Details of the Target
General Information of Target
| Target ID | LDTP09126 | |||||
|---|---|---|---|---|---|---|
| Target Name | Probable aminopeptidase NPEPL1 (NPEPL1) | |||||
| Gene Name | NPEPL1 | |||||
| Gene ID | 79716 | |||||
| Synonyms |
KIAA1974; Probable aminopeptidase NPEPL1; EC 3.4.11.-; Aminopeptidase-like 1 |
|||||
| 3D Structure | ||||||
| Sequence |
MANVGLQFQASAGDSDPQSRPLLLLGQLHHLHRVPWSHVRGKLQPRVTEELWQAALSTLN
PNPTDSCPLYLNYATVAALPCRVSRHNSPSAAHFITRLVRTCLPPGAHRCIVMVCEQPEV FASACALARAFPLFTHRSGASRRLEKKTVTVEFFLVGQDNGPVEVSTLQCLANATDGVRL AARIVDTPCNEMNTDTFLEEINKVGKELGIIPTIIRDEELKTRGFGGIYGVGKAALHPPA LAVLSHTPDGATQTIAWVGKGIVYDTGGLSIKGKTTMPGMKRDCGGAAAVLGAFRAAIKQ GFKDNLHAVFCLAENSVGPNATRPDDIHLLYSGKTVEINNTDAEGRLVLADGVSYACKDL GADIILDMATLTGAQGIATGKYHAAVLTNSAEWEAACVKAGRKCGDLVHPLVYCPELHFS EFTSAVADMKNSVADRDNSPSSCAGLFIASHIGFDWPGVWVHLDIAAPVHAGERATGFGV ALLLALFGRASEDPLLNLVSPLGCEVDVEEGDLGRDSKRRRLV |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Peptidase M17 family
|
|||||
| Function | Probably catalyzes the removal of unsubstituted N-terminal amino acids from various peptides. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| COLO800 | SNV: p.V412G; p.E416G | DBIA Probe Info | |||
| HCT116 | SNV: p.A449T | . | |||
| HEC1B | SNV: p.A481T | . | |||
| HUH7 | SNV: p.P278Q | DBIA Probe Info | |||
| KP4 | SNV: p.R142Q | DBIA Probe Info | |||
| NCIH1944 | SNV: p.D359V | DBIA Probe Info | |||
| NCIH2172 | SNV: p.R282P | DBIA Probe Info | |||
| SW403 | Insertion: p.K399Ter | DBIA Probe Info | |||
| TC71 | SNV: p.G232S | DBIA Probe Info | |||
| TOV21G | SNV: p.P278Q | DBIA Probe Info | |||
| UMUC3 | SNV: p.D435N | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
12.26 | LDD0402 | [1] | |
|
STPyne Probe Info |
 |
K274(6.48) | LDD0277 | [2] | |
|
DBIA Probe Info |
 |
C357(0.76) | LDD3310 | [3] | |
|
THZ1-DTB Probe Info |
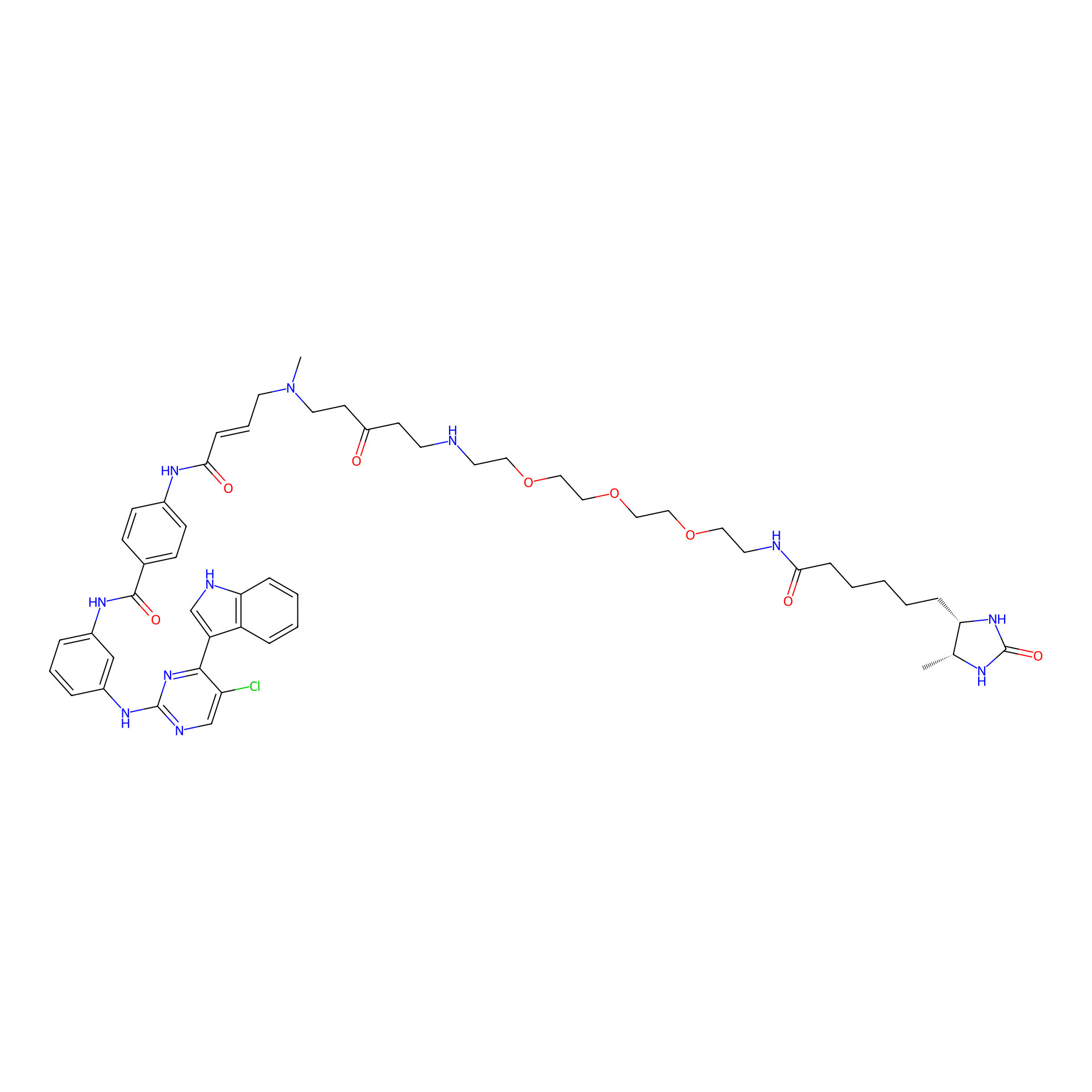 |
C67(1.03) | LDD0460 | [4] | |
|
AHL-Pu-1 Probe Info |
 |
C81(2.13) | LDD0170 | [5] | |
|
HHS-482 Probe Info |
 |
Y382(0.80) | LDD0285 | [6] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0162 | [7] | |
|
IPM Probe Info |
 |
N.A. | LDD0025 | [8] | |
|
TFBX Probe Info |
 |
C102(0.00); C284(0.00) | LDD0148 | [8] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [9] | |
