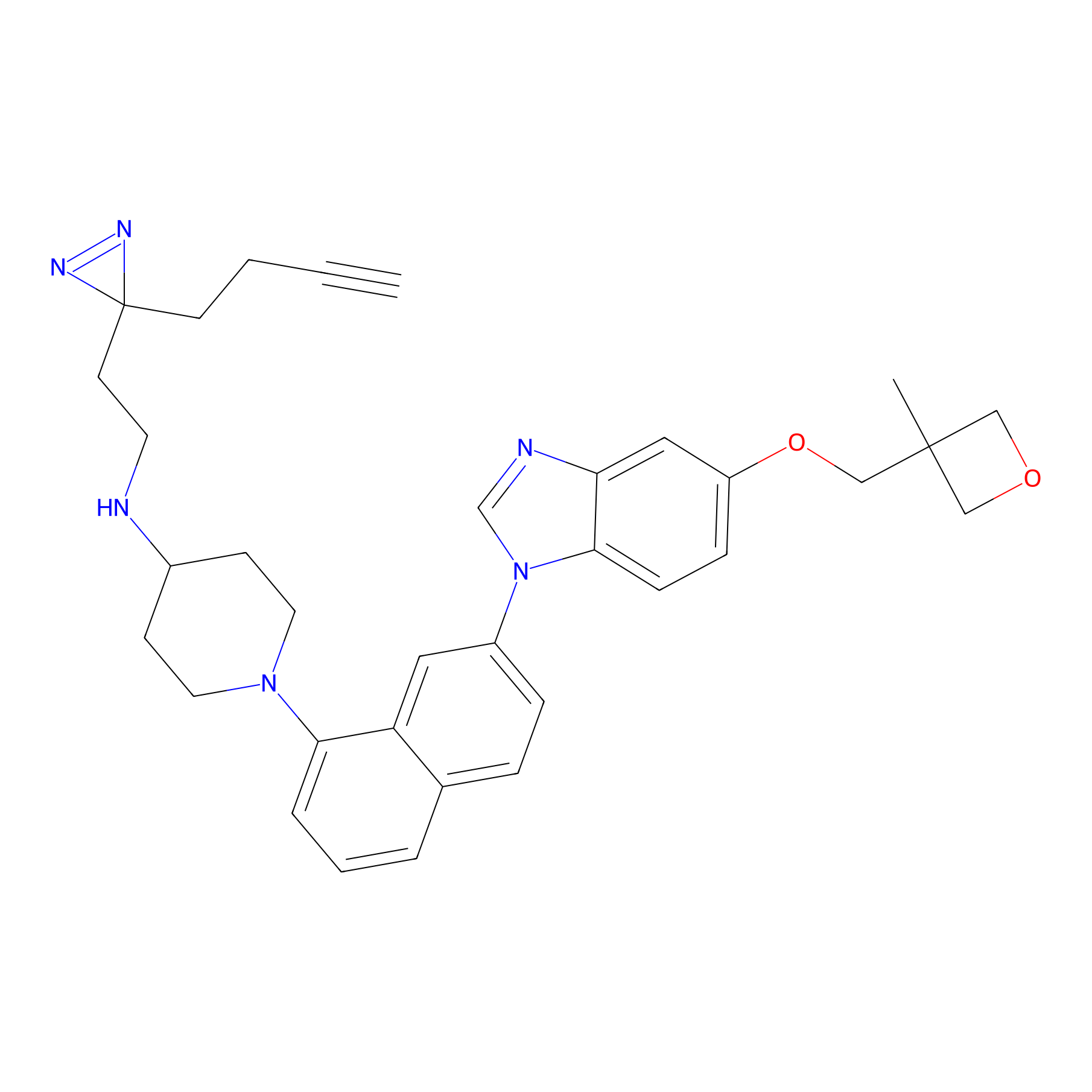 |
3.03 | LDD0429 | [13] | |
|
Diazir Probe Info |