|
NAIA_5 Probe Info |
 |
C284(0.00); C189(0.00); C357(0.00); C102(0.00) | LDD2223 | [10] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | DM93 | C81(2.13) | LDD0170 | [5] |
| LDCM0156 | Aniline | NCI-H1299 | 11.58 | LDD0403 | [1] |
| LDCM0632 | CL-Sc | Hep-G2 | C189(1.51); C284(1.36); C357(0.89); C397(0.82) | LDD2227 | [10] |
| LDCM0634 | CY-0357 | Hep-G2 | C357(0.99) | LDD2228 | [10] |
| LDCM0572 | Fragment10 | MDA-MB-231 | C81(5.70) | LDD1465 | [11] |
| LDCM0574 | Fragment12 | MDA-MB-231 | C81(20.00) | LDD1468 | [11] |
| LDCM0576 | Fragment14 | MDA-MB-231 | C81(1.35) | LDD1471 | [11] |
| LDCM0580 | Fragment21 | MDA-MB-231 | C81(1.55) | LDD1473 | [11] |
| LDCM0589 | Fragment31 | MDA-MB-231 | C81(0.95) | LDD1477 | [11] |
| LDCM0596 | Fragment38 | MDA-MB-231 | C81(2.01) | LDD1480 | [11] |
| LDCM0566 | Fragment4 | MDA-MB-231 | C81(20.00) | LDD1461 | [11] |
| LDCM0614 | Fragment56 | MDA-MB-231 | C81(0.99) | LDD1484 | [11] |
| LDCM0570 | Fragment8 | MDA-MB-231 | C81(20.00) | LDD1462 | [11] |
| LDCM0123 | JWB131 | DM93 | Y382(0.80) | LDD0285 | [6] |
| LDCM0124 | JWB142 | DM93 | Y382(1.28) | LDD0286 | [6] |
| LDCM0125 | JWB146 | DM93 | Y382(0.97) | LDD0287 | [6] |
| LDCM0126 | JWB150 | DM93 | Y382(3.28) | LDD0288 | [6] |
| LDCM0127 | JWB152 | DM93 | Y382(1.66) | LDD0289 | [6] |
| LDCM0128 | JWB198 | DM93 | Y382(0.11) | LDD0290 | [6] |
| LDCM0129 | JWB202 | DM93 | Y382(0.88) | LDD0291 | [6] |
| LDCM0130 | JWB211 | DM93 | Y382(1.52) | LDD0292 | [6] |
| LDCM0022 | KB02 | MDA-MB-231 | C81(20.00) | LDD1460 | [11] |
| LDCM0023 | KB03 | 22RV1 | C74(1.48) | LDD2660 | [3] |
| LDCM0024 | KB05 | COLO792 | C357(0.76) | LDD3310 | [3] |
| LDCM0131 | RA190 | MM1.R | C189(2.43) | LDD0304 | [12] |
| LDCM0021 | THZ1 | HeLa S3 | C67(1.03) | LDD0460 | [4] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Caspase-6 (CASP6) | Peptidase C14A family | P55212 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| MyoD family inhibitor (MDFI) | MDFI family | Q99750 | |||
| Huntingtin-interacting protein 1 (HIP1) | SLA2 family | O00291 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Alpha-globin transcription factor CP2 (TFCP2) | Grh/CP2 family | Q12800 | |||
References