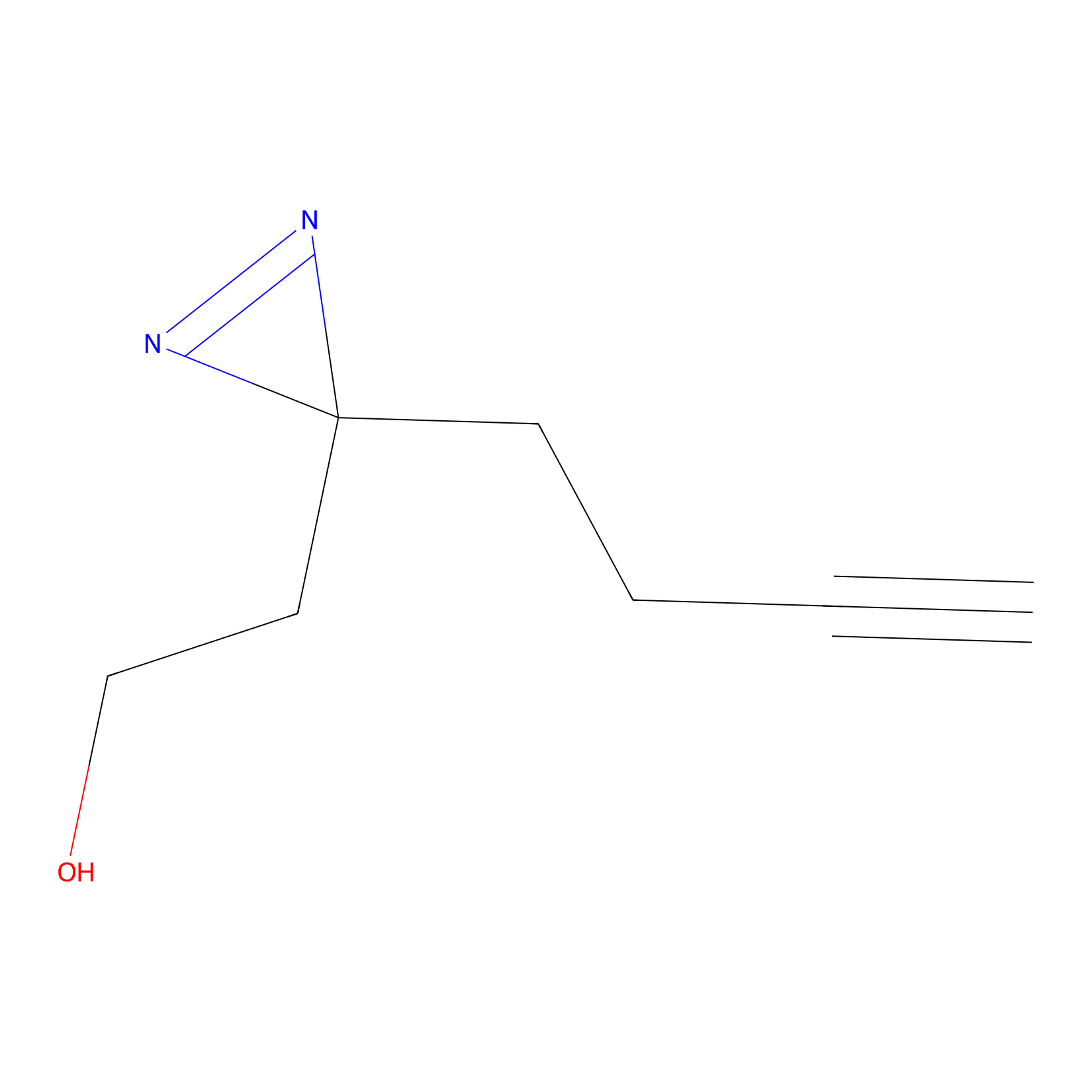 |
E3(0.00); E16(0.00) | LDD0011 | [8] | |
Competitor(s) Related to This Target
References
